Structural Basis of Targeting the Exportin CRM1 in Cancer
Abstract
:1. Introduction
2. Observation: Alteration of Distribution of Proteins Related to Cancer
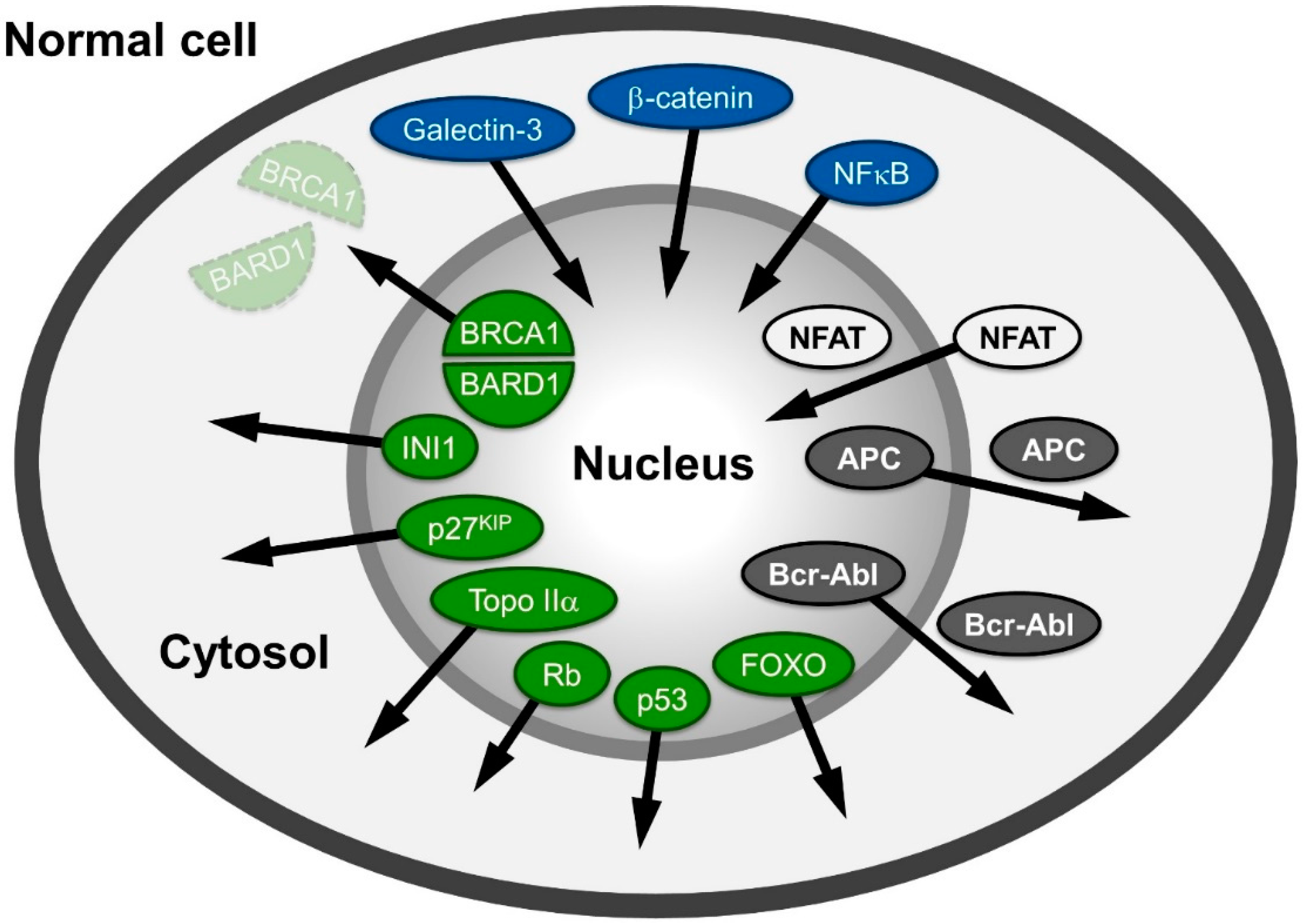
3. The Nucleocytoplasmic Transport Machinery

3.1. Bi-Functional CRM1: Discovery as an Export Receptor and Cell Cycle Control Factor
3.2. Conformational States of CRM1 during Nucleocytoplasmic Transport
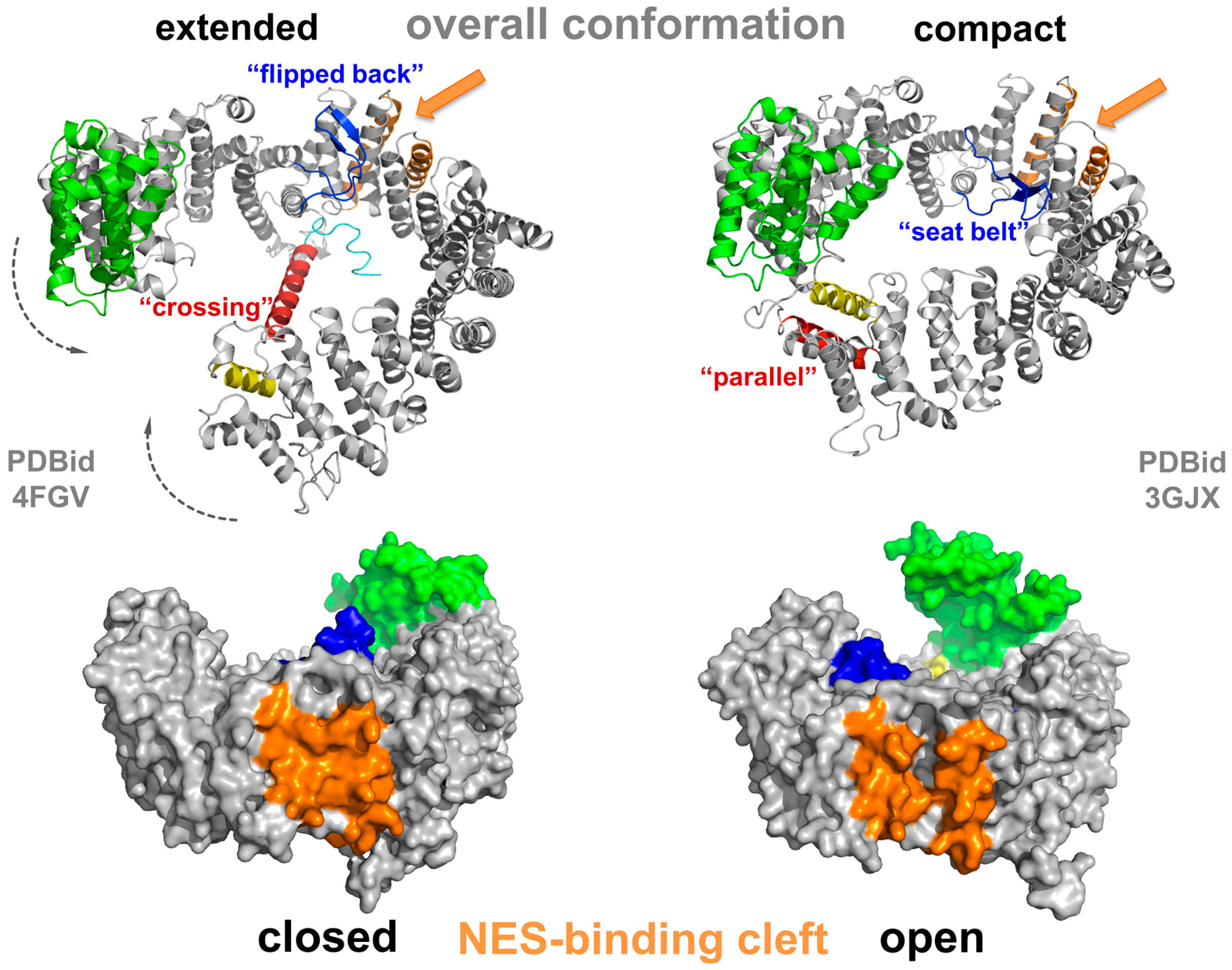
3.3. NES Recognition by CRM1 and Export of (Proto-) Oncoproteins or Tumor Suppressors

3.4. Drug Binding to CRM1

4. Conclusions, Outlook, Pending Issues
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Dobbelstein, M.; Moll, U. Targeting tumour-supportive cellular machineries in anticancer drug development. Nat. Rev. Drug Discov. 2014, 13, 179–196. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, I.; Madar, S.; Rotter, V. Cancer research, a field on the verge of a paradigm shift? Trends Mol. Med. 2012, 18, 299–303. [Google Scholar] [CrossRef] [PubMed]
- Weinberg, R.A. Coming full circle-from endless complexity to simplicity and back again. Cell 2014, 157, 267–271. [Google Scholar] [CrossRef] [PubMed]
- Faustino, R.S.; Nelson, T.J.; Terzic, A.; Perez-Terzic, C. Nuclear transport: Target for therapy. Clin. Pharmacol. Ther. 2007, 81, 880–886. [Google Scholar] [CrossRef] [PubMed]
- Turner, J.G.; Dawson, J.; Sullivan, D.M. Nuclear export of proteins and drug resistance in cancer. Biochem. Pharmacol. 2012, 83, 1021–1032. [Google Scholar] [CrossRef] [PubMed]
- Hill, R.; Cautain, B.; de Pedro, N.; Link, W. Targeting nucleocytoplasmic transport in cancer therapy. Oncotarget 2014, 5, 11–28. [Google Scholar] [CrossRef] [PubMed]
- Stommel, J.M.; Marchenko, N.D.; Jimenez, G.S.; Moll, U.M.; Hope, T.J.; Wahl, G.M. A leucine-rich nuclear export signal in the p53 tetramerization domain: Regulation of subcellular localization and p53 activity by nes masking. EMBO J. 1999, 18, 1660–1672. [Google Scholar] [CrossRef] [PubMed]
- Foo, R.S.; Nam, Y.J.; Ostreicher, M.J.; Metzl, M.D.; Whelan, R.S.; Peng, C.F.; Ashton, A.W.; Fu, W.; Mani, K.; Chin, S.F.; et al. Regulation of p53 tetramerization and nuclear export by arc. Proc. Natl. Acad. Sci. USA 2007, 104, 20826–20831. [Google Scholar] [CrossRef] [PubMed]
- Muller, P.A.; Vousden, K.H. Mutant p53 in cancer: New functions and therapeutic opportunities. Cancer Cell 2014, 25, 304–317. [Google Scholar] [CrossRef] [PubMed]
- Vousden, K.H.; Prives, C. P53 and prognosis: New insights and further complexity. Cell 2005, 120, 7–10. [Google Scholar] [CrossRef] [PubMed]
- Jiao, W.; Datta, J.; Lin, H.M.; Dundr, M.; Rane, S.G. Nucleocytoplasmic shuttling of the retinoblastoma tumor suppressor protein via cdk phosphorylation-dependent nuclear export. J. Biol. Chem. 2006, 281, 38098–38108. [Google Scholar] [CrossRef] [PubMed]
- Kowalik, A.; Kopczynski, J.; Wypiorkiewicz, E.; Gozdz, S.; Mezyk, R.; Siedlecki, J.A. Active transport of rb protein from the nucleus to the cytoplasm as one of the development mechanisms of her2-positive breast cancer. Pol. J. Pathol. Off. J. Pol. Soc. Pathol. 2013, 64, 9–14. [Google Scholar] [CrossRef]
- Mittnacht, S.; Lees, J.A.; Desai, D.; Harlow, E.; Morgan, D.O.; Weinberg, R.A. Distinct sub-populations of the retinoblastoma protein show a distinct pattern of phosphorylation. EMBO J. 1994, 13, 118–127. [Google Scholar] [PubMed]
- Stokke, T.; Erikstein, B.K.; Smedshammer, L.; Boye, E.; Steen, H.B. The retinoblastoma gene product is bound in the nucleus in early g1 phase. Exp. Cell Res. 1993, 204, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Neufeld, K.L.; Nix, D.A.; Bogerd, H.; Kang, Y.; Beckerle, M.C.; Cullen, B.R.; White, R.L. Adenomatous polyposis coli protein contains two nuclear export signals and shuttles between the nucleus and cytoplasm. Proc. Natl. Acad. Sci. USA 2000, 97, 12085–12090. [Google Scholar] [CrossRef] [PubMed]
- Neufeld, K.L.; White, R.L. Nuclear and cytoplasmic localizations of the adenomatous polyposis coli protein. Proc. Natl. Acad. Sci. USA 1997, 94, 3034–3039. [Google Scholar] [CrossRef] [PubMed]
- Powell, S.M.; Zilz, N.; Beazer-Barclay, Y.; Bryan, T.M.; Hamilton, S.R.; Thibodeau, S.N.; Vogelstein, B.; Kinzler, K.W. APC mutations occur early during colorectal tumorigenesis. Nature 1992, 359, 235–237. [Google Scholar] [CrossRef] [PubMed]
- Henderson, B.R.; Fagotto, F. The ins and outs of APC and β-catenin nuclear transport. EMBO Rep. 2002, 3, 834–839. [Google Scholar] [CrossRef] [PubMed]
- Jamieson, C.; Sharma, M.; Henderson, B.R. Targeting the beta-catenin nuclear transport pathway in cancer. Semin. Cancer Biol. 2014, 27, 20–29. [Google Scholar] [CrossRef] [PubMed]
- Crabtree, G.R.; Olson, E.N. Nfat signaling: Choreographing the social lives of cells. Cell 2002, 109, S67–S79. [Google Scholar] [CrossRef]
- Mancini, M.; Toker, A. Nfat proteins: Emerging roles in cancer progression. Nat. Rev. Cancer 2009, 9, 810–820. [Google Scholar] [CrossRef] [PubMed]
- Muller, M.R.; Rao, A. Nfat, immunity and cancer: A transcription factor comes of age. Nat. Rev. Immunol. 2010, 10, 645–656. [Google Scholar] [CrossRef] [PubMed]
- Pan, M.G.; Xiong, Y.; Chen, F. Nfat gene family in inflammation and cancer. Curr. Mol. Med. 2013, 13, 543–554. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.J.; Nag, S.; Wang, W.; Zhou, J.; Zhang, W.D.; Wang, H.; Zhang, R. Nfat as cancer target: Mission possible? Biochim. Biophys. Acta 2014, 1846, 297–311. [Google Scholar] [CrossRef] [PubMed]
- Lugo, T.G.; Pendergast, A.M.; Muller, A.J.; Witte, O.N. Tyrosine kinase activity and transformation potency of Bcr-Abl oncogene products. Science 1990, 247, 1079–1082. [Google Scholar] [CrossRef] [PubMed]
- Cilloni, D.; Saglio, G. Molecular pathways: Bcr-Abl. Clin. Cancer Res. 2012, 18, 930–937. [Google Scholar] [CrossRef] [PubMed]
- Drake, J.M.; Lee, J.K.; Witte, O.N. Clinical targeting of mutated and wild-type protein tyrosine kinases in cancer. Mol. Cell Biol. 2014, 34, 1722–1732. [Google Scholar] [CrossRef] [PubMed]
- Brohawn, S.G.; Partridge, J.R.; Whittle, J.R.; Schwartz, T.U. The nuclear pore complex has entered the atomic age. Structure 2009, 17, 1156–1168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hurt, E.; Beck, M. Towards understanding nuclear pore complex architecture and dynamics in the age of integrative structural analysis. Curr. Opin. Cell Biol. 2015, 34, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Kabachinski, G.; Schwartz, T.U. The nuclear pore complex—Structure and function at a glance. J. Cell Sci. 2015, 128, 423–429. [Google Scholar] [CrossRef] [PubMed]
- Andrade, M.A.; Bork, P. Heat repeats in the huntington’s disease protein. Nat. Genet. 1995, 11, 115–116. [Google Scholar] [CrossRef] [PubMed]
- Andrade, M.A.; Perez-Iratxeta, C.; Ponting, C.P. Protein repeats: Structures, functions, and evolution. J. Struct. Biol. 2001, 134, 117–131. [Google Scholar] [CrossRef] [PubMed]
- Izaurralde, E.; Kutay, U.; von Kobbe, C.; Mattaj, I.W.; Gorlich, D. The asymmetric distribution of the constituents of the ran system is essential for transport into and out of the nucleus. EMBO J. 1997, 16, 6535–6547. [Google Scholar] [CrossRef] [PubMed]
