Ubiquitination Regulates the Morphogenesis and Function of Sperm Organelles
Abstract
:1. Introduction
2. Ubiquitinating and Deubiquitinating Enzymes during Spermatogenesis
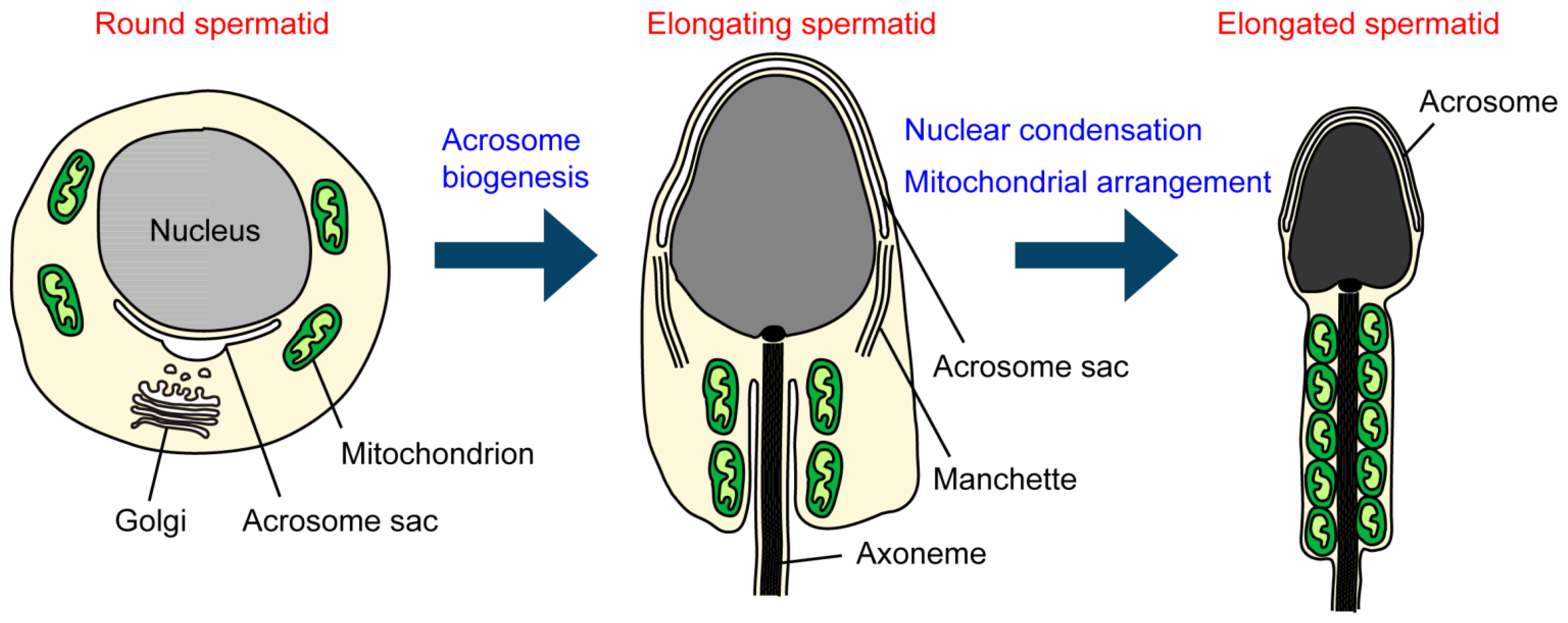
3. Roles of Ubiquitination in Sperm Organelles
3.1. Histone Ubiquitination for the Purpose of Nuclear Condensation
3.2. Acrosome Formation and Membrane Trafficking
3.2.1. TMF/ARA160—A Mediator of Fusion of Golgi-Derived Vesicles
3.2.2. MARCH11—Regulation of Golgi-to-Endosome Transport
3.2.3. USP8/UBPy—The Retrograde Pathway for Acrosome Formation
3.2.4. RNF19a and MARCH7—Acroplaxome-Localized E3s
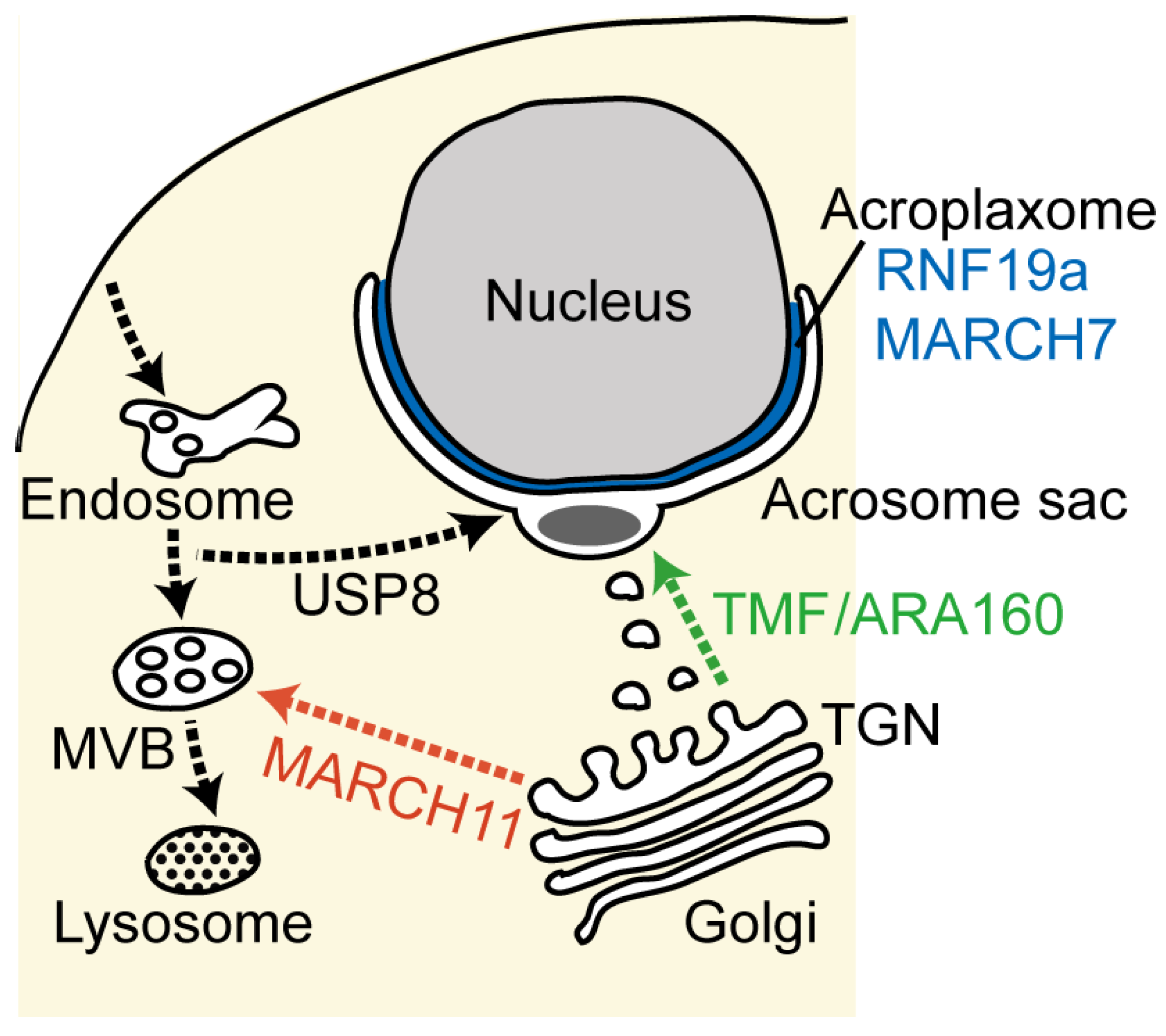
3.3. Maternal Inheritance of Mitochondrial DNA
3.3.1. Ubiquitination of Sperm Mitochondria
3.3.2. Does “Mitophagy” Eliminate Paternal Mitochondria?
4. Conclusions
Acknowledgments
Conflicts of Interest
References
- Glickman, M.H.; Ciechanover, A. The ubiquitin-proteasome proteolytic pathway: Destruction for the sake of construction. Physiol. Rev. 2002, 82, 373–428. [Google Scholar]
- Li, W.; Bengtson, M.H.; Ulbrich, A.; Matsuda, A.; Reddy, V.A.; Orth, A.; Chanda, S.K.; Batalov, S.; Joazeiro, C.A. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle's dynamics and signaling. PLoS One 2008, 3, e1487. [Google Scholar] [CrossRef]
- Ikeda, F.; Dikic, I. Atypical ubiquitin chains: New molecular signals. ‘Protein Modifications: Beyond the Usual Suspects’ review series. EMBO Rep. 2008, 9, 536–542. [Google Scholar] [CrossRef]
- Kulathu, Y.; Komander, D. Atypical ubiquitylation—the unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nat. Rev. Mol. Cell Biol. 2012, 13, 508–523. [Google Scholar] [CrossRef]
- Reyes-Turcu, F.E.; Ventii, K.H.; Wilkinson, K.D. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu. Rev. Biochem. 2009, 78, 363–397. [Google Scholar] [CrossRef]
- Nakamura, N. The role of the transmembrane RING finger proteins in cellular and organelle function. Membranes 2011, 1, 354–393. [Google Scholar] [CrossRef]
- Vembar, S.S.; Brodsky, J.L. One step at a time: Endoplasmic reticulum-associated degradation. Nat. Rev. Mol. Cell Biol. 2008, 9, 944–957. [Google Scholar] [CrossRef]
- Shields, S.B.; Piper, R.C. How ubiquitin functions with ESCRTs. Traffic 2011, 12, 1306–1317. [Google Scholar] [CrossRef]
- Clermont, Y.; Oko, R.; Hermo, L. Cell Biology of Mammalian Spermatogenesis. In Desjarjins C and Ewing LL (ed) Cell. and Molecular Biology of the Testis; Oxford University Press: New York, NY, USA, 1993; pp. 332–376. [Google Scholar]
- Schultz, N.; Hamra, F.K.; Garbers, D.L. A multitude of genes expressed solely in meiotic or postmeiotic spermatogenic cells offers a myriad of contraceptive targets. Proc. Natl. Acad. Sci. USA 2003, 100, 12201–12206. [Google Scholar] [CrossRef]
- Shima, J.E.; McLean, D.J.; McCarrey, J.R.; Griswold, M.D. The murine testicular transcriptome: Characterizing gene expression in the testis during the progression of spermatogenesis. Biol. Reprod. 2004, 71, 319–330. [Google Scholar] [CrossRef]
- Pang, A.L.; Johnson, W.; Ravindranath, N.; Dym, M.; Rennert, O.M.; Chan, W.Y. Expression profiling of purified male germ cells: Stage-specific expression patterns related to meiosis and postmeiotic development. Physiol. Genomics 2006, 24, 75–85. [Google Scholar]
- Wang, G.; Guo, Y.; Zhou, T.; Shi, X.; Yu, J.; Yang, Y.; Wu, Y.; Wang, J.; Liu, M.; Chen, X.; et al. In-depth proteomic analysis of the human sperm reveals complex protein compositions. J. Proteomics 2013, 79, 114–122. [Google Scholar] [CrossRef]
- Roth, G.; Himstedt, W. Response characteristics of neurons in the tectum opticum of Salamandra. Naturwissenschaften 1978, 65, 657–658. [Google Scholar] [CrossRef]
- Loir, M.; Caraty, A.; Lanneau, M.; Menezo, Y.; Muh, J.P.; Sautiere, P. Purification and characterization of ubiquitin from mammalian testis. FEBS Lett. 1984, 169, 199–204. [Google Scholar] [CrossRef]
