Complex Commingling: Nucleoporins and the Spindle Assembly Checkpoint
Abstract
:1. Introduction

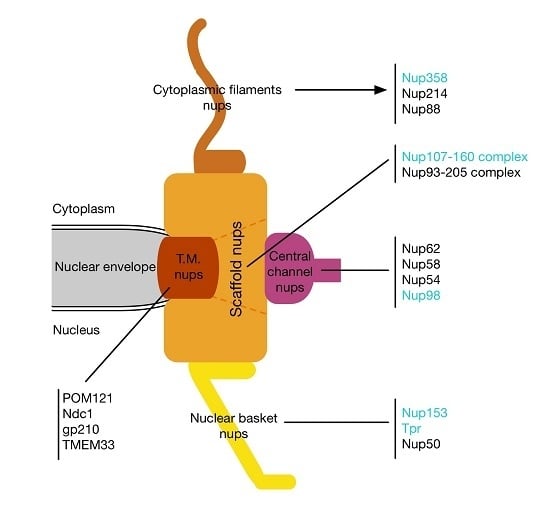
2. Mitotic Functions of Nucleoporins and the Spindle Assembly Checkpoint
2.1. Nucleoporins at Kinetochores
2.2. Nucleoporins, the SAC, and the APC/C
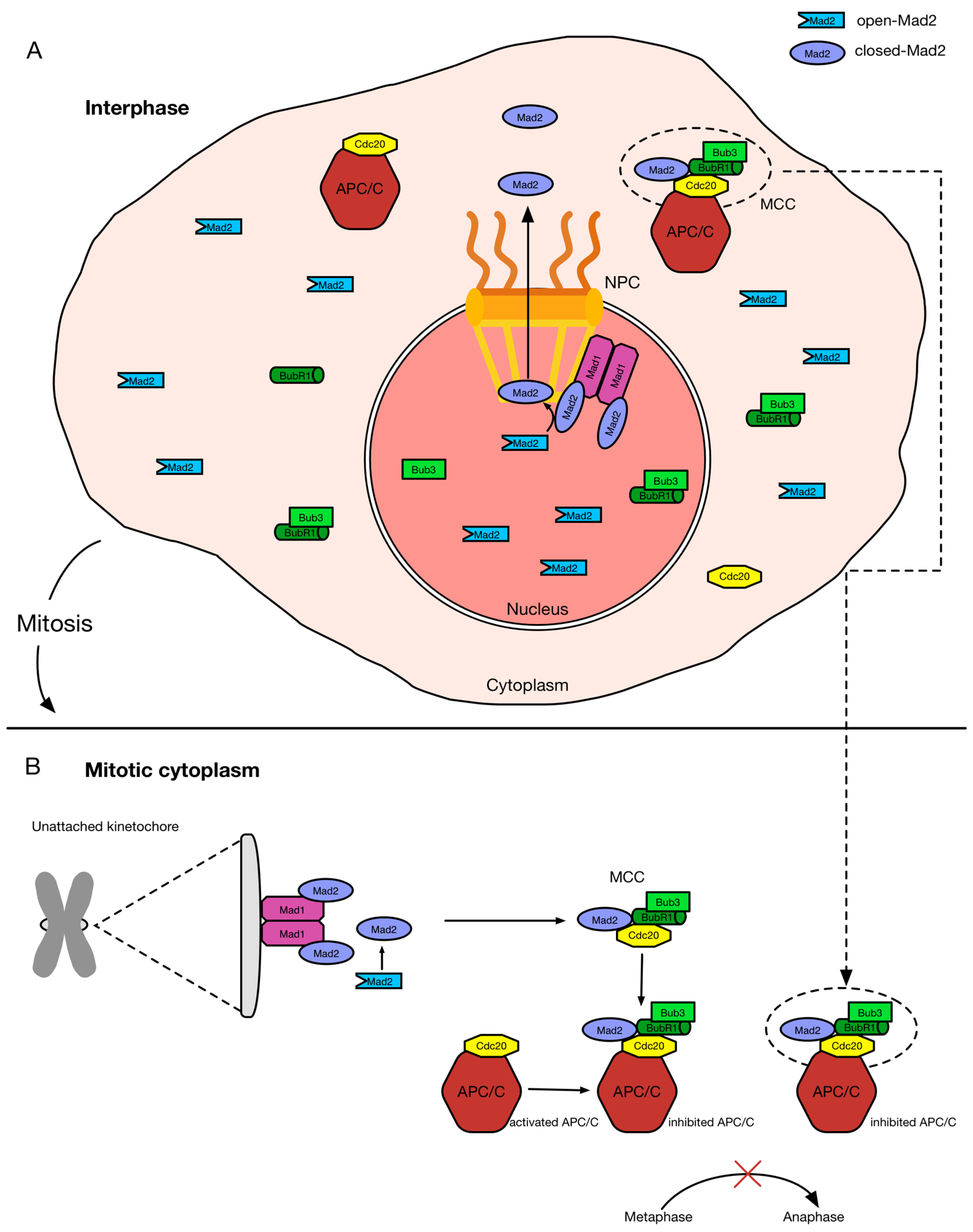
3. The SAC Regulators Mad1 and Mad2 during Interphase
3.1. Mad1 and Mad2 Localization at NPCs
| Species | Localization of Mad1 and Mad2 at NPCs | Nucleoporins Partners |
|---|---|---|
| Arabidopsis | √ | NUA (Tpr) |
| Saccharomyces cerevisiae | √ | Mlp1p/Mlp2p (Tpr) |
| Nup53p | ||
| Schizosaccharomyces pombe | √ | No partners identified yet |
| Aspergillus nidulans | √ | Mlp1 (Tpr) |
| Drosophila melanogaster | √ | Mtor (Tpr) |
| Caenorhabditis elegans | √ | Npp-5 |
| Human | √ | Tpr |
| Nup153 |
3.2. Function of SAC Proteins at NPCs
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Fahrenkrog, B.; Aebi, U. The nuclear pore complex: Nucleocytoplasmic transport and beyond. Nat. Rev. Mol. Cell Biol. 2003, 4, 757–766. [Google Scholar] [CrossRef] [PubMed]
- Ori, A.; Banterle, N.; Iskar, M.; Andres-Pons, A.; Escher, C.; Khanh Bui, H.; Sparks, L.; Solis-Mezarino, V.; Rinner, O.; Bork, P.; et al. Cell type-specific nuclear pores: A case in point for context-dependent stoichiometry of molecular machines. Mol. Syst. Biol. 2013, 9, 648. [Google Scholar] [CrossRef] [PubMed]
- Reichelt, R.; Holzenburg, A.; Buhle, E.L., Jr.; Jarnik, M.; Engel, A.; Aebi, U. Correlation between structure and mass distribution of the nuclear pore complex and of distinct pore complex components. J. Cell Biol. 1990, 110, 883–894. [Google Scholar] [CrossRef] [PubMed]
- Rout, M.P.; Blobel, G. Isolation of the yeast nuclear pore complex. J. Cell Biol. 1993, 123, 771–783. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Rout, M.P.; Akey, C.W. Three-dimensional architecture of the isolated yeast nuclear pore complex: Functional and evolutionary implications. Mol. Cell 1998, 1, 223–234. [Google Scholar] [CrossRef]
- Gerace, L.; Burke, B. Functional organization of the nuclear envelope. Annu. Rev. Cell Biol. 1988, 4, 335–374. [Google Scholar] [CrossRef] [PubMed]
- Lim, R.Y.; Aebi, U.; Fahrenkrog, B. Towards reconciling structure and function in the nuclear pore complex. Histochem. Cell Biol. 2008, 129, 105–116. [Google Scholar] [CrossRef] [PubMed]
- Fahrenkrog, B.; Hurt, E.C.; Aebi, U.; Pante, N. Molecular architecture of the yeast nuclear pore complex: Localization of nsp1p subcomplexes. J. Cell Biol. 1998, 143, 577–588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elad, N.; Maimon, T.; Frenkiel-Krispin, D.; Lim, R.Y.; Medalia, O. Structural analysis of the nuclear pore complex by integrated approaches. Curr. Opin. Struct. Biol. 2009, 19, 226–232. [Google Scholar] [CrossRef] [PubMed]