- Ohtsubo, M.; Okazaki, H.; Nishimoto, T. The rcc1 protein, a regulator for the onset of chromosome condensation locates in the nucleus and binds to DNA. J. Cell Biol. 1989, 109, 1389–1397. [Google Scholar] [CrossRef] [PubMed]
- Bischoff, F.R.; Maier, G.; Tilz, G.; Ponstingl, H. A 47-kda human nuclear protein recognized by antikinetochore autoimmune sera is homologous with the protein encoded by rcc1, a gene implicated in onset of chromosome condensation. Proc. Natl. Acad. Sci. USA 1990, 87, 8617–8621. [Google Scholar] [CrossRef] [PubMed]
- Uchida, S.; Sekiguchi, T.; Nishitani, H.; Miyauchi, K.; Ohtsubo, M.; Nishimoto, T. Premature chromosome condensation is induced by a point mutation in the hamster rcc1 gene. Mol. Cell Biol. 1990, 10, 577–584. [Google Scholar] [PubMed]
- Smith, A.E.; Slepchenko, B.M.; Schaff, J.C.; Loew, L.M.; Macara, I.G. Systems analysis of ran transport. Science 2002, 295, 488–491. [Google Scholar] [CrossRef] [PubMed]
- Kalab, P.; Weis, K.; Heald, R. Visualization of a ran-gtp gradient in interphase and mitotic xenopus egg extracts. Science 2002, 295, 2452–2456. [Google Scholar] [CrossRef] [PubMed]
- Bischoff, F.R.; Klebe, C.; Kretschmer, J.; Wittinghofer, A.; Ponstingl, H. RanGAP1 induces GTPase activity of nuclear ras-related ran. Proc. Natl. Acad. Sci. USA 1994, 91, 2587–2591. [Google Scholar] [CrossRef] [PubMed]
- Mahajan, R.; Delphin, C.; Guan, T.; Gerace, L.; Melchior, F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell 1997, 88, 97–107. [Google Scholar] [CrossRef]
- Matunis, M.J.; Coutavas, E.; Blobel, G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell Biol. 1996, 135, 1457–1470. [Google Scholar] [CrossRef] [PubMed]
- Culjkovic-Kraljacic, B.; Baguet, A.; Volpon, L.; Amri, A.; Borden, K.L. The oncogene eIF4e reprograms the nuclear pore complex to promote mrna export and oncogenic transformation. Cell Rep. 2012, 2, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Culjkovic-Kraljacic, B.; Borden, K.L. Aiding and abetting cancer: Mrna export and the nuclear pore. Trends Cell Biol. 2013, 23, 328–335. [Google Scholar] [CrossRef] [PubMed]
- Adachi, Y.; Yanagida, M. Higher order chromosome structure is affected by cold-sensitive mutations in a schizosaccharomyces pombe gene CRM1+ which encodes a 115-kd protein preferentially localized in the nucleus and its periphery. J. Cell Biol. 1989, 108, 1195–1207. [Google Scholar] [CrossRef] [PubMed]
- Fornerod, M.; Boer, J.; van Baal, S.; Morreau, H.; Grosveld, G. Interaction of cellular proteins with the leukemia specific fusion proteins dek-can and set-can and their normal counterpart, the nucleoporin can. Oncogene 1996, 13, 1801–1808. [Google Scholar] [PubMed]
- Fornerod, M.; van Deursen, J.; van Baal, S.; Reynolds, A.; Davis, D.; Murti, K.G.; Fransen, J.; Grosveld, G. The human homologue of yeast CRM1 is in a dynamic subcomplex with can/Nup214 and a novel nuclear pore component Nup88. EMBO J. 1997, 16, 807–816. [Google Scholar] [CrossRef] [PubMed]
- Fornerod, M.; Ohno, M.; Yoshida, M.; Mattaj, I.W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 1997, 90, 1051–1060. [Google Scholar] [CrossRef]
- Fukuda, M.; Asano, S.; Nakamura, T.; Adachi, M.; Yoshida, M.; Yanagida, M.; Nishida, E. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature 1997, 390, 308–311. [Google Scholar] [PubMed]
- Neville, M.; Stutz, F.; Lee, L.; Davis, L.I.; Rosbash, M. The importin-beta family member CRM1p bridges the interaction between rev and the nuclear pore complex during nuclear export. Curr. Biol. 1997, 7, 767–775. [Google Scholar] [CrossRef]
- Ossareh-Nazari, B.; Bachelerie, F.; Dargemont, C. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science 1997, 278, 141–144. [Google Scholar] [CrossRef] [PubMed]
- Stade, K.; Ford, C.S.; Guthrie, C.; Weis, K. Exportin 1 (CRM1p) is an essential nuclear export factor. Cell 1997, 90, 1041–1050. [Google Scholar] [CrossRef]
- Kehlenbach, R.H.; Dickmanns, A.; Gerace, L. Nucleocytoplasmic shuttling factors including ran and CRM1 mediate nuclear export of nfat in vitro. J. Cell Biol. 1998, 141, 863–874. [Google Scholar] [CrossRef] [PubMed]
- Fischer, U.; Huber, J.; Boelens, W.C.; Mattaj, I.W.; Luhrmann, R. The HIV-1 rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular rnas. Cell 1995, 82, 475–483. [Google Scholar] [CrossRef]
- Wen, W.; Meinkoth, J.L.; Tsien, R.Y.; Taylor, S.S. Identification of a signal for rapid export of proteins from the nucleus. Cell 1995, 82, 463–473. [Google Scholar] [CrossRef]
- Fu, S.C.; Huang, H.C.; Horton, P.; Juan, H.F. Validness: A database of validated leucine-rich nuclear export signals. Nucleic Acids Res. 2013, 41, D338–D343. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Farmer, A.; Collett, G.; Grishin, N.V.; Chook, Y.M. Sequence and structural analyses of nuclear export signals in the nesdb database. Mol. Biol. Cell 2012, 23, 3677–3693. [Google Scholar] [CrossRef] [PubMed]
- Ohno, M.; Segref, A.; Bachi, A.; Wilm, M.; Mattaj, I.W. Phax, a mediator of u snrna nuclear export whose activity is regulated by phosphorylation. Cell 2000, 101, 187–198. [Google Scholar] [CrossRef]
- Segref, A.; Mattaj, I.W.; Ohno, M. The evolutionarily conserved region of the U snRNA export mediator phax is a novel rna-binding domain that is essential for u snrna export. RNA 2001, 7, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Wolff, B.; Sanglier, J.J.; Wang, Y. Leptomycin b is an inhibitor of nuclear export: Inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) rev protein and rev-dependent mrna. Chem. Biol. 1997, 4, 139–147. [Google Scholar] [CrossRef]
- Booth, D.S.; Cheng, Y.; Frankel, A.D. The export receptor CRM1 forms a dimer to promote nuclear export of HIV RNA. eLife 2014, 3, e04121. [Google Scholar] [CrossRef] [PubMed]
- Roscioli, E.; di Francesco, L.; Bolognesi, A.; Giubettini, M.; Orlando, S.; Harel, A.; Schinina, M.E.; Lavia, P. Importin-beta negatively regulates multiple aspects of mitosis including RanGAP1 recruitment to kinetochores. J. Cell Biol. 2012, 196, 435–450. [Google Scholar] [CrossRef] [PubMed]
- Di Fiore, B.; Ciciarello, M.; Lavia, P. Mitotic functions of the Ran-GTPase network: The importance of being in the right place at the right time. Cell Cycle 2004, 3, 305–313. [Google Scholar] [CrossRef] [PubMed]
- Arnaoutov, A.; Azuma, Y.; Ribbeck, K.; Joseph, J.; Boyarchuk, Y.; Karpova, T.; McNally, J.; Dasso, M. CRM1 is a mitotic effector of ran-gtp in somatic cells. Nat. Cell Biol. 2005, 7, 626–632. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Budhu, A.; Forgues, M.; Wang, X.W. Temporal and spatial control of nucleophosmin by the ran-CRM1 complex in centrosome duplication. Nat. Cell Biol. 2005, 7, 823–830. [Google Scholar] [CrossRef] [PubMed]
- Torosantucci, L.; de Luca, M.; Guarguaglini, G.; Lavia, P.; Degrassi, F. Localized rangtp accumulation promotes microtubule nucleation at kinetochores in somatic mammalian cells. Mol. Biol. Cell 2008, 19, 1873–1882. [Google Scholar] [CrossRef] [PubMed]
- Roscioli, E.; Bolognesi, A.; Guarguaglini, G.; Lavia, P. Ran control of mitosis in human cells: Gradients and local signals. Biochem. Soc. Trans. 2010, 38, 1709–1714. [Google Scholar] [CrossRef] [PubMed]