- Hou, C.C.; Yang, W.X. New insights to the ubiquitin-proteasome pathway (UPP) mechanism during spermatogenesis. Mol. Biol. Rep. 2013, 40, 3213–3230. [Google Scholar] [CrossRef]
- Hou, X.; Zhang, W.; Xiao, Z.; Gan, H.; Lin, X.; Liao, S.; Han, C. Mining and characterization of ubiquitin E3 ligases expressed in the mouse testis. BMC Genomics 2012, 13, 495. [Google Scholar] [CrossRef]
- Clague, M.J.; Barsukov, I.; Coulson, J.M.; Liu, H.; Rigden, D.J.; Urbe, S. Deubiquitylases from genes to organism. Physiol. Rev. 2013, 93, 1289–1315. [Google Scholar] [CrossRef]
- Sun, C.; Skaletsky, H.; Birren, B.; Devon, K.; Tang, Z.; Silber, S.; Oates, R.; Page, D.C. An azoospermic man with a de novo point mutation in the Y-chromosomal gene USP9Y. Nat. Genet. 1999, 23, 429–432. [Google Scholar] [CrossRef]
- Foresta, C.; Ferlin, A.; Moro, E. Deletion and expression analysis of AZFa genes on the human Y chromosome revealed a major role for DBY in male infertility. Hum. Mol. Genet. 2000, 9, 1161–1169. [Google Scholar] [CrossRef]
- Stouffs, K.; Lissens, W.; Tournaye, H.; van Steirteghem, A.; Liebaers, I. Possible role of USP26 in patients with severely impaired spermatogenesis. Eur. J. Hum. Genet. 2005, 13, 336–340. [Google Scholar] [CrossRef]
- Wright, A.; Reiley, W.W.; Chang, M.; Jin, W.; Lee, A.J.; Zhang, M.; Sun, S.C. Regulation of early wave of germ cell apoptosis and spermatogenesis by deubiquitinating enzyme CYLD. Dev. Cell 2007, 13, 705–716. [Google Scholar] [CrossRef]
- Crimmins, S.; Sutovsky, M.; Chen, P.C.; Huffman, A.; Wheeler, C.; Swing, D.A.; Roth, K.; Wilson, J.; Sutovsky, P.; Wilson, S. Transgenic rescue of ataxia mice reveals a male-specific sterility defect. Dev. Biol. 2009, 325, 33–42. [Google Scholar] [CrossRef]
- Kim, N.; Xiao, R.; Choi, H.; Jo, H.; Kim, J.H.; Uhm, S.J.; Park, C. Abnormal sperm development in pcd(3J)-/- mice: The importance of Agtpbp1 in spermatogenesis. Mol. Cells 2011, 31, 39–48. [Google Scholar] [CrossRef]
- Bedard, N.; Yang, Y.; Gregory, M.; Cyr, D.G.; Suzuki, J.; Yu, X.; Chian, R.C.; Hermo, L.; O’Flaherty, C.; Smith, C.E.; et al. Mice lacking the USP2 deubiquitinating enzyme have severe male subfertility associated with defects in fertilization and sperm motility. Biol. Reprod. 2011, 85, 594–604. [Google Scholar] [CrossRef]
- Manku, G.; Wing, S.S.; Culty, M. Expression of the ubiquitin proteasome system in neonatal rat gonocytes and spermatogonia: Role in gonocyte differentiation. Biol. Reprod. 2012, 87, 44. [Google Scholar] [CrossRef]
- Braun, R.E. Packaging paternal chromosomes with protamine. Nat. Genet. 2001, 28, 10–12. [Google Scholar]
- Oliva, R. Protamines and male infertility. Hum. Reprod. Update 2006, 12, 417–435. [Google Scholar] [CrossRef]
- Kierszenbaum, A.L.; Tres, L.L. The acrosome-acroplaxome-manchette complex and the shaping of the spermatid head. Arch. Histol. Cytol. 2004, 67, 271–284. [Google Scholar] [CrossRef]
- Ward, W.S.; Coffey, D.S. DNA packaging and organization in mammalian spermatozoa: Comparison with somatic cells. Biol. Reprod. 1991, 44, 569–574. [Google Scholar] [CrossRef]
- Baarends, W.M.; Hoogerbrugge, J.W.; Roest, H.P.; Ooms, M.; Vreeburg, J.; Hoeijmakers, J.H.; Grootegoed, J.A. Histone ubiquitination and chromatin remodeling in mouse spermatogenesis. Dev. Biol. 1999, 207, 322–333. [Google Scholar] [CrossRef]
- Sung, P.; Prakash, S.; Prakash, L. The RAD6 protein of Saccharomyces cerevisiae polyubiquitinates histones, and its acidic domain mediates this activity. Genes Dev. 1988, 2, 1476–1485. [Google Scholar] [CrossRef]
- Jentsch, S.; McGrath, J.P.; Varshavsky, A. The yeast DNA repair gene RAD6 encodes a ubiquitin-conjugating enzyme. Nature 1987, 329, 131–134. [Google Scholar] [CrossRef]
- Robzyk, K.; Recht, J.; Osley, M.A. Rad6-dependent ubiquitination of histone H2B in yeast. Science 2000, 287, 501–504. [Google Scholar] [CrossRef]