- Beck, M.; Forster, F.; Ecke, M.; Plitzko, J.M.; Melchior, F.; Gerisch, G.; Baumeister, W.; Medalia, O. Nuclear pore complex structure and dynamics revealed by cryoelectron tomography. Science 2004, 306, 1387–1390. [Google Scholar] [CrossRef] [PubMed]
- Fiserova, J.; Kiseleva, E.; Goldberg, M.W. Nuclear envelope and nuclear pore complex structure and organization in tobacco by-2 cells. Plant J. 2009, 59, 243–255. [Google Scholar] [CrossRef] [PubMed]
- Beck, M.; Lucic, V.; Forster, F.; Baumeister, W.; Medalia, O. Snapshots of nuclear pore complexes in action captured by cryo-electron tomography. Nature 2007, 449, 611–615. [Google Scholar] [CrossRef] [PubMed]
- Stoffler, D.; Feja, B.; Fahrenkrog, B.; Walz, J.; Typke, D.; Aebi, U. Cryo-electron tomography provides novel insights into nuclear pore architecture: Implications for nucleocytoplasmic transport. J. Mol. Biol. 2003, 328, 119–130. [Google Scholar] [CrossRef]
- Cronshaw, J.M.; Krutchinsky, A.N.; Zhang, W.; Chait, B.T.; Matunis, M.J. Proteomic analysis of the mammalian nuclear pore complex. J. Cell Biol. 2002, 158, 915–927. [Google Scholar] [CrossRef] [PubMed]
- Rout, M.P.; Aitchison, J.D.; Suprapto, A.; Hjertaas, K.; Zhao, Y.; Chait, B.T. The yeast nuclear pore complex: Composition, architecture, and transport mechanism. J. Cell Biol. 2000, 148, 635–651. [Google Scholar] [CrossRef] [PubMed]
- Asakawa, H.; Yang, H.J.; Yamamoto, T.G.; Ohtsuki, C.; Chikashige, Y.; Sakata-Sogawa, K.; Tokunaga, M.; Iwamoto, M.; Hiraoka, Y.; Haraguchi, T. Characterization of nuclear pore complex components in fission yeast schizosaccharomyces pombe. Nucleus 2014, 5, 149–162. [Google Scholar] [CrossRef] [PubMed]
- Neumann, N.; Jeffares, D.C.; Poole, A.M. Outsourcing the nucleus: Nuclear pore complex genes are no longer encoded in nucleomorph genomes. Evol. Bioinform. Online 2006, 2, 23–34. [Google Scholar]
- Tamura, K.; Fukao, Y.; Iwamoto, M.; Haraguchi, T.; Hara-Nishimura, I. Identification and characterization of nuclear pore complex components in arabidopsis thaliana. Plant Cell 2010, 22, 4084–4097. [Google Scholar] [CrossRef] [PubMed]
- Hallberg, E.; Wozniak, R.W.; Blobel, G. An integral membrane protein of the pore membrane domain of the nuclear envelope contains a nucleoporin-like region. Plant Cell 1993, 122, 513–521. [Google Scholar] [CrossRef]
- Mansfeld, J.; Guttinger, S.; Hawryluk-Gara, L.A.; Pante, N.; Mall, M.; Galy, V.; Haselmann, U.; Muhlhausser, P.; Wozniak, R.W.; Mattaj, I.W.; et al. The conserved transmembrane nucleoporin Ndc1 is required for nuclear pore complex assembly in vertebrate cells. Mol. Cell 2006, 22, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Stavru, F.; Hulsmann, B.B.; Spang, A.; Hartmann, E.; Cordes, V.C.; Gorlich, D. Ndc1: A crucial membrane-integral nucleoporin of metazoan nuclear pore complexes. J. Cell Biol. 2006, 173, 509–519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chadrin, A.; Hess, B.; San Roman, M.; Gatti, X.; Lombard, B.; Loew, D.; Barral, Y.; Palancade, B.; Doye, V. Pom33, a novel transmembrane nucleoporin required for proper nuclear pore complex distribution. J. Cell Biol. 2010, 189, 795–811. [Google Scholar] [CrossRef] [PubMed]