- Neuber, A.; Franke, J.; Wittstruck, A.; Schlenstedt, G.; Sommer, T.; Stade, K. Nuclear export receptor XPO1/CRM1 is physically and functionally linked to the spindle pole body in budding yeast. Mol. Cell Biol. 2008, 28, 5348–5358. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Biswas, A.; Suel, K.E.; Jackson, L.K.; Martinez, R.; Gu, H.; Chook, Y.M. Structural basis for leucine-rich nuclear export signal recognition by CRM1. Nature 2009, 458, 1136–1141. [Google Scholar] [CrossRef] [PubMed]
- Monecke, T.; Guttler, T.; Neumann, P.; Dickmanns, A.; Gorlich, D.; Ficner, R. Crystal structure of the nuclear export receptor CRM1 in complex with snurportin1 and rangtp. Science 2009, 324, 1087–1091. [Google Scholar] [CrossRef] [PubMed]
- Monecke, T.; Dickmanns, A.; Ficner, R. Allosteric control of the exportin CRM1 unraveled by crystal structure analysis. FEBS J. 2014, 281, 4179–4194. [Google Scholar] [CrossRef] [PubMed]
- Zachariae, U.; Grubmuller, H. A highly strained nuclear conformation of the exportin CSE1P revealed by molecular dynamics simulations. Structure 2006, 14, 1469–1478. [Google Scholar] [CrossRef] [PubMed]
- Zachariae, U.; Grubmuller, H. Importin-beta: Structural and dynamic determinants of a molecular spring. Structure 2008, 16, 906–915. [Google Scholar] [CrossRef] [PubMed]
- Monecke, T.; Haselbach, D.; Voβ, B.; Russek, A.; Neumann, A.; Thomson, E.; Hurt, E.; Zachariae, U.; Stark, H.; Grubmüller, H.; et al. Structural basis for cooperativity of CRM1 export complex formation. Proc. Natl. Acad. Sci. USA 2013, 110, 960–965. [Google Scholar] [CrossRef] [PubMed]
- Dolker, N.; Blanchet, C.E.; Voss, B.; Haselbach, D.; Kappel, C.; Monecke, T.; Svergun, D.I.; Stark, H.; Ficner, R.; Zachariae, U.; et al. Structural determinants and mechanism of mammalian CRM1 allostery. Structure 2013, 21, 1350–1360. [Google Scholar] [CrossRef] [PubMed]
- Saito, N.; Matsuura, Y. A 2.1-A-resolution crystal structure of unliganded CRM1 reveals the mechanism of autoinhibition. J. Mol. Biol. 2013, 425, 350–364. [Google Scholar] [CrossRef] [PubMed]
- Fox, A.M.; Ciziene, D.; McLaughlin, S.H.; Stewart, M. Electrostatic interactions involving the extreme C terminus of nuclear export factor CRM1 modulate its affinity for cargo. J. Biol. Chem. 2011, 286, 29325–29335. [Google Scholar] [CrossRef] [PubMed]
- Petosa, C.; Schoehn, G.; Askjaer, P.; Bauer, U.; Moulin, M.; Steuerwald, U.; Soler-Lopez, M.; Baudin, F.; Mattaj, I.W.; Muller, C.W. Architecture of CRM1/exportin1 suggests how cooperativity is achieved during formation of a nuclear export complex. Mol. Cell 2004, 16, 761–775. [Google Scholar] [CrossRef] [PubMed]
- Guttler, T.; Madl, T.; Neumann, P.; Deichsel, D.; Corsini, L.; Monecke, T.; Ficner, R.; Sattler, M.; Gorlich, D. Nes consensus redefined by structures of pki-type and rev-type nuclear export signals bound to CRM1. Nat. Struct. Mol. Biol. 2010, 17, 1367–1376. [Google Scholar] [CrossRef] [PubMed]
- Gorlich, D.; Dabrowski, M.; Bischoff, F.R.; Kutay, U.; Bork, P.; Hartmann, E.; Prehn, S.; Izaurralde, E. A novel class of rangtp binding proteins. J. Cell Biol. 1997, 138, 65–80. [Google Scholar] [CrossRef] [PubMed]
- Koyama, M.; Matsuura, Y. An allosteric mechanism to displace nuclear export cargo from CRM1 and rangtp by ranbp1. EMBO J. 2010, 29, 2002–2013. [Google Scholar] [CrossRef] [PubMed]
- Matunis, M.J.; Wu, J.; Blobel, G. Sumo-1 modification and its role in targeting the Ran-GTPase-activating protein, RanGAP1, to the nuclear pore complex. J. Cell Biol. 1998, 140, 499–509. [Google Scholar] [CrossRef] [PubMed]
- Strasser, A.; Dickmanns, A.; Luhrmann, R.; Ficner, R. Structural basis for M3G-cap-mediated nuclear import of spliceosomal usnrnps by snurportin1. EMBO J. 2005, 24, 2235–2243. [Google Scholar] [CrossRef] [PubMed]
- Paraskeva, E.; Izaurralde, E.; Bischoff, F.R.; Huber, J.; Kutay, U.; Hartmann, E.; Luhrmann, R.; Gorlich, D. CRM1-mediated recycling of snurportin 1 to the cytoplasm. J. Cell Biol. 1999, 145, 255–264. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, J.A.; Henderson, B.R. Identification of a functional nuclear export sequence in BRCA1. J. Biol. Chem. 2000, 275, 38589–38596. [Google Scholar] [CrossRef] [PubMed]
- Hwang, C.Y.; Kim, I.Y.; Kwon, K.S. Cytoplasmic localization and ubiquitination of p21(cip1) by reactive oxygen species. Biochem. Biophys. Res. Commun. 2007, 358, 219–225. [Google Scholar] [CrossRef] [PubMed]
- Benzeno, S.; Diehl, J.A. C-terminal sequences direct cyclin d1-CRM1 binding. J. Biol. Chem. 2004, 279, 56061–56066. [Google Scholar] [CrossRef] [PubMed]
- Henderson, B.R. Nuclear-cytoplasmic shuttling of APC regulates β-catenin subcellular localization and turnover. Nat. Cell Biol. 2000, 2, 653–660. [Google Scholar] [CrossRef] [PubMed]
- Rosin-Arbesfeld, R.; Townsley, F.; Bienz, M. The APC tumour suppressor has a nuclear export function. Nature 2000, 406, 1009–1012. [Google Scholar] [PubMed]
- Bartholomeusz, G.; Wu, Y.; Ali Seyed, M.; Xia, W.; Kwong, K.Y.; Hortobagyi, G.; Hung, M.C. Nuclear translocation of the pro-apoptotic BCL-2 family member bok induces apoptosis. Mol. Carcinog. 2006, 45, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Brunet, A.; Kanai, F.; Stehn, J.; Xu, J.; Sarbassova, D.; Frangioni, J.V.; Dalal, S.N.; de Caprio, J.A.; Greenberg, M.E.; Yaffe, M.B. 14-3-3 transits to the nucleus and participates in dynamic nucleocytoplasmic transport. J. Cell Biol. 2002, 156, 817–828. [Google Scholar] [CrossRef] [PubMed]
- Latre de Late, P.; Pepin, A.; Assaf-Vandecasteele, H.; Espinasse, C.; Nicolas, V.; Asselin-Labat, M.L.; Bertoglio, J.; Pallardy, M.; Biola-Vidamment, A. Glucocorticoid-induced leucine zipper (GILZ) promotes the nuclear exclusion of FOXO3 in a CRM1-dependent manner. J. Biol. Chem. 2010, 285, 5594–5605. [Google Scholar] [CrossRef] [PubMed]
- Howell, J.J.; Stoffel, M. Nuclear export-independent inhibition of FOXA2 by insulin. J. Biol. Chem. 2009, 284, 24816–24824. [Google Scholar] [CrossRef] [PubMed]
- Falini, B.; Bolli, N.; Shan, J.; Martelli, M.P.; Liso, A.; Pucciarini, A.; Bigerna, B.; Pasqualucci, L.; Mannucci, R.; Rosati, R.; et al. Both carboxy-terminus nes motif and mutated tryptophan(s) are crucial for aberrant nuclear export of nucleophosmin leukemic mutants in NPMC+ AML. Blood 2006, 107, 4514–4523. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Maggi, L.B., Jr.; Brady, S.N.; Apicelli, A.J.; Dai, M.S.; Lu, H.; Weber, J.D. Nucleophosmin is essential for ribosomal protein l5 nuclear export. Mol. Cell Biol. 2006, 26, 3798–3809. [Google Scholar] [CrossRef] [PubMed]
- Suetsugu, S.; Takenawa, T. Translocation of n-wasp by nuclear localization and export signals into the nucleus modulates expression of HSP90. J. Biol. Chem. 2003, 278, 42515–42523. [Google Scholar] [CrossRef] [PubMed]
- Mirski, S.E.; Bielawski, J.C.; Cole, S.P. Identification of functional nuclear export sequences in human topoisomerase IIα and β. Biochem. Biophys. Res. Commun. 2003, 306, 905–911. [Google Scholar] [CrossRef]
- Turner, J.G.; Engel, R.; Derderian, J.A.; Jove, R.; Sullivan, D.M. Human topoisomerase IIα nuclear export is mediated by two CRM-1-dependent nuclear export signals. J. Cell Sci. 2004, 117, 3061–3071. [Google Scholar] [CrossRef] [PubMed]