- Hwang, W.W.; Venkatasubrahmanyam, S.; Ianculescu, A.G.; Tong, A.; Boone, C.; Madhani, H.D. A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol. Cell 2003, 11, 261–266. [Google Scholar] [CrossRef]
- Wood, A.; Krogan, N.J.; Dover, J.; Schneider, J.; Heidt, J.; Boateng, M.A.; Dean, K.; Golshani, A.; Zhang, Y.; Greenblatt, J.F.; et al. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol. Cell 2003, 11, 267–274. [Google Scholar] [CrossRef]
- Dover, J.; Schneider, J.; Tawiah-Boateng, M.A.; Wood, A.; Dean, K.; Johnston, M.; Shilatifard, A. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J. Biol. Chem. 2002, 277, 28368–28371. [Google Scholar] [CrossRef]
- Sun, Z.W.; Allis, C.D. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 2002, 418, 104–108. [Google Scholar] [CrossRef]
- Briggs, S.D.; Xiao, T.; Sun, Z.W.; Caldwell, J.A.; Shabanowitz, J.; Hunt, D.F.; Allis, C.D.; Strahl, B.D. Gene silencing: Trans-histone regulatory pathway in chromatin. Nature 2002, 418, 498. [Google Scholar]
- Henry, K.W.; Wyce, A.; Lo, W.S.; Duggan, L.J.; Emre, N.C.; Kao, C.F.; Pillus, L.; Shilatifard, A.; Osley, M.A.; Berger, S.L. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 2003, 17, 2648–2663. [Google Scholar] [CrossRef]
- Kao, C.F.; Hillyer, C.; Tsukuda, T.; Henry, K.; Berger, S.; Osley, M.A. Rad6 plays a role in transcriptional activation through ubiquitylation of histone H2B. Genes Dev. 2004, 18, 184–195. [Google Scholar] [CrossRef]
- Wood, A.; Schneider, J.; Shilatifard, A. Cross-talking histones: Implications for the regulation of gene expression and DNA repair. Biochem. Cell Biol. 2005, 83, 460–467. [Google Scholar] [CrossRef]
- Pavri, R.; Zhu, B.; Li, G.; Trojer, P.; Mandal, S.; Shilatifard, A.; Reinberg, D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell 2006, 125, 703–717. [Google Scholar] [CrossRef]
- Osley, M.A. Regulation of histone H2A and H2B ubiquitylation. Brief. Funct. Genomics Proteomics 2006, 5, 179–189. [Google Scholar] [CrossRef]
- Weake, V.M.; Workman, J.L. Histone ubiquitination: Triggering gene activity. Mol. Cell 2008, 29, 653–663. [Google Scholar] [CrossRef]
- Koken, M.H.; Hoogerbrugge, J.W.; Jasper-Dekker, I.; de Wit, J.; Willemsen, R.; Roest, H.P.; Grootegoed, J.A.; Hoeijmakers, J.H. Expression of the ubiquitin-conjugating DNA repair enzymes HHR6A and B suggests a role in spermatogenesis and chromatin modification. Dev. Biol. 1996, 173, 119–132. [Google Scholar] [CrossRef]
- Roest, H.P.; van Klaveren, J.; de Wit, J.; van Gurp, C.G.; Koken, M.H.; Vermey, M.; van Roijen, J.H.; Hoogerbrugge, J.W.; Vreeburg, J.T.; Baarends, W.M.; et al. Inactivation of the HR6B ubiquitin-conjugating DNA repair enzyme in mice causes male sterility associated with chromatin modification. Cell 1996, 86, 799–810. [Google Scholar] [CrossRef]
- Gaucher, J.; Reynoird, N.; Montellier, E.; Boussouar, F.; Rousseaux, S.; Khochbin, S. From meiosis to postmeiotic events: The secrets of histone disappearance. FEBS J. 2010, 277, 599–604. [Google Scholar] [CrossRef]
- Qian, M.X.; Pang, Y.; Liu, C.H.; Haratake, K.; Du, B.Y.; Ji, D.Y.; Wang, G.F.; Zhu, Q.Q.; Song, W.; Yu, Y.; et al. Acetylation-mediated proteasomal degradation of core histones during DNA repair and spermatogenesis. Cell 2013, 153, 1012–1024. [Google Scholar] [CrossRef]
- Savulescu, A.F.; Glickman, M.H. Proteasome activator 200: The heat is on. Mol. Cell. Proteomics 2011, 10, R110.006890. [Google Scholar]
- Khor, B.; Bredemeyer, A.L.; Huang, C.Y.; Turnbull, I.R.; Evans, R.; Maggi, L.B., Jr.; White, J.M.; Walker, L.M.; Carnes, K.; Hess, R.A.; et al. Proteasome activator PA200 is required for normal spermatogenesis. Mol. Cell. Biol. 2006, 26, 2999–3007. [Google Scholar] [CrossRef]