- Vasu, S.; Shah, S.; Orjalo, A.; Park, M.; Fischer, W.H.; Forbes, D.J. Novel vertebrate nucleoporins Nup133 and Nup160 play a role in mRNA export. J. Cell Biol. 2001, 155, 339–354. [Google Scholar] [CrossRef] [PubMed]
- Harel, A.; Orjalo, A.V.; Vincent, T.; Lachish-Zalait, A.; Vasu, S.; Shah, S.; Zimmerman, E.; Elbaum, M.; Forbes, D.J. Removal of a single pore subcomplex results in vertebrate nuclei devoid of nuclear pores. Mol. Cell 2003, 11, 853–864. [Google Scholar] [CrossRef]
- Loiodice, I.; Alves, A.; Rabut, G.; van Overbeek, M.; Ellenberg, J.; Sibarita, J.B.; Doye, V. The entire Nup107-160 complex, including three new members, is targeted as one entity to kinetochores in mitosis. Mol. Biol. Cell 2004, 15, 3333–3344. [Google Scholar] [CrossRef] [PubMed]
- Boehmer, T.; Enninga, J.; Dales, S.; Blobel, G.; Zhong, H. Depletion of a single nucleoporin, Nup107, prevents the assembly of a subset of nucleoporins into the nuclear pore complex. Proc. Natl. Acad. Sci. USA 2003, 100, 981–985. [Google Scholar] [CrossRef] [PubMed]
- Belgareh, N.; Rabut, G.; Bai, S.W.; van Overbeek, M.; Beaudouin, J.; Daigle, N.; Zatsepina, O.V.; Pasteau, F.; Labas, V.; Fromont-Racine, M.; et al. An evolutionarily conserved NPC subcomplex, which redistributes in part to kinetochores in mammalian cells. J. Cell Biol. 2001, 154, 1147–1160. [Google Scholar] [CrossRef] [PubMed]
- Grandi, P.; Dang, T.; Pane, N.; Shevchenko, A.; Mann, M.; Forbes, D.; Hurt, E. Nup93, a vertebrate homologue of yeast Nic96p, forms a complex with a novel 205-kDa protein and is required for correct nuclear pore assembly. Mol. Biol. Cell 1997, 8, 2017–2038. [Google Scholar] [CrossRef] [PubMed]
- Miller, B.R.; Powers, M.; Park, M.; Fischer, W.; Forbes, D.J. Identification of a new vertebrate nucleoporin, Nup188, with the use of a novel organelle trap assay. Mol. Biol. Cell 2000, 11, 3381–3396. [Google Scholar] [CrossRef] [PubMed]
- Hawryluk-Gara, L.A.; Shibuya, E.K.; Wozniak, R.W. Vertebrate Nup53 interacts with the nuclear lamina and is required for the assembly of a Nup93-containing complex. Mol. Biol. Cell 2005, 16, 2382–2394. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Matunis, M.J.; Kraemer, D.; Blobel, G.; Coutavas, E. Nup358, a cytoplasmically exposed nucleoporin with peptide repeats, Ran-GTP binding sites, zinc fingers, a cyclophilin a homologous domain, and a leucine-rich region. J. Biol. Chem. 1995, 270, 14209–14213. [Google Scholar] [CrossRef] [PubMed]