- Mirski, S.E.; Sparks, K.E.; Friedrich, B.; Kohler, M.; Mo, Y.Y.; Beck, W.T.; Cole, S.P. Topoisomerase II binds importin α isoforms and exportin/CRM1 but does not shuttle between the nucleus and cytoplasm in proliferating cells. Exp. Cell Res. 2007, 313, 627–637. [Google Scholar] [CrossRef] [PubMed]
- Arregi, I.; Falces, J.; Olazabal-Herrero, A.; Alonso-Marino, M.; Taneva, S.G.; Rodriguez, J.A.; Urbaneja, M.A.; Banuelos, S. Leukemia-associated mutations in nucleophosmin alter recognition by CRM1: Molecular basis of aberrant transport. PLoS ONE 2015, 10, e0130610. [Google Scholar] [CrossRef] [PubMed]
- Ishida, N.; Hara, T.; Kamura, T.; Yoshida, M.; Nakayama, K.; Nakayama, K.I. Phosphorylation of p27kip1 on serine 10 is required for its binding to CRM1 and nuclear export. J. Biol. Chem. 2002, 277, 14355–14358. [Google Scholar] [CrossRef] [PubMed]
- Ishida, N.; Hara, T.; Kamura, T.; Yoshida, M.; Nakayama, K.; Nakayama, K.I. Phosphorylation of p27kip1 on serine 10 is required for its binding to CRM1 and nuclear export. J. Biol. Chem. 2015, 290, 6754. [Google Scholar] [CrossRef] [PubMed]
- Connor, M.K.; Kotchetkov, R.; Cariou, S.; Resch, A.; Lupetti, R.; Beniston, R.G.; Melchior, F.; Hengst, L.; Slingerland, J.M. CRM1/ran-mediated nuclear export of p27(kip1) involves a nuclear export signal and links p27 export and proteolysis. Mol. Biol. Cell 2003, 14, 201–213. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xiong, Y. A p53 amino-terminal nuclear export signal inhibited by DNA damage-induced phosphorylation. Science 2001, 292, 1910–1915. [Google Scholar] [CrossRef] [PubMed]
- Santiago, A.; Li, D.; Zhao, L.Y.; Godsey, A.; Liao, D. P53 sumoylation promotes its nuclear export by facilitating its release from the nuclear export receptor CRM1. Mol. Biol. Cell 2013, 24, 2739–2752. [Google Scholar] [CrossRef] [PubMed]
- Lohrum, M.A.; Woods, D.B.; Ludwig, R.L.; Balint, E.; Vousden, K.H. C-terminal ubiquitination of p53 contributes to nuclear export. Mol. Cell Biol. 2001, 21, 8521–8532. [Google Scholar] [CrossRef] [PubMed]
- Inoue, T.; Geyer, R.K.; Howard, D.; Yu, Z.K.; Maki, C.G. MDM2 can promote the ubiquitination, nuclear export, and degradation of p53 in the absence of direct binding. J. Biol. Chem. 2001, 276, 45255–45260. [Google Scholar] [CrossRef] [PubMed]
- Craig, E.; Zhang, Z.K.; Davies, K.P.; Kalpana, G.V. A masked nes in INI1/HSNF5 mediates HCRM1-dependent nuclear export: Implications for tumorigenesis. EMBO J. 2002, 21, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; McKeon, F. NF-AT activation requires suppression of CRM1-dependent export by calcineurin (see comments). Nature 1999, 398, 256–260. [Google Scholar] [PubMed]
- Jeyasekharan, A.D.; Liu, Y.; Hattori, H.; Pisupati, V.; Jonsdottir, A.B.; Rajendra, E.; Lee, M.; Sundaramoorthy, E.; Schlachter, S.; Kaminski, C.F.; et al. A cancer-associated BRCA2 mutation reveals masked nuclear export signals controlling localization. Nat. Struct. Mol. Biol. 2013, 20, 1191–1198. [Google Scholar] [CrossRef] [PubMed]
- Nitiss, J.L. Targeting DNA topoisomerase II in cancer chemotherapy. Nat. Rev. Cancer 2009, 9, 338–350. [Google Scholar] [CrossRef] [PubMed]
- Nitiss, J.L. DNA topoisomerase II and its growing repertoire of biological functions. Nat. Rev. Cancer 2009, 9, 327–337. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.C. Cellular roles of DNA topoisomerases: A molecular perspective. Nat. Rev. Mol. Cell Biol. 2002, 3, 430–440. [Google Scholar] [CrossRef] [PubMed]
- Engel, R.; Valkov, N.I.; Gump, J.L.; Hazlehurst, L.; Dalton, W.S.; Sullivan, D.M. The cytoplasmic trafficking of DNA topoisomerase IIα correlates with etoposide resistance in human myeloma cells. Exp. Cell Res. 2004, 295, 421–431. [Google Scholar] [CrossRef] [PubMed]
- Valkov, N.I.; Sullivan, D.M. Drug resistance to DNA topoisomerase I and II inhibitors in human leukemia, lymphoma, and multiple myeloma. Semin. Hematol. 1997, 34, 48–62. [Google Scholar] [PubMed]
- Turner, J.G.; Marchion, D.C.; Dawson, J.L.; Emmons, M.F.; Hazlehurst, L.A.; Washausen, P.; Sullivan, D.M. Human multiple myeloma cells are sensitized to topoisomerase II inhibitors by CRM1 inhibition. Cancer Res. 2009, 69, 6899–6905. [Google Scholar] [CrossRef] [PubMed]
- Turner, J.G.; Dawson, J.; Emmons, M.F.; Cubitt, C.L.; Kauffman, M.; Shacham, S.; Hazlehurst, L.A.; Sullivan, D.M. CRM1 inhibition sensitizes drug resistant human myeloma cells to topoisomerase II and proteasome inhibitors both in vitro and ex vivo. J. Cancer 2013, 4, 614–625. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Xu, X.L.; Yang, M.C.; Wei, F.; Ayi, T.C.; Bowcock, A.M.; Baer, R. Cell cycle-dependent colocalization of BARD1 and BRCA1 proteins in discrete nuclear domains. Proc. Natl. Acad. Sci. USA 1997, 94, 12075–12080. [Google Scholar] [CrossRef] [PubMed]
- Baer, R.; Ludwig, T. The BRCA1/BARD1 heterodimer, a tumor suppressor complex with ubiquitin E3 ligase activity. Curr. Opin. Genet. Dev. 2002, 12, 86–91. [Google Scholar] [CrossRef]
- Fabbro, M.; Rodriguez, J.A.; Baer, R.; Henderson, B.R. BARD1 induces BRCA1 intranuclear foci formation by increasing ring-dependent BRCA1 nuclear import and inhibiting BRCA1 nuclear export. J. Biol. Chem. 2002, 277, 21315–21324. [Google Scholar] [CrossRef] [PubMed]
- Scully, R.; Chen, J.; Ochs, R.L.; Keegan, K.; Hoekstra, M.; Feunteun, J.; Livingston, D.M. Dynamic changes of BRCA1 subnuclear location and phosphorylation state are initiated by DNA damage. Cell 1997, 90, 425–435. [Google Scholar] [CrossRef]
- Rodriguez, J.A.; Schuchner, S.; Au, W.W.; Fabbro, M.; Henderson, B.R. Nuclear-cytoplasmic shuttling of BARD1 contributes to its proapoptotic activity and is regulated by dimerization with BRCA1. Oncogene 2004, 23, 1809–1820. [Google Scholar] [CrossRef] [PubMed]
- Thompson, M.E.; Robinson-Benion, C.L.; Holt, J.T. An amino-terminal motif functions as a second nuclear export sequence in BRCA1. J. Biol. Chem. 2005, 280, 21854–21857. [Google Scholar] [CrossRef] [PubMed]
- Fabbro, M.; Henderson, B.R. Regulation of tumor suppressors by nuclear-cytoplasmic shuttling. Exp. Cell Res. 2003, 282, 59–69. [Google Scholar] [CrossRef]
- Rodriguez, J.A.; Span, S.W.; Ferreira, C.G.; Kruyt, F.A.; Giaccone, G. CRM1-mediated nuclear export determines the cytoplasmic localization of the antiapoptotic protein survivin. Exp. Cell Res. 2002, 275, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Knauer, S.K.; Kramer, O.H.; Knosel, T.; Engels, K.; Rodel, F.; Kovacs, A.F.; Dietmaier, W.; Klein-Hitpass, L.; Habtemichael, N.; Schweitzer, A.; et al. Nuclear export is essential for the tumor-promoting activity of survivin. FASEB J. 2007, 21, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Conway, E.M.; Pollefeyt, S.; Cornelissen, J.; DeBaere, I.; Steiner-Mosonyi, M.; Ong, K.; Baens, M.; Collen, D.; Schuh, A.C. Three differentially expressed survivin cdna variants encode proteins with distinct antiapoptotic functions. Blood 2000, 95, 1435–1442. [Google Scholar] [PubMed]
- Uren, A.G.; Wong, L.; Pakusch, M.; Fowler, K.J.; Burrows, F.J.; Vaux, D.L.; Choo, K.H. Survivin and the inner centromere protein incenp show similar cell-cycle localization and gene knockout phenotype. Curr. Biol. 2000, 10, 1319–1328. [Google Scholar] [CrossRef]