- Lu, L.Y.; Wu, J.; Ye, L.; Gavrilina, G.B.; Saunders, T.L.; Yu, X. RNF8-dependent histone modifications regulate nucleosome removal during spermatogenesis. Dev. Cell 2010, 18, 371–384. [Google Scholar] [CrossRef]
- Baarends, W.M.; Wassenaar, E.; van der Laan, R.; Hoogerbrugge, J.; Sleddens-Linkels, E.; Hoeijmakers, J.H.; de Boer, P.; Grootegoed, J.A. Silencing of unpaired chromatin and histone H2A ubiquitination in mammalian meiosis. Mol. Cell. Biol. 2005, 25, 1041–1053. [Google Scholar] [CrossRef]
- Turner, J.M.; Mahadevaiah, S.K.; Fernandez-Capetillo, O.; Nussenzweig, A.; Xu, X.; Deng, C.X.; Burgoyne, P.S. Silencing of unsynapsed meiotic chromosomes in the mouse. Nat. Genet. 2005, 37, 41–47. [Google Scholar]
- Kwon, Y.T.; Xia, Z.; An, J.Y.; Tasaki, T.; Davydov, I.V.; Seo, J.W.; Sheng, J.; Xie, Y.; Varshavsky, A. Female lethality and apoptosis of spermatocytes in mice lacking the UBR2 ubiquitin ligase of the N-end rule pathway. Mol. Cell. Biol. 2003, 23, 8255–8271. [Google Scholar] [CrossRef]
- An, J.Y.; Kim, E.A.; Jiang, Y.; Zakrzewska, A.; Kim, D.E.; Lee, M.J.; Mook-Jung, I.; Zhang, Y.; Kwon, Y.T. UBR2 mediates transcriptional silencing during spermatogenesis via histone ubiquitination. Proc. Natl. Acad. Sci. USA 2010, 107, 1912–1917. [Google Scholar] [CrossRef]
- Mulugeta Achame, E.; Wassenaar, E.; Hoogerbrugge, J.W.; Sleddens-Linkels, E.; Ooms, M.; Sun, Z.W.; van, I.W.F.; Grootegoed, J.A.; Baarends, W.M. The ubiquitin-conjugating enzyme HR6B is required for maintenance of X chromosome silencing in mouse spermatocytes and spermatids. BMC Genomics 2010, 11, 367. [Google Scholar] [CrossRef]
- Baarends, W.M.; Wassenaar, E.; Hoogerbrugge, J.W.; Schoenmakers, S.; Sun, Z.W.; Grootegoed, J.A. Increased phosphorylation and dimethylation of XY body histones in the Hr6b-knockout mouse is associated with derepression of the X chromosome. J. Cell Sci. 2007, 120, 1841–1851. [Google Scholar] [CrossRef]
- Baarends, W.M.; Wassenaar, E.; Hoogerbrugge, J.W.; van Cappellen, G.; Roest, H.P.; Vreeburg, J.; Ooms, M.; Hoeijmakers, J.H.; Grootegoed, J.A. Loss of HR6B ubiquitin-conjugating activity results in damaged synaptonemal complex structure and increased crossing-over frequency during the male meiotic prophase. Mol. Cell. Biol. 2003, 23, 1151–1162. [Google Scholar] [CrossRef]
- van der Laan, R.; Uringa, E.J.; Wassenaar, E.; Hoogerbrugge, J.W.; Sleddens, E.; Odijk, H.; Roest, H.P.; de Boer, P.; Hoeijmakers, J.H.; Grootegoed, J.A.; et al. Ubiquitin ligase Rad18Sc localizes to the XY body and to other chromosomal regions that are unpaired and transcriptionally silenced during male meiotic prophase. J. Cell Sci. 2004, 117, 5023–5033. [Google Scholar] [CrossRef]
- Kim, J.; Hake, S.B.; Roeder, R.G. The human homolog of yeast BRE1 functions as a transcriptional coactivator through direct activator interactions. Mol. Cell 2005, 20, 759–770. [Google Scholar] [CrossRef]
- Liu, Z.; Oughtred, R.; Wing, S.S. Characterization of E3Histone, a novel testis ubiquitin protein ligase which ubiquitinates histones. Mol. Cell. Biol. 2005, 25, 2819–2831. [Google Scholar] [CrossRef]
- Liu, Z.; Miao, D.; Xia, Q.; Hermo, L.; Wing, S.S. Regulated expression of the ubiquitin protein ligase, E3(Histone)/LASU1/Mule/ARF-BP1/HUWE1, during spermatogenesis. Dev. Dyn. 2007, 236, 2889–2898. [Google Scholar] [CrossRef]
- Moreno, R.D.; Ramalho-Santos, J.; Chan, E.K.; Wessel, G.M.; Schatten, G. The Golgi apparatus segregates from the lysosomal/acrosomal vesicle during rhesus spermiogenesis: Structural alterations. Dev. Biol. 2000, 219, 334–349. [Google Scholar] [CrossRef]
- Ramalho-Santos, J.; Moreno, R.D.; Wessel, G.M.; Chan, E.K.; Schatten, G. Membrane trafficking machinery components associated with the mammalian acrosome during spermiogenesis. Exp. Cell Res. 2001, 267, 45–60. [Google Scholar] [CrossRef]
- Ramalho-Santos, J.; Moreno, R.D. Targeting and fusion proteins during mammalian spermiogenesis. Biol. Res. 2001, 34, 147–152. [Google Scholar]