- Kraemer, D.; Wozniak, R.W.; Blobel, G.; Radu, A. The human can protein, a putative oncogene product associated with myeloid leukemogenesis, is a nuclear pore complex protein that faces the cytoplasm. Proc. Natl. Acad. Sci. USA 1994, 91, 1519–1523. [Google Scholar] [CrossRef] [PubMed]
- Bastos, R.; de Pouplana, L.R.; Enarson, M.; Bodoor, K.; Burke, B. Nup84, a novel nucleoporin that is associated with CAN/Nup214 on the cytoplasmic face of the nuclear pore complex. J. Cell Biol. 1997, 137, 989–1000. [Google Scholar] [CrossRef] [PubMed]
- Fornerod, M.; van Deursen, J.; van Baal, S.; Reynolds, A.; Davis, D.; Murti, K.G.; Fransen, J.; Grosveld, G. The human homologue of yeast CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component Nup88. EMBO J. 1997, 16, 807–816. [Google Scholar] [CrossRef] [PubMed]
- Cordes, V.C.; Reidenbach, S.; Rackwitz, H.R.; Franke, W.W. Identification of protein p270/Tpr as a constitutive component of the nuclear pore complex-attached intranuclear filaments. J. Cell Biol. 1997, 136, 515–529. [Google Scholar] [CrossRef] [PubMed]
- Frosst, P.; Guan, T.; Subauste, C.; Hahn, K.; Gerace, L. Tpr is localized within the nuclear basket of the pore complex and has a role in nuclear protein export. J. Cell Biol. 2002, 156, 617–630. [Google Scholar] [CrossRef] [PubMed]
- Hase, M.E.; Cordes, V.C. Direct interaction with Nup153 mediates binding of Tpr to the periphery of the nuclear pore complex. Mol. Biol. Cell 2003, 14, 1923–1940. [Google Scholar] [CrossRef] [PubMed]
- Guan, T.; Kehlenbach, R.H.; Schirmer, E.C.; Kehlenbach, A.; Fan, F.; Clurman, B.E.; Arnheim, N.; Gerace, L. Nup50, a nucleoplasmically oriented nucleoporin with a role in nuclear protein export. Mol. Cell Biol. 2000, 20, 5619–5630. [Google Scholar] [CrossRef] [PubMed]
- Sukegawa, J.; Blobel, G. A nuclear pore complex protein that contains zinc finger motifs, binds DNA, and faces the nucleoplasm. Cell 1993, 72, 29–38. [Google Scholar] [CrossRef]
- Pante, N.; Thomas, F.; Aebi, U.; Burke, B.; Bastos, R. Recombinant Nup153 incorporates in vivo into xenopus oocyte nuclear pore complexes. J. Struct. Biol. 2000, 129, 306–312. [Google Scholar] [CrossRef] [PubMed]
- Pante, N.; Bastos, R.; McMorrow, I.; Burke, B.; Aebi, U. Interactions and three-dimensional localization of a group of nuclear pore complex proteins. J. Cell Biol. 1994, 126, 603–617. [Google Scholar] [CrossRef] [PubMed]
- Fahrenkrog, B.; Maco, B.; Fager, A.M.; Koser, J.; Sauder, U.; Ullman, K.S.; Aebi, U. Domain-specific antibodies reveal multiple-site topology of Nup153 within the nuclear pore complex. J. Struct. Biol. 2002, 140, 254–267. [Google Scholar] [CrossRef]
- Walther, T.C.; Fornerod, M.; Pickersgill, H.; Goldberg, M.; Allen, T.D.; Mattaj, I.W. The nucleoporin Nup153 is required for nuclear pore basket formation, nuclear pore complex anchoring and import of a subset of nuclear proteins. EMBO J. 2001, 20, 5703–5714. [Google Scholar] [CrossRef] [PubMed]