- Knauer, S.K.; Bier, C.; Habtemichael, N.; Stauber, R.H. The survivin-CRM1 interaction is essential for chromosomal passenger complex localization and function. EMBO Rep. 2006, 7, 1259–1265. [Google Scholar] [PubMed]
- Polyak, K.; Lee, M.H.; Erdjument-Bromage, H.; Koff, A.; Roberts, J.M.; Tempst, P.; Massague, J. Cloning of p27kip1, a cyclin-dependent kinase inhibitor and a potential mediator of extracellular antimitogenic signals. Cell 1994, 78, 59–66. [Google Scholar] [CrossRef]
- Rivard, N.; L’Allemain, G.; Bartek, J.; Pouyssegur, J. Abrogation of p27kip1 by cdna antisense suppresses quiescence (G0 state) in fibroblasts. J. Biol. Chem. 1996, 271, 18337–18341. [Google Scholar] [CrossRef] [PubMed]
- Coats, S.; Flanagan, W.M.; Nourse, J.; Roberts, J.M. Requirement of p27kip1 for restriction point control of the fibroblast cell cycle. Science 1996, 272, 877–880. [Google Scholar] [CrossRef] [PubMed]
- Bouchard, C.; Thieke, K.; Maier, A.; Saffrich, R.; Hanley-Hyde, J.; Ansorge, W.; Reed, S.; Sicinski, P.; Bartek, J.; Eilers, M. Direct induction of cyclin D2 by myc contributes to cell cycle progression and sequestration of p27. EMBO J. 1999, 18, 5321–5333. [Google Scholar] [CrossRef] [PubMed]
- Perez-Roger, I.; Kim, S.H.; Griffiths, B.; Sewing, A.; Land, H. Cyclins D1 and D2 mediate MYC-induced proliferation via sequestration of p27(kip1) and p21(cip1). EMBO J. 1999, 18, 5310–5320. [Google Scholar] [CrossRef] [PubMed]
- Serres, M.P.; Zlotek-Zlotkiewicz, E.; Concha, C.; Gurian-West, M.; Daburon, V.; Roberts, J.M.; Besson, A. Cytoplasmic p27 is oncogenic and cooperates with RAS both in vivo and in vitro. Oncogene 2011, 30, 2846–2858. [Google Scholar] [CrossRef] [PubMed]
- Sherr, C.J.; Roberts, J.M. Cdk inhibitors: Positive and negative regulators of G1-phase progression. Genes Dev. 1999, 13, 1501–1512. [Google Scholar] [CrossRef] [PubMed]
- Besson, A.; Assoian, R.K.; Roberts, J.M. Regulation of the cytoskeleton: An oncogenic function for CDK inhibitors? Nat. Rev. Cancer 2004, 4, 948–955. [Google Scholar] [CrossRef] [PubMed]
- Etchin, J.; Sanda, T.; Mansour, M.R.; Kentsis, A.; Montero, J.; Le, B.T.; Christie, A.L.; McCauley, D.; Rodig, S.J.; Kauffman, M.; et al. Kpt-330 inhibitor of CRM1 (XPO1)-mediated nuclear export has selective anti-leukaemic activity in preclinical models of T-cell acute lymphoblastic leukaemia and acute myeloid leukaemia. Br. J. Haematol. 2013, 161, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, Y.; Xiang, J.; Ji, F.; Deng, Y.; Tang, C.; Yang, S.; Xi, Q.; Liu, R.; Di, W. Knockdown of CRM1 inhibits the nuclear export of p27(kip1) phosphorylated at serine 10 and plays a role in the pathogenesis of epithelial ovarian cancer. Cancer Lett. 2014, 343, 6–13. [Google Scholar] [CrossRef] [PubMed]
- Noske, A.; Weichert, W.; Niesporek, S.; Roske, A.; Buckendahl, A.C.; Koch, I.; Sehouli, J.; Dietel, M.; Denkert, C. Expression of the nuclear export protein chromosomal region maintenance/exportin 1/XPO1 is a prognostic factor in human ovarian cancer. Cancer 2008, 112, 1733–1743. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.Y.; Yue, L.; Qiu, W.S.; Wang, L.W.; Zhou, X.H.; Sun, Y.J. Prognostic value of CRM1 in pancreas cancer. Clin. Investig. Med. 2009, 32, 315–321. [Google Scholar]
- Azmi, A.S.; Aboukameel, A.; Bao, B.; Sarkar, F.H.; Philip, P.A.; Kauffman, M.; Shacham, S.; Mohammad, R.M. Selective inhibitors of nuclear export block pancreatic cancer cell proliferation and reduce tumor growth in mice. Gastroenterology 2013, 144, 447–456. [Google Scholar] [CrossRef] [PubMed]
- Inoue, H.; Kauffman, M.; Shacham, S.; Landesman, Y.; Yang, J.; Evans, C.P.; Weiss, R.H. CRM1 blockade by selective inhibitors of nuclear export (sine) attenuates kidney cancer growth. J. Urol. 2012, 189, 2317–2326. [Google Scholar] [CrossRef] [PubMed]
- Van der Watt, P.J.; Maske, C.P.; Hendricks, D.T.; Parker, M.I.; Denny, L.; Govender, D.; Birrer, M.J.; Leaner, V.D. The karyopherin proteins, CRM1 and karyopherin β1, are overexpressed in cervical cancer and are critical for cancer cell survival and proliferation. Intern. J. Cancer 2009, 124, 1829–1840. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Qiu, W.; Yao, R.; Xiang, J.; Sun, X.; Liu, S.; Lv, J.; Yue, L. CRM1 is a novel independent prognostic factor for the poor prognosis of gastric carcinomas. Med. Oncol. 2013, 30, 726. [Google Scholar] [CrossRef] [PubMed]
- Yao, H. The expression of CRM1 is associated with prognosis in human osteosarcoma. Oncol. Rep. 2009, 21, 229–235. [Google Scholar] [CrossRef] [PubMed]
- Shen, A.; Wang, Y.; Zhao, Y.; Zou, L.; Sun, L.; Cheng, C. Expression of CRM1 in human gliomas and its significance in p27 expression and clinical prognosis. Neurosurgery 2009, 65, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Lapalombella, R.; Sun, Q.; Williams, K.; Tangeman, L.; Jha, S.; Zhong, Y.; Goettl, V.; Mahoney, E.; Berglund, C.; Gupta, S.; et al. Selective inhibitors of nuclear export show that CRM1/XPO1 is a target in chronic lymphocytic leukemia. Blood 2012, 120, 4621–4634. [Google Scholar] [CrossRef] [PubMed]
- Kojima, K.; Kornblau, S.M.; Ruvolo, V.; Dilip, A.; Duvvuri, S.; Davis, R.E.; Zhang, M.; Wang, Z.; Coombes, K.R.; Zhang, N.; et al. Prognostic impact and targeting of CRM1 in acute myeloid leukemia. Blood 2013, 121, 4166–4174. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Wang, M.; Tamayo, A.T.; Shacham, S.; Kauffman, M.; Lee, J.; Zhang, L.; Ou, Z.; Li, C.; Sun, L.; et al. Novel selective inhibitors of nuclear export CRM1 antagonists for therapy in mantle cell lymphoma. Exp. Hematol. 2013, 41, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, J.; Braggio, E.; Kortuem, K.M.; Egan, J.B.; Zhu, Y.X.; Xin, C.S.; Tiedemann, R.E.; Palmer, S.E.; Garbitt, V.M.; McCauley, D.; et al. Genome-wide studies in multiple myeloma identify XPO1/CRM1 as a critical target validated using the selective nuclear export inhibitor KPT-276. Leukemia 2013, 27, 2357–2365. [Google Scholar] [CrossRef] [PubMed]
- Tai, Y.T.; Landesman, Y.; Acharya, C.; Calle, Y.; Zhong, M.Y.; Cea, M.; Tannenbaum, D.; Cagnetta, A.; Reagan, M.; Munshi, A.A.; et al. CRM1 inhibition induces tumor cell cytotoxicity and impairs osteoclastogenesis in multiple myeloma: Molecular mechanisms and therapeutic implications. Leukemia 2014, 28, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Pathria, G.; Wagner, C.; Wagner, S.N. Inhibition of CRM1-mediated nucleocytoplasmic transport: Triggering human melanoma cell apoptosis by perturbing multiple cellular pathways. J. Investig. Dermatol. 2012, 132, 2780–2790. [Google Scholar] [CrossRef] [PubMed]
- Ranganathan, P.; Yu, X.; Na, C.; Santhanam, R.; Shacham, S.; Kauffman, M.; Walker, A.; Klisovic, R.; Blum, W.; Caligiuri, M.; et al. Preclinical activity of a novel CRM1 inhibitor in acute myeloid leukemia. Blood 2012, 120, 1765–1773. [Google Scholar] [CrossRef] [PubMed]