- Kang-Decker, N.; Mantchev, G.T.; Juneja, S.C.; McNiven, M.A.; van Deursen, J.M. Lack of acrosome formation in Hrb-deficient mice. Science 2001, 294, 1531–1533. [Google Scholar] [CrossRef]
- Yao, R.; Ito, C.; Natsume, Y.; Sugitani, Y.; Yamanaka, H.; Kuretake, S.; Yanagida, K.; Sato, A.; Toshimori, K.; Noda, T. Lack of acrosome formation in mice lacking a Golgi protein, GOPC. Proc. Natl. Acad. Sci. USA 2002, 99, 11211–11216. [Google Scholar] [CrossRef]
- Kierszenbaum, A.L.; Tres, L.L.; Rivkin, E.; Kang-Decker, N.; van Deursen, J.M. The acroplaxome is the docking site of Golgi-derived myosin Va/Rab27a/b- containing proacrosomal vesicles in wild-type and Hrb mutant mouse spermatids. Biol. Reprod. 2004, 70, 1400–1410. [Google Scholar] [CrossRef]
- Xiao, N.; Kam, C.; Shen, C.; Jin, W.; Wang, J.; Lee, K.M.; Jiang, L.; Xia, J. PICK1 deficiency causes male infertility in mice by disrupting acrosome formation. J. Clin. Invest. 2009, 119, 802–812. [Google Scholar] [CrossRef]
- Roqueta-Rivera, M.; Abbott, T.L.; Sivaguru, M.; Hess, R.A.; Nakamura, M.T. Deficiency in the omega-3 fatty acid pathway results in failure of acrosome biogenesis in mice. Biol. Reprod. 2011, 85, 721–732. [Google Scholar] [CrossRef]
- Lerer-Goldshtein, T.; Bel, S.; Shpungin, S.; Pery, E.; Motro, B.; Goldstein, R.S.; Bar-Sheshet, S.I.; Breitbart, H.; Nir, U. TMF/ARA160: A key regulator of sperm development. Dev. Biol. 2010, 348, 12–21. [Google Scholar] [CrossRef]
- Fridmann-Sirkis, Y.; Siniossoglou, S.; Pelham, H.R. TMF is a golgin that binds Rab6 and influences Golgi morphology. BMC Cell Biol. 2004, 5, 18. [Google Scholar] [CrossRef] [Green Version]
- Yamane, J.; Kubo, A.; Nakayama, K.; Yuba-Kubo, A.; Katsuno, T.; Tsukita, S. Functional involvement of TMF/ARA160 in Rab6-dependent retrograde membrane traffic. Exp. Cell Res. 2007, 313, 3472–3485. [Google Scholar] [CrossRef]
- Miller, V.J.; Sharma, P.; Kudlyk, T.A.; Frost, L.; Rofe, A.P.; Watson, I.J.; Duden, R.; Lowe, M.; Lupashin, V.V.; Ungar, D. Molecular insights into vesicle tethering at the Golgi by the conserved oligomeric Golgi (COG) complex and the golgin TATA element modulatory factor (TMF). J. Biol. Chem. 2013, 288, 4229–4240. [Google Scholar] [CrossRef]
- Perry, E.; Tsruya, R.; Levitsky, P.; Pomp, O.; Taller, M.; Weisberg, S.; Parris, W.; Kulkarni, S.; Malovani, H.; Pawson, T.; et al. TMF/ARA160 is a BC-box-containing protein that mediates the degradation of Stat3. Oncogene 2004, 23, 8908–8919. [Google Scholar] [CrossRef]
- Abrham, G.; Volpe, M.; Shpungin, S.; Nir, U. TMF/ARA160 downregulates proangiogenic genes and attenuates the progression of PC3 xenografts. Int. J. Cancer 2009, 125, 43–53. [Google Scholar] [CrossRef]
- Piper, R.C.; Katzmann, D.J. Biogenesis and function of multivesicular bodies. Annu. Rev. Cell Dev. Biol. 2007, 23, 519–547. [Google Scholar] [CrossRef]
- Haraguchi, C.M.; Mabuchi, T.; Hirata, S.; Shoda, T.; Hoshi, K.; Yokota, S. Ubiquitin signals in the developing acrosome during spermatogenesis of rat testis: An immunoelectron microscopic study. J. Histochem. Cytochem. 2004, 52, 1393–1403. [Google Scholar] [CrossRef]
- Morokuma, Y.; Nakamura, N.; Kato, A.; Notoya, M.; Yamamoto, Y.; Sakai, Y.; Fukuda, H.; Yamashina, S.; Hirata, Y.; Hirose, S. MARCH-XI, a novel transmembrane ubiquitin ligase implicated in ubiquitin-dependent protein sorting in developing spermatids. J. Biol. Chem. 2007, 282, 24806–24815. [Google Scholar] [CrossRef]
- Yogo, K.; Tojima, H.; Ohno, J.Y.; Ogawa, T.; Nakamura, N.; Hirose, S.; Takeya, T.; Kohsaka, T. Identification of SAMT family proteins as substrates of MARCH11 in mouse spermatids. Histochem. Cell Biol. 2012, 137, 53–65. [Google Scholar] [CrossRef]
- Nathan, J.A.; Lehner, P.J. The trafficking and regulation of membrane receptors by the RING-CH ubiquitin E3 ligases. Exp. Cell Res. 2009, 315, 1593–1600. [Google Scholar] [CrossRef]