- Guan, T.; Muller, S.; Klier, G.; Pante, N.; Blevitt, J.M.; Haner, M.; Paschal, B.; Aebi, U.; Gerace, L. Structural analysis of the p62 complex, an assembly of O-linked glycoproteins that localizes near the central gated channel of the nuclear pore complex. Mol. Biol. Cell 1995, 6, 1591–1603. [Google Scholar] [CrossRef] [PubMed]
- Schwarz-Herion, K.; Maco, B.; Sauder, U.; Fahrenkrog, B. Domain topology of the p62 complex within the 3-D architecture of the nuclear pore complex. J. Mol. Biol. 2007, 370, 796–806. [Google Scholar] [CrossRef] [PubMed]
- Powers, M.A.; Forbes, D.J.; Dahlberg, J.E.; Lund, E. The vertebrate glfg nucleoporin, Nup98, is an essential component of multiple RNA export pathways. J. Cell Biol. 1997, 136, 241–250. [Google Scholar] [CrossRef] [PubMed]
- Krull, S.; Thyberg, J.; Bjorkroth, B.; Rackwitz, H.R.; Cordes, V.C. Nucleoporins as components of the nuclear pore complex core structure and Tpr as the architectural element of the nuclear basket. Mol. Biol. Cell 2004, 15, 4261–4277. [Google Scholar] [CrossRef] [PubMed]
- Chatel, G.; Desai, S.H.; Mattheyses, A.L.; Powers, M.A.; Fahrenkrog, B. Domain topology of nucleoporin Nup98 within the nuclear pore complex. J. Struct. Biol. 2012, 177, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Mor, A.; White, M.A.; Fontoura, B.M. Nuclear trafficking in health and disease. Curr. Opin. Cell Biol. 2014, 28, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Chook, Y.M.; Suel, K.E. Nuclear import by karyopherin-betas: Recognition and inhibition. Biochim. Biophys. Acta 2011, 1813, 1593–1606. [Google Scholar] [CrossRef] [PubMed]
- Tran, E.J.; King, M.C.; Corbett, A.H. Macromolecular transport between the nucleus and the cytoplasm: Advances in mechanism and emerging links to disease. Biochim. Biophys. Acta 2014, 1843, 2784–2795. [Google Scholar] [CrossRef] [PubMed]
- Gorlich, D.; Pante, N.; Kutay, U.; Aebi, U.; Bischoff, F.R. Identification of different roles for rangdp and rangtp in nuclear protein import. EMBO J. 1996, 15, 5584–5594. [Google Scholar] [PubMed]
- Hoelz, A.; Blobel, G. Cell biology: Popping out of the nucleus. Nature 2004, 432, 815–816. [Google Scholar] [CrossRef] [PubMed]
- Melchior, F.; Paschal, B.; Evans, J.; Gerace, L. Inhibition of nuclear protein import by nonhydrolyzable analogues of gtp and identification of the small gtpase Ran/TC4 as an essential transport factor. J. Cell Biol. 1993, 123, 1649–1659. [Google Scholar] [CrossRef] [PubMed]
- Moore, M.S.; Blobel, G. The GTP-binding protein Ran/TC4 is required for protein import into the nucleus. Nature 1993, 365, 661–663. [Google Scholar] [CrossRef] [PubMed]
- Fried, H.; Kutay, U. Nucleocytoplasmic transport: Taking an inventory. Cell. Mol. Life Sci. 2003, 60, 1659–1688. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Hetzer, M.W. Functional interactions between nucleoporins and chromatin. Curr. Opin. Cell Biol. 2011, 23, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Capelson, M.; Liang, Y.; Schulte, R.; Mair, W.; Wagner, U.; Hetzer, M.W. Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell 2010, 140, 372–383. [Google Scholar] [CrossRef] [PubMed]