- Etchin, J.; Sun, Q.; Kentsis, A.; Farmer, A.; Zhang, Z.C.; Sanda, T.; Mansour, M.R.; Barcelo, C.; McCauley, D.; Kauffman, M.; et al. Antileukemic activity of nuclear export inhibitors that spare normal hematopoietic cells. Leukemia 2013, 27, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Caceres-Gorriti, K.Y.; Carmona, E.; Barres, V.; Rahimi, K.; Letourneau, I.J.; Tonin, P.N.; Provencher, D.; Mes-Masson, A.M. Ran nucleo-cytoplasmic transport and mitotic spindle assembly partners xpo7 and tpx2 are new prognostic biomarkers in serous epithelial ovarian cancer. PLoS ONE 2014, 9, e91000. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Wang, Y.; Huang, H.; Yang, Q.; Cai, J.; Wang, Q.; Gu, X.; Xu, P.; Zhang, S.; Li, M.; et al. Upregulation of kpnbeta1 in gastric cancer cell promotes tumor cell proliferation and predicts poor prognosis. Tumour Biol. 2015. [Google Scholar] [CrossRef]
- Zhang, P.; Yang, X.; Ma, X.; Ingram, D.R.; Lazar, A.J.; Torres, K.E.; Pollock, R.E. Antitumor effects of pharmacological EZH2 inhibition on malignant peripheral nerve sheath tumor through the miR-30a and KPNB1 pathway. Mol. Cancer 2015, 14, 55. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Garnett, J.; Creighton, C.J.; Al Sannaa, G.A.; Igram, D.R.; Lazar, A.; Liu, X.; Liu, C.; Pollock, R.E. EZH2-miR-30d-KPNB1 pathway regulates malignant peripheral nerve sheath tumour cell survival and tumourigenesis. J. Pathol. 2014, 232, 308–318. [Google Scholar] [CrossRef] [PubMed]
- Martens-de Kemp, S.R.; Nagel, R.; Stigter-van Walsum, M.; van der Meulen, I.H.; van Beusechem, V.W.; Braakhuis, B.J.; Brakenhoff, R.H. Functional genetic screens identify genes essential for tumor cell survival in head and neck and lung cancer. Clin. Cancer Res. 2013, 19, 1994–2003. [Google Scholar] [CrossRef] [PubMed]
- Kuusisto, H.V.; Jans, D.A. Hyper-dependence of breast cancer cell types on the nuclear transporter importin β1. Biochim. Biophys. Acta 2015, 1853, 1870–1878. [Google Scholar] [CrossRef] [PubMed]
- Christiansen, A.; Dyrskjot, L. The functional role of the novel biomarker karyopherin α2 (KPNA2) in cancer. Cancer Lett. 2013, 331, 18–23. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.; Lu, Y.; Qin, H.; Zhou, Y.; Gu, Y.; Zhou, J.; Wang, X.; Fan, D. High ran level is correlated with poor prognosis in patients with colorectal cancer. Int J. Clin. Oncol. 2013, 18, 856–863. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Zhao, X.; Li, K.; Luo, G.; Nie, Y.; Shi, Y.; Zhou, Y.; Ren, G.; Feng, B.; Liu, Z.; et al. Thioredoxin-like protein 2 is overexpressed in colon cancer and promotes cancer cell metastasis by interaction with ran. Antioxid. Redox Signal. 2013, 19, 899–911. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Lu, Y.; Zhao, X.; Sun, Y.; Shi, Y.; Fan, H.; Liu, C.; Zhou, J.; Nie, Y.; Wu, K.; et al. Ran-GTPase protein promotes human pancreatic cancer proliferation by deregulating the expression of survivin and cell cycle proteins. Biochem. Biophys. Res. Commun. 2013, 440, 322–329. [Google Scholar] [CrossRef] [PubMed]
- Chang, K.C.; Chang, W.C.; Chang, Y.; Hung, L.Y.; Lai, C.H.; Yeh, Y.M.; Chou, Y.W.; Chen, C.H. Ran-GTPase-activating protein 1 is a therapeutic target in diffuse large B-cell lymphoma. PLoS ONE 2013, 8, e79863. [Google Scholar] [CrossRef] [PubMed]
- Yuen, H.F.; Chan, K.K.; Grills, C.; Murray, J.T.; Platt-Higgins, A.; Eldin, O.S.; O’Byrne, K.; Janne, P.; Fennell, D.A.; Johnston, P.G.; et al. Ran is a potential therapeutic target for cancer cells with molecular changes associated with activation of the PI3K/Akt/MTORC1 and Ras/MEK/ERK pathways. Clin. Cancer Res. 2012, 18, 380–391. [Google Scholar] [CrossRef] [PubMed]
- Soderholm, J.F.; Bird, S.L.; Kalab, P.; Sampathkumar, Y.; Hasegawa, K.; Uehara-Bingen, M.; Weis, K.; Heald, R. Importazole, a small molecule inhibitor of the transport receptor importin-β. ACS Chem. Biol. 2011, 6, 700–708. [Google Scholar] [CrossRef] [PubMed]
- Kudo, N.; Khochbin, S.; Nishi, K.; Kitano, K.; Yanagida, M.; Yoshida, M.; Horinouchi, S. Molecular cloning and cell cycle-dependent expression of mammalian CRM1, a protein involved in nuclear export of proteins. J. Biol. Chem. 1997, 272, 29742–29751. [Google Scholar] [CrossRef] [PubMed]
- Murakami, N.; Ye, Y.; Kawanishi, M.; Aoki, S.; Kudo, N.; Yoshida, M.; Nakayama, E.E.; Shioda, T.; Kobayashi, M. New rev-transport inhibitor with anti-HIV activity from valerianae radix. Bioorg. Med. Chem. Lett. 2002, 12, 2807–2810. [Google Scholar] [CrossRef]
- Kudo, N.; Matsumori, N.; Taoka, H.; Fujiwara, D.; Schreiner, E.P.; Wolff, B.; Yoshida, M.; Horinouchi, S. Leptomycin b inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc. Natl. Acad. Sci. USA 1999, 96, 9112–9117. [Google Scholar] [CrossRef] [PubMed]
- Kudo, N.; Wolff, B.; Sekimoto, T.; Schreiner, E.P.; Yoneda, Y.; Yanagida, M.; Horinouchi, S.; Yoshida, M. Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp. Cell Res. 1998, 242, 540–547. [Google Scholar] [CrossRef] [PubMed]
- Hamamoto, T.; Gunji, S.; Tsuji, H.; Beppu, T. Leptomycins a and b, new antifungal antibiotics. I. Taxonomy of the producing strain and their fermentation, purification and characterization. J. Antibiot. (Tokyo) 1983, 36, 639–645. [Google Scholar] [CrossRef] [PubMed]
- Hamamoto, T.; Seto, H.; Beppu, T. Leptomycins A and B, new antifungal antibiotics. II. Structure elucidation. J. Antibiot. (Tokyo) 1983, 36, 646–650. [Google Scholar] [CrossRef] [PubMed]
- Hayakawa, Y.; Adachi, K.; Komeshima, N. New antitumor antibiotics. J. Antibiot. (Tokyo) 1987, 40, 1349–1352. [Google Scholar] [CrossRef] [PubMed]
- Hayakawa, Y.; Sohda, K.Y.; Shin-Ya, K.; Hidaka, T.; Seto, H. Anguinomycins C and D, new antitumor antibiotics with selective cytotoxicity against transformed cells. J. Antibiot. (Tokyo) 1995, 48, 954–961. [Google Scholar] [CrossRef] [PubMed]
- Bonazzi, S.; Eidam, O.; Guttinger, S.; Wach, J.Y.; Zemp, I.; Kutay, U.; Gademann, K. Anguinomycins and derivatives: Total syntheses, modeling, and biological evaluation of the inhibition of nucleocytoplasmic transport. J. Am. Chem. Soc. 2010, 132, 1432–1442. [Google Scholar] [CrossRef] [PubMed]
- Bonazzi, S.; Guttinger, S.; Zemp, I.; Kutay, U.; Gademann, K. Total synthesis, configuration, and biological evaluation of anguinomycin C. Angew. Chem. Int. Ed. 2007, 46, 8707–8710. [Google Scholar] [CrossRef] [PubMed]
- Schummer, D.; Gerth, K.; Reichenbach, H.; Hofle, G. Antibiotics from gliding bacteria 63. Ratjadone—A new antifungal metabolite from sorangium-cellulosum. Liebigs Ann. 1995, 685–688. [Google Scholar] [CrossRef]
- Gerth, K.; Schummer, D.; Hofle, G.; Irschik, H.; Reichenbach, H. Ratjadon: A new antifungal compound from sorangium cellulosum (myxobacteria) production, physio-chemical and biological properties. J. Antibiot. (Tokyo) 1995, 48, 973–976. [Google Scholar] [CrossRef] [PubMed]
- Kalesse, M.; Christmann, M.; Bhatt, U.; Quitschalle, M.; Claus, E.; Saeed, A.; Burzlaff, A.; Kasper, C.; Haustedt, L.O.; Hofer, E.; et al. The chemistry and biology of ratjadone. ChemBioChem 2001, 2, 709–714. [Google Scholar] [CrossRef]
- Williams, D.R.; Ihle, D.C.; Plummer, S.V. Total synthesis of (−)-ratjadone. Org. Lett. 2001, 3, 1383–1386. [Google Scholar] [CrossRef] [PubMed]
- Koster, M.; Lykke-Andersen, S.; Elnakady, Y.A.; Gerth, K.; Washausen, P.; Hofle, G.; Sasse, F.; Kjems, J.; Hauser, H. Ratjadones inhibit nuclear export by blocking CRM1/exportin 1. Exp. Cell Res. 2003, 286, 321–331. [Google Scholar] [CrossRef]