- Tang, X.M.; Lalli, M.F.; Clermont, Y. A cytochemical study of the Golgi apparatus of the spermatid during spermiogenesis in the rat. Am. J. Anat. 1982, 163, 283–294. [Google Scholar] [CrossRef]
- Martinez-Menarguez, J.A.; Aviles, M.; Madrid, J.F.; Castells, M.T.; Ballesta, J. Glycosylation in Golgi apparatus of early spermatids of rat. A high resolution lectin cytochemical study. Eur. J. Cell Biol. 1993, 61, 21–33. [Google Scholar]
- Iyengar, P.V.; Hirota, T.; Hirose, S.; Nakamura, N. Membrane-associated RING-CH 10 (MARCH10 protein) is a microtubule-associated E3 ubiquitin ligase of the spermatid flagella. J. Biol. Chem. 2011, 286, 39082–39090. [Google Scholar] [CrossRef]
- Zhao, B.; Ito, K.; Iyengar, P.V.; Hirose, S.; Nakamura, N. MARCH7 E3 ubiquitin ligase is highly expressed in developing spermatids of rats and its possible involvement in head and tail formation. Histochem. Cell Biol. 2013, 139, 447–460. [Google Scholar] [CrossRef]
- West, A.P.; Willison, K.R. Brefeldin A and mannose 6-phosphate regulation of acrosomic related vesicular trafficking. Eur. J. Cell Biol. 1996, 70, 315–321. [Google Scholar]
- Li, Y.C.; Hu, X.Q.; Zhang, K.Y.; Guo, J.; Hu, Z.Y.; Tao, S.X.; Xiao, L.J.; Wang, Q.Z.; Han, C.S.; Liu, Y.X. Afaf, a novel vesicle membrane protein, is related to acrosome formation in murine testis. FEBS Lett. 2006, 580, 4266–4273. [Google Scholar] [CrossRef]
- Li, S.; Qiao, Y.; Di, Q.; Le, X.; Zhang, L.; Zhang, X.; Zhang, C.; Cheng, J.; Zong, S.; Koide, S.S.; et al. Interaction of SH3P13 and DYDC1 protein: A germ cell component that regulates acrosome biogenesis during spermiogenesis. Eur. J. Cell Biol. 2009, 88, 509–520. [Google Scholar] [CrossRef]
- Berruti, G.; Ripolone, M.; Ceriani, M. USP8, a regulator of endosomal sorting, is involved in mouse acrosome biogenesis through interaction with the spermatid ESCRT-0 complex and microtubules. Biol. Reprod. 2010, 82, 930–939. [Google Scholar] [CrossRef]
- Berruti, G.; Martegani, E. The deubiquitinating enzyme mUBPy interacts with the sperm-specific molecular chaperone MSJ-1: The relation with the proteasome, acrosome, and centrosome in mouse male germ cells. Biol. Reprod. 2005, 72, 14–21. [Google Scholar] [CrossRef]
- Wright, M.H.; Berlin, I.; Nash, P.D. Regulation of endocytic sorting by ESCRT-DUB-mediated deubiquitination. Cell Biochem. Biophys. 2011, 60, 39–46. [Google Scholar] [CrossRef]
- Bonifacino, J.S.; Hierro, A. Transport according to GARP: Receiving retrograde cargo at the trans-Golgi network. Trends Cell Biol. 2011, 21, 159–167. [Google Scholar] [CrossRef]
- Heimann, P.; Laage, S.; Jockusch, H. Defect of sperm assembly in a neurological mutant of the mouse, wobbler (WR). Differentiation 1991, 47, 77–83. [Google Scholar] [CrossRef]
- Leestma, J.E.; Sepsenwol, S. Sperm tail axoneme alterations in the Wobbler mouse. J. Reprod. Fertil. 1980, 58, 267–270. [Google Scholar] [CrossRef]
- Schmitt-John, T.; Drepper, C.; Mussmann, A.; Hahn, P.; Kuhlmann, M.; Thiel, C.; Hafner, M.; Lengeling, A.; Heimann, P.; Jones, J.M.; et al. Mutation of Vps54 causes motor neuron disease and defective spermiogenesis in the wobbler mouse. Nat. Genet. 2005, 37, 1213–1215. [Google Scholar] [CrossRef]
- Paiardi, C.; Pasini, M.E.; Gioria, M.; Berruti, G. Failure of acrosome formation and globozoospermia in the wobbler mouse, a Vps54 spontaneous recessive mutant. Spermatogenesis 2011, 1, 52–62. [Google Scholar] [CrossRef]
- Kierszenbaum, A.L.; Rivkin, E.; Tres, L.L. Acroplaxome, an F-actin-keratin-containing plate, anchors the acrosome to the nucleus during shaping of the spermatid head. Mol. Biol. Cell 2003, 14, 4628–4640. [Google Scholar]
- Yang, W.X.; Jefferson, H.; Sperry, A.O. The molecular motor KIFC1 associates with a complex containing nucleoporin NUP62 that is regulated during development and by the small GTPase RAN. Biol. Reprod. 2006, 74, 684–690. [Google Scholar] [CrossRef]