- De Strambio, C.C.; Niepel, M.; Rout, M.P. The nuclear pore complex: Bridging nuclear transport and gene regulation. Nat. Rev. Mol. Cell Biol. 2010, 11, 490–501. [Google Scholar] [CrossRef] [PubMed]
- Kalverda, B.; Pickersgill, H.; Shloma, V.V.; Fornerod, M. Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell 2010, 140, 360–371. [Google Scholar] [CrossRef] [PubMed]
- Vaquerizas, J.M.; Suyama, R.; Kind, J.; Miura, K.; Luscombe, N.M.; Akhtar, A. Nuclear pore proteins Nup153 and megator define transcriptionally active regions in the drosophila genome. PLoS Genet. 2010, 6, e1000846. [Google Scholar] [CrossRef] [PubMed]
- Galy, V.; Olivo-Marin, J.C.; Scherthan, H.; Doye, V.; Rascalou, N.; Nehrbass, U. Nuclear pore complexes in the organization of silent telomeric chromatin. Nature 2000, 403, 108–112. [Google Scholar] [CrossRef] [PubMed]
- Lemaitre, C.; Fischer, B.; Kalousi, A.; Hoffbeck, A.S.; Guirouilh-Barbat, J.; Shahar, O.D.; Genet, D.; Goldberg, M.; Betrand, P.; Lopez, B.; et al. The nucleoporin 153, a novel factor in double-strand break repair and DNA damage response. Oncogene 2012, 31, 4803–4809. [Google Scholar] [CrossRef] [PubMed]
- Nakano, H.; Wang, W.; Hashizume, C.; Funasaka, T.; Sato, H.; Wong, R.W. Unexpected role of nucleoporins in coordination of cell cycle progression. Cell Cycle 2011, 10, 425–433. [Google Scholar] [CrossRef] [PubMed]
- Chatel, G.; Fahrenkrog, B. Dynamics and diverse functions of nuclear pore complex proteins. Nucleus 2012, 3, 162–171. [Google Scholar] [CrossRef] [PubMed]
- Wozniak, R.; Burke, B.; Doye, V. Nuclear transport and the mitotic apparatus: An evolving relationship. Cell. Mol. Life Sci. 2010, 67, 2215–2230. [Google Scholar] [CrossRef] [PubMed]
- Gascoigne, K.E.; Cheeseman, I.M. Kinetochore assembly: If you build it, they will come. Curr. Opin. Cell Biol. 2011, 23, 102–108. [Google Scholar] [CrossRef] [PubMed]
- Foley, E.A.; Kapoor, T.M. Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat. Rev. Mol. Cell Biol. 2013, 14, 25–37. [Google Scholar] [CrossRef] [PubMed]
- Walczak, C.E.; Heald, R. Mechanisms of mitotic spindle assembly and function. Int. Rev. Cytol. 2008, 265, 111–158. [Google Scholar] [PubMed]
- Antonin, W.; Ellenberg, J.; Dultz, E. Nuclear pore complex assembly through the cell cycle: Regulation and membrane organization. FEBS Lett. 2008, 582, 2004–2016. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.U. Kinetochore-microtubule interactions: Steps towards bi-orientation. EMBO J. 2010, 29, 4070–4082. [Google Scholar] [CrossRef] [PubMed]