- Meissner, T.; Krause, E.; Vinkemeier, U. Ratjadone and leptomycin B block CRM1-dependent nuclear export by identical mechanisms. FEBS Lett. 2004, 576, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Fleta-Soriano, E.; Martinez, J.P.; Hinkelmann, B.; Gerth, K.; Washausen, P.; Diez, J.; Frank, R.; Sasse, F.; Meyerhans, A. The myxobacterial metabolite ratjadone a inhibits HIV infection by blocking the rev/CRM1-mediated nuclear export pathway. Microb. Cell Factories 2014, 13, 17. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Carrasco, Y.P.; Hu, Y.; Guo, X.; Mirzaei, H.; Macmillan, J.; Chook, Y.M. Nuclear export inhibition through covalent conjugation and hydrolysis of leptomycin B by CRM1. Proc. Natl. Acad. Sci. USA 2013, 110, 1303–1308. [Google Scholar] [CrossRef] [PubMed]
- Roberts, B.J.; Hamelehle, K.L.; Sebolt, J.S.; Leopold, W.R. In vivo and in vitro anticancer activity of the structurally novel and highly potent antibiotic Ci-940 and its hydroxy analog (pd 114,721). Cancer Chemother. Pharmacol. 1986, 16, 95–101. [Google Scholar] [CrossRef] [PubMed]
- Newlands, E.S.; Rustin, G.J.; Brampton, M.H. Phase i trial of elactocin. Br. J.Cancer 1996, 74, 648–649. [Google Scholar] [CrossRef] [PubMed]
- Mutka, S.C.; Yang, W.Q.; Dong, S.D.; Ward, S.L.; Craig, D.A.; Timmermans, P.B.; Murli, S. Identification of nuclear export inhibitors with potent anticancer activity in vivo. Cancer Res. 2009, 69, 510–517. [Google Scholar] [CrossRef] [PubMed]
- Daelemans, D.; Afonina, E.; Nilsson, J.; Werner, G.; Kjems, J.; De Clercq, E.; Pavlakis, G.N.; Vandamme, A.M. A synthetic HIV-1 Rev inhibitor interfering with the CRM1-mediated nuclear export. Proc. Natl. Acad. Sci. USA 2002, 99, 14440–14445. [Google Scholar] [CrossRef] [PubMed]
- Van Neck, T.; Pannecouque, C.; Vanstreels, E.; Stevens, M.; Dehaen, W.; Daelemans, D. Inhibition of the CRM1-mediated nucleocytoplasmic transport by N-azolylacrylates: Structure-activity relationship and mechanism of action. Bioorg. Med. Chem. 2008, 16, 9487–9497. [Google Scholar] [CrossRef] [PubMed]
- Haines, J.D.; Herbin, O.; de la Hera, B.; Vidaurre, O.G.; Moy, G.A.; Sun, Q.; Fung, H.Y.; Albrecht, S.; Alexandropoulos, K.; McCauley, D.; et al. Nuclear export inhibitors avert progression in preclinical models of inflammatory demyelination. Nat. Neurosci. 2015, 18, 511–520. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Han, X.; Wang, J.; Yao, J.; Shi, Y. Antitumor effects of a novel chromosome region maintenance 1 (CRM1) inhibitor on non-small cell lung cancer cells in vitro and in mouse tumor xenografts. PLoS ONE 2014, 9, e89848. [Google Scholar] [CrossRef] [PubMed]
- Walker, C.J.; Oaks, J.J.; Santhanam, R.; Neviani, P.; Harb, J.G.; Ferenchak, G.; Ellis, J.J.; Landesman, Y.; Eisfeld, A.K.; Gabrail, N.Y.; et al. Preclinical and clinical efficacy of XPO1/CRM1 inhibition by the karyopherin inhibitor KPT-330 in pH+ leukemias. Blood 2013, 122, 3034–3044. [Google Scholar] [CrossRef] [PubMed]
- London, C.A.; Bernabe, L.F.; Barnard, S.; Kisseberth, W.C.; Borgatti, A.; Henson, M.; Wilson, H.; Jensen, K.; Ito, D.; Modiano, J.F.; et al. Preclinical evaluation of the novel, orally bioavailable selective inhibitor of nuclear export (sine) KPT-335 in spontaneous canine cancer: Results of a phase I study. PLoS ONE 2014, 9, e87585. [Google Scholar] [CrossRef] [PubMed]
- Hilliard, M.; Frohnert, C.; Spillner, C.; Marcone, S.; Nath, A.; Lampe, T.; Fitzgerald, D.J.; Kehlenbach, R.H. The anti-inflammatory prostaglandin 15-deoxy-δ(12,14)-pgj2 inhibits CRM1-dependent nuclear protein export. J. Biol. Chem. 2010, 285, 22202–22210. [Google Scholar] [CrossRef] [PubMed]
- Niu, M.; Wu, S.; Mao, L.; Yang, Y. CRM1 is a cellular target of curcumin: New insights for the myriad of biological effects of an ancient spice. Traffic 2013, 14, 1042–1052. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, B.B.; Kumar, A.; Bharti, A.C. Anticancer potential of curcumin: Preclinical and clinical studies. Anticancer Res. 2003, 23, 363–398. [Google Scholar] [PubMed]
- Sharma, R.A.; Euden, S.A.; Platton, S.L.; Cooke, D.N.; Shafayat, A.; Hewitt, H.R.; Marczylo, T.H.; Morgan, B.; Hemingway, D.; Plummer, S.M.; et al. Phase I clinical trial of oral curcumin: Biomarkers of systemic activity and compliance. Clin. Cancer Res. 2004, 10, 6847–6854. [Google Scholar]
- Silva, A.L.; Rech, S.B.; von Poser, G.L. Quantitative determination of valepotriates from valeriana native to south brazil. Planta Medica 2002, 68, 570–572. [Google Scholar] [CrossRef] [PubMed]
- Tamura, S.; Shimizu, N.; Fujiwara, K.; Kaneko, M.; Kimura, T.; Murakami, N. Bioisostere of valtrate, anti-HIV principle by inhibition for nuclear export of rev. Bioorg. Med. Chem. Lett. 2010, 20, 2159–2162. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Niu, M.; Xu, X.; Cai, W.; Zeng, L.; Zhou, X.; Yu, R.; Xu, K. CRM1 is a direct cellular target of the natural anti-cancer agent plumbagin. J. Pharmacol. Sci. 2014, 124, 486–493. [Google Scholar] [CrossRef] [PubMed]
- Niu, M.; Xu, X.; Shen, Y.; Yao, Y.; Qiao, J.; Zhu, F.; Zeng, L.; Liu, X.; Xu, K. Piperlongumine is a novel nuclear export inhibitor with potent anticancer activity. Chem. Biol. Interact. 2015, 237, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.Y.; Wu, C.C.; Lan, Y.H.; Chang, F.R.; Teng, C.M.; Wu, Y.C. Goniothalamin induces cell cycle-specific apoptosis by modulating the redox status in MDA-MB-231 cells. Eur. J. Pharmacol. 2005, 522, 20–29. [Google Scholar] [CrossRef] [PubMed]
- Wach, J.Y.; Guttinger, S.; Kutay, U.; Gademann, K. The cytotoxic styryl lactone goniothalamin is an inhibitor of nucleocytoplasmic transport. Bioorg. Med. Chem. Lett. 2010, 20, 2843–2846. [Google Scholar] [CrossRef] [PubMed]
- De Fatima, A.; Kohn, L.K.; Antonio, M.A.; de Carvalho, J.E.; Pilli, R.A. (R)-goniothalamin: Total syntheses and cytotoxic activity against cancer cell lines. Bioorg. Med. Chem. 2005, 13, 2927–2933. [Google Scholar] [CrossRef] [PubMed]
- Al-Qubaisi, M.; Rozita, R.; Yeap, S.K.; Omar, A.R.; Ali, A.M.; Alitheen, N.B. Selective cytotoxicity of goniothalamin against hepatoblastoma HEPG2 cells. Molecules 2011, 16, 2944–2959. [Google Scholar] [CrossRef] [PubMed]
- Chiu, C.C.; Liu, P.L.; Huang, K.J.; Wang, H.M.; Chang, K.F.; Chou, C.K.; Chang, F.R.; Chong, I.W.; Fang, K.; Chen, J.S.; et al. Goniothalamin inhibits growth of human lung cancer cells through DNA damage, apoptosis, and reduced migration ability. J. Agric. Food Chem. 2011, 59, 4288–4293. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Li, B. 1ʹs-1ʹ-acetoxychavicol acetate isolated from alpinia galanga inhibits human immunodeficiency virus type 1 replication by blocking rev transport. J. Gen. Virol. 2006, 87, 2047–2053. [Google Scholar] [CrossRef] [PubMed]
- Tamura, S.; Shiomi, A.; Kaneko, M.; Ye, Y.; Yoshida, M.; Yoshikawa, M.; Kimura, T.; Kobayashi, M.; Murakami, N. New rev-export inhibitor from alpinia galanga and structure-activity relationship. Bioorg. Med. Chem. Lett. 2009, 19, 2555–2557. [Google Scholar] [CrossRef] [PubMed]
- Murakami, N.; Sugimoto, M.; Kobayashi, M. Participation of the β-hydroxyketone part for potent cytotoxicity of callystatin a, a spongean polyketide. Bioorg. Med. Chem. 2001, 9, 57–67. [Google Scholar] [CrossRef]