- Saade, M.; Irla, M.; Govin, J.; Victorero, G.; Samson, M.; Nguyen, C. Dynamic distribution of Spatial during mouse spermatogenesis and its interaction with the kinesin KIF17b. Exp. Cell Res. 2007, 313, 614–626. [Google Scholar] [CrossRef] [Green Version]
- Lehti, M.S.; Kotaja, N.; Sironen, A. KIF3A is essential for sperm tail formation and manchette function. Mol. Cell. Endocrinol. 2013, 377, 44–55. [Google Scholar] [CrossRef]
- Hall, E.S.; Eveleth, J.; Jiang, C.; Redenbach, D.M.; Boekelheide, K. Distribution of the microtubule-dependent motors cytoplasmic dynein and kinesin in rat testis. Biol. Reprod. 1992, 46, 817–828. [Google Scholar] [CrossRef]
- Yoshida, T.; Ioshii, S.O.; Imanaka-Yoshida, K.; Izutsu, K. Association of cytoplasmic dynein with manchette microtubules and spermatid nuclear envelope during spermiogenesis in rats. J. Cell. Sci. 1994, 107 ( Pt.3), 625–633. [Google Scholar]
- Rivkin, E.; Kierszenbaum, A.L.; Gil, M.; Tres, L.L. Rnf19a, a ubiquitin protein ligase, and Psmc3, a component of the 26S proteasome, tether to the acrosome membranes and the head-tail coupling apparatus during rat spermatid development. Dev. Dyn. 2009, 238, 1851–1861. [Google Scholar] [CrossRef]
- Federico, A.; Cardaioli, E.; Da Pozzo, P.; Formichi, P.; Gallus, G.N.; Radi, E. Mitochondria, oxidative stress and neurodegeneration. J. Neurol. Sci. 2012, 322, 254–262. [Google Scholar] [CrossRef]
- Ramalho-Santos, J.; Varum, S.; Amaral, S.; Mota, P.C.; Sousa, A.P.; Amaral, A. Mitochondrial functionality in reproduction: From gonads and gametes to embryos and embryonic stem cells. Hum. Reprod. Update 2009, 15, 553–572. [Google Scholar] [CrossRef]
- Piomboni, P.; Focarelli, R.; Stendardi, A.; Ferramosca, A.; Zara, V. The role of mitochondria in energy production for human sperm motility. Int. J. Androl. 2012, 35, 109–124. [Google Scholar] [CrossRef]
- Amaral, A.; Lourenco, B.; Marques, M.; Ramalho-Santos, J. Mitochondria functionality and sperm quality. Reproduction 2013, 146, R163–R174. [Google Scholar] [CrossRef]
- Gyllensten, U.; Wharton, D.; Josefsson, A.; Wilson, A.C. Paternal inheritance of mitochondrial DNA in mice. Nature 1991, 352, 255–257. [Google Scholar] [CrossRef]
- Kaneda, H.; Hayashi, J.; Takahama, S.; Taya, C.; Lindahl, K.F.; Yonekawa, H. Elimination of paternal mitochondrial DNA in intraspecific crosses during early mouse embryogenesis. Proc. Natl. Acad. Sci. USA 1995, 92, 4542–4546. [Google Scholar] [CrossRef]
- Shitara, H.; Hayashi, J.I.; Takahama, S.; Kaneda, H.; Yonekawa, H. Maternal inheritance of mouse mtDNA in interspecific hybrids: Segregation of the leaked paternal mtDNA followed by the prevention of subsequent paternal leakage. Genetics 1998, 148, 851–857. [Google Scholar]
- Piko, L.; Taylor, K.D. Amounts of mitochondrial DNA and abundance of some mitochondrial gene transcripts in early mouse embryos. Dev. Biol. 1987, 123, 364–374. [Google Scholar] [CrossRef]
- Hecht, N.B.; Liem, H.; Kleene, K.C.; Distel, R.J.; Ho, S.M. Maternal inheritance of the mouse mitochondrial genome is not mediated by a loss or gross alteration of the paternal mitochondrial DNA or by methylation of the oocyte mitochondrial DNA. Dev. Biol. 1984, 102, 452–461. [Google Scholar] [CrossRef]
- Shitara, H.; Kaneda, H.; Sato, A.; Inoue, K.; Ogura, A.; Yonekawa, H.; Hayashi, J.I. Selective and continuous elimination of mitochondria microinjected into mouse eggs from spermatids, but not from liver cells, occurs throughout embryogenesis. Genetics 2000, 156, 1277–1284. [Google Scholar]
- Sutovsky, P.; Moreno, R.D.; Ramalho-Santos, J.; Dominko, T.; Simerly, C.; Schatten, G. Ubiquitin tag for sperm mitochondria. Nature 1999, 402, 371–372. [Google Scholar] [CrossRef]
- Sutovsky, P.; Moreno, R.D.; Ramalho-Santos, J.; Dominko, T.; Simerly, C.; Schatten, G. Ubiquitinated sperm mitochondria, selective proteolysis, and the regulation of mitochondrial inheritance in mammalian embryos. Biol. Reprod. 2000, 63, 582–590. [Google Scholar] [CrossRef]
- Luo, S.M.; Ge, Z.J.; Wang, Z.W.; Jiang, Z.Z.; Wang, Z.B.; Ouyang, Y.C.; Hou, Y.; Schatten, H.; Sun, Q.Y. Unique insights into maternal mitochondrial inheritance in mice. Proc. Natl. Acad. Sci. USA 2013, 110, 13038–13043. [Google Scholar] [CrossRef]
- Yonashiro, R.; Ishido, S.; Kyo, S.; Fukuda, T.; Goto, E.; Matsuki, Y.; Ohmura-Hoshino, M.; Sada, K.; Hotta, H.; Yamamura, H.; et al. A novel mitochondrial ubiquitin ligase plays a critical role in mitochondrial dynamics. EMBO J. 2006, 25, 3618–3626. [Google Scholar] [CrossRef]
- Nakamura, N.; Kimura, Y.; Tokuda, M.; Honda, S.; Hirose, S. MARCH-V is a novel mitofusin 2- and Drp1-binding protein able to change mitochondrial morphology. EMBO Rep. 2006, 7, 1019–1022. [Google Scholar] [CrossRef]
- Ashrafi, G.; Schwarz, T.L. The pathways of mitophagy for quality control and clearance of mitochondria. Cell Death Differ. 2013, 20, 31–42. [Google Scholar] [CrossRef]
- Feng, D.; Liu, L.; Zhu, Y.; Chen, Q. Molecular signaling toward mitophagy and its physiological significance. Exp. Cell Res. 2013, 319, 1697–1705. [Google Scholar] [CrossRef]
- Sato, M.; Sato, K. Degradation of paternal mitochondria by fertilization-triggered autophagy in C. elegans embryos. Science 2011, 334, 1141–1144. [Google Scholar] [CrossRef]
- Al Rawi, S.; Louvet-Vallee, S.; Djeddi, A.; Sachse, M.; Culetto, E.; Hajjar, C.; Boyd, L.; Legouis, R.; Galy, V. Postfertilization autophagy of sperm organelles prevents paternal mitochondrial DNA transmission. Science 2011, 334, 1144–1147. [Google Scholar] [CrossRef]
- Tsukamoto, S.; Kuma, A.; Murakami, M.; Kishi, C.; Yamamoto, A.; Mizushima, N. Autophagy is essential for preimplantation development of mouse embryos. Science 2008, 321, 117–120. [Google Scholar] [CrossRef]
- Okatsu, K.; Saisho, K.; Shimanuki, M.; Nakada, K.; Shitara, H.; Sou, Y.S.; Kimura, M.; Sato, S.; Hattori, N.; Komatsu, M.; et al. p62/SQSTM1 cooperates with Parkin for perinuclear clustering of depolarized mitochondria. Genes Cells 2010, 15, 887–900. [Google Scholar]
- Narendra, D.; Kane, L.A.; Hauser, D.N.; Fearnley, I.M.; Youle, R.J. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy 2010, 6, 1090–1106. [Google Scholar] [CrossRef]
- Escalier, D. New insights into the assembly of the periaxonemal structures in mammalian spermatozoa. Biol. Reprod. 2003, 69, 373–378. [Google Scholar] [CrossRef]
- Hermo, L.; Pelletier, R.M.; Cyr, D.G.; Smith, C.E. Surfing the wave, cycle, life history, and genes/proteins expressed by testicular germ cells. Part 4: Intercellular bridges, mitochondria, nuclear envelope, apoptosis, ubiquitination, membrane/voltage-gated channels, methylation/acetylation, and transcription factors. Microsc. Res. Tech. 2010, 73, 364–408. [Google Scholar]
- Sutovsky, P. Ubiquitin-dependent proteolysis in mammalian spermatogenesis, fertilization, and sperm quality control: Killing three birds with one stone. Microsc. Res. Tech. 2003, 61, 88–102. [Google Scholar] [CrossRef]
- Sutovsky, P. Sperm proteasome and fertilization. Reproduction 2011, 142, 1–14. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Nakamura, N. Ubiquitination Regulates the Morphogenesis and Function of Sperm Organelles. Cells 2013, 2, 732-750. https://doi.org/10.3390/cells2040732
Nakamura N. Ubiquitination Regulates the Morphogenesis and Function of Sperm Organelles. Cells. 2013; 2(4):732-750. https://doi.org/10.3390/cells2040732
Chicago/Turabian StyleNakamura, Nobuhiro. 2013. "Ubiquitination Regulates the Morphogenesis and Function of Sperm Organelles" Cells 2, no. 4: 732-750. https://doi.org/10.3390/cells2040732