- Kalesse, M.; Chary, K.P.; Quitschalle, M.; Burzlaff, A.; Kasper, C.; Scheper, T. The total synthesis of (−)-callystatin a. Chemistry 2003, 9, 1129–1136. [Google Scholar] [CrossRef] [PubMed]
- Langille, N.F.; Panek, J.S. Total synthesis of (−)-callystatin a. Org. Lett. 2004, 6, 3203–3206. [Google Scholar] [CrossRef] [PubMed]
- Hayakawa, Y.; Sohda, K.; Furihata, K.; Kuzuyama, T.; Shin-ya, K.; Seto, H. Studies on new antitumor antibiotics, leptofuranins A, B, C and D.I. Taxonomy, fermentation, isolation and biological activities. J. Antibiot. (Tokyo) 1996, 49, 974–979. [Google Scholar] [CrossRef] [PubMed]
- Hayakawa, Y.; Sohda, K.; Seto, H. Studies on new antitumor antibiotics, leptofuranins A, B, C and D II. Physiocochemical properties and structure elucidation. J. Antibiot. (Tokyo) 1996, 49, 980–984. [Google Scholar] [CrossRef] [PubMed]
- Sakakibara, K.; Saito, N.; Sato, T.; Suzuki, A.; Hasegawa, Y.; Friedman, J.M.; Kufe, D.W.; Vonhoff, D.D.; Iwami, T.; Kawabe, T. Cbs9106 is a novel reversible oral CRM1 inhibitor with CRM1 degrading activity. Blood 2011, 118, 3922–3931. [Google Scholar] [CrossRef] [PubMed]
- Almholt, D.L.; Loechel, F.; Nielsen, S.J.; Krog-Jensen, C.; Terry, R.; Bjorn, S.P.; Pedersen, H.C.; Praestegaard, M.; Moller, S.; Heide, M.; et al. Nuclear export inhibitors and kinase inhibitors identified using a mapk-activated protein kinase 2 redistribution screen. Assay Drug Dev. Technol. 2004, 2, 7–20. [Google Scholar] [CrossRef] [PubMed]
- Kau, T.R.; Schroeder, F.; Ramaswamy, S.; Wojciechowski, C.L.; Zhao, J.J.; Roberts, T.M.; Clardy, J.; Sellers, W.R.; Silver, P.A. A chemical genetic screen identifies inhibitors of regulated nuclear export of a forkhead transcription factor in pten-deficient tumor cells. Cancer Cell 2003, 4, 463–476. [Google Scholar] [CrossRef]
- Salas Fragomeni, R.A.; Chung, H.W.; Landesman, Y.; Senapedis, W.; Saint-Martin, J.R.; Tsao, H.; Flaherty, K.T.; Shacham, S.; Kauffman, M.; Cusack, J.C. CRM1 and braf inhibition synergize and induce tumor regression in braf-mutant melanoma. Mol. Cancer Ther. 2013, 12, 1171–1179. [Google Scholar] [CrossRef] [PubMed]
- Abraham, S.A.; Holyoake, T.L. Redirecting traffic using the XPO1 police. Blood 2013, 122, 2926–2928. [Google Scholar] [CrossRef] [PubMed]
- Wettersten, H.I.; Landesman, Y.; Friedlander, S.; Shacham, S.; Kauffman, M.; Weiss, R.H. Specific inhibition of the nuclear exporter exportin-1 attenuates kidney cancer growth. PLoS ONE 2014, 9, e113867. [Google Scholar] [CrossRef] [PubMed]
- De Cesare, M.; Cominetti, D.; Doldi, V.; Lopergolo, A.; Deraco, M.; Gandellini, P.; Friedlander, S.; Landesman, Y.; Kauffman, M.G.; Shacham, S.; et al. Anti-tumor activity of selective inhibitors of XPO1/CRM1-mediated nuclear export in diffuse malignant peritoneal mesothelioma: The role of survivin. Oncotarget 2015, 6, 13119–13132. [Google Scholar] [CrossRef] [PubMed]
- Niu, M.; Chong, Y.; Han, Y.; Liu, X. Novel reversible selective inhibitor of nuclear export shows that CRM1 is a target in colorectal cancer cells. Cancer Biol. Ther. 2015, 16, 1110–1118. [Google Scholar] [CrossRef] [PubMed]
- Senapedis, W.T.; Baloglu, E.; Landesman, Y. Clinical translation of nuclear export inhibitors in cancer. Semin. Cancer Biol. 2014, 27, 74–86. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Wei, G.; Parikh, K.; Liu, D. Selective inhibitors of nuclear export (sine) in hematological malignancies. Exp. Hematol. Oncol. 2015, 4, 7. [Google Scholar] [CrossRef] [PubMed]
- Ishizawa, J.; Kojima, K.; Hail, N., Jr.; Tabe, Y.; Andreeff, M. Expression, function, and targeting of the nuclear exporter chromosome region maintenance 1 (CRM1) protein. Pharmacol. Ther. 2015, 153, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Parikh, K.; Cang, S.; Sekhri, A.; Liu, D. Selective inhibitors of nuclear export (sine)—A novel class of anti-cancer agents. J. Hematol. Oncol. 2014, 7, 78. [Google Scholar] [CrossRef] [PubMed]
- Kazim, S.; Malafa, M.P.; Coppola, D.; Husain, K.; Zibadi, S.; Kashyap, T.; Crochiere, M.; Landesman, Y.; Rashal, T.; Sullivan, D.M.; et al. Selective nuclear export inhibitor KPT-330 enhances the antitumor activity of gemcitabine in human pancreatic cancer. Mol. Cancer Ther. 2015, 14, 1570–1581. [Google Scholar] [CrossRef] [PubMed]
- Hing, Z.A.; Mantel, R.; Beckwith, K.A.; Guinn, D.; Williams, E.; Smith, L.L.; Williams, K.; Johnson, A.J.; Lehman, A.M.; Byrd, J.C.; et al. Selinexor is effective in acquired resistance to ibrutinib and synergizes with ibrutinib in chronic lymphocytic leukemia. Blood 2015, 125, 3128–3132. [Google Scholar] [CrossRef] [PubMed]
- Bernad, R.; van der Velde, H.; Fornerod, M.; Pickersgill, H. Nup358/ RanBP2 attaches to the nuclear pore complex via association with Nup88 and Nup214/can and plays a supporting role in CRM1-mediated nuclear protein export. Mol. Cell Biol. 2004, 24, 2373–2384. [Google Scholar] [CrossRef] [PubMed]
- Hutten, S.; Kehlenbach, R.H. Nup214 is required for CRM1-dependent nuclear protein export in vivo. Mol. Cell Biol. 2006, 26, 6772–6785. [Google Scholar] [CrossRef] [PubMed]
- Roth, P.; Xylourgidis, N.; Sabri, N.; Uv, A.; Fornerod, M.; Samakovlis, C. The drosophila nucleoporin DNUP88 localizes DNUP214 and CRM1 on the nuclear envelope and attenuates nes-mediated nuclear export. J. Cell Biol. 2003, 163, 701–706. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.L.; Mahboobi, S.H.; Moussavi-Baygi, R.; Mofrad, M.R. The interaction of CRM1 and the nuclear pore protein tpr. PLoS ONE 2014, 9, e93709. [Google Scholar] [CrossRef] [PubMed]
- Waldmann, I.; Spillner, C.; Kehlenbach, R.H. The nucleoporin-like protein NLP1 (HCG1) promotes CRM1-dependent nuclear protein export. J. Cell Sci. 2012, 125, 144–154. [Google Scholar] [CrossRef] [PubMed]
- Takeda, A.; Sarma, N.J.; Abdul-Nabi, A.M.; Yaseen, N.R. Inhibition of CRM1-mediated nuclear export of transcription factors by leukemogenic Nup98 fusion proteins. J. Biol. Chem. 2010, 285, 16248–16257. [Google Scholar] [CrossRef] [PubMed]
- Roloff, S.; Spillner, C.; Kehlenbach, R.H. Several phenylalanine-glycine motives in the nucleoporin Nup214 are essential for binding of the nuclear export receptor CRM1. J. Biol. Chem. 2013, 288, 3952–3963. [Google Scholar] [PubMed]
- Oka, M.; Asally, M.; Yasuda, Y.; Ogawa, Y.; Tachibana, T.; Yoneda, Y. The mobile fg nucleoporin NUP98 is a cofactor for CRM1-dependent protein export. Mol. Biol. Cell 2010, 21, 1885–1896. [Google Scholar] [CrossRef] [PubMed]
- Bernad, R.; Engelsma, D.; Sanderson, H.; Pickersgill, H.; Fornerod, M. Nup214- Nup88 nucleoporin subcomplex is required for CRM1-mediated 60 s preribosomal nuclear export. J. Biol. Chem. 2006, 281, 19378–19386. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dickmanns, A.; Monecke, T.; Ficner, R. Structural Basis of Targeting the Exportin CRM1 in Cancer. Cells 2015, 4, 538-568. https://doi.org/10.3390/cells4030538
Dickmanns A, Monecke T, Ficner R. Structural Basis of Targeting the Exportin CRM1 in Cancer. Cells. 2015; 4(3):538-568. https://doi.org/10.3390/cells4030538
Chicago/Turabian StyleDickmanns, Achim, Thomas Monecke, and Ralf Ficner. 2015. "Structural Basis of Targeting the Exportin CRM1 in Cancer" Cells 4, no. 3: 538-568. https://doi.org/10.3390/cells4030538