- Rasala, B.A.; Orjalo, A.V.; Shen, Z.; Briggs, S.; Forbes, D.J. Elys is a dual nucleoporin/kinetochore protein required for nuclear pore assembly and proper cell division. Proc. Natl. Acad. Sci. USA 2006, 103, 17801–17806. [Google Scholar] [CrossRef] [PubMed]
- Franz, C.; Walczak, R.; Yavuz, S.; Santarella, R.; Gentzel, M.; Askjaer, P.; Galy, V.; Hetzer, M.; Mattaj, I.W.; Antonin, W. Mel-28/elys is required for the recruitment of nucleoporins to chromatin and postmitotic nuclear pore complex assembly. EMBO Rep. 2007, 8, 165–172. [Google Scholar] [CrossRef] [PubMed]
- Walther, T.C.; Alves, A.; Pickersgill, H.; Loiodice, I.; Hetzer, M.; Galy, V.; Hulsmann, B.B.; Kocher, T.; Wilm, M.; Allen, T.; et al. The conserved Nup107-160 complex is critical for nuclear pore complex assembly. Cell 2003, 113, 195–206. [Google Scholar] [CrossRef]
- Zuccolo, M.; Alves, A.; Galy, V.; Bolhy, S.; Formstecher, E.; Racine, V.; Sibarita, J.B.; Fukagawa, T.; Shiekhattar, R.; Yen, T.; et al. The human Nup107-160 nuclear pore subcomplex contributes to proper kinetochore functions. EMBO J. 2007, 26, 1853–1864. [Google Scholar] [CrossRef] [PubMed]
- Platani, M.; Santarella-Mellwig, R.; Posch, M.; Walczak, R.; Swedlow, J.R.; Mattaj, I.W. The Nup107-160 nucleoporin complex promotes mitotic events via control of the localization state of the chromosome passenger complex. Mol. Biol. Cell 2009, 20, 5260–5275. [Google Scholar] [CrossRef] [PubMed]
- Mishra, R.K.; Chakraborty, P.; Arnaoutov, A.; Fontoura, B.M.; Dasso, M. The Nup107-160 complex and gamma-turc regulate microtubule polymerization at kinetochores. Nat. Cell Biol. 2010, 12, 164–169. [Google Scholar] [CrossRef] [PubMed]
- Pichler, A.; Gast, A.; Seeler, J.S.; Dejean, A.; Melchior, F. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell 2002, 108, 109–120. [Google Scholar] [CrossRef]
- Matunis, M.J.; Coutavas, E.; Blobel, G. A novel ubiquitin-like modification modulates the partitioning of the ran-gtpase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell Biol. 1996, 135, 1457–1470. [Google Scholar] [CrossRef] [PubMed]
- Mahajan, R.; Delphin, C.; Guan, T.; Gerace, L.; Melchior, F. A small ubiquitin-related polypeptide involved in targeting rangap1 to nuclear pore complex protein RanBP2. Cell 1997, 88, 97–107. [Google Scholar] [CrossRef]
- Lee, G.W.; Melchior, F.; Matunis, M.J.; Mahajan, R.; Tian, Q.; Anderson, P. Modification of Ran GTpase-activating protein by the small ubiquitin-related modifier SUMO-1 requires UBC9, an E2-type ubiquitin-conjugating enzyme homologue. J. Biol. Chem. 1998, 273, 6503–6507. [Google Scholar] [CrossRef] [PubMed]
- Saitoh, H.; Pu, R.; Cavenagh, M.; Dasso, M. RanBP2 associates with UBC9p and a modified form of rangap1. Proc. Natl. Acad. Sci. USA 1997, 94, 3736–3741. [Google Scholar] [CrossRef] [PubMed]
- Salina, D.; Enarson, P.; Rattner, J.B.; Burke, B. Nup358 integrates nuclear envelope breakdown with kinetochore assembly. J. Cell Biol. 2003, 162, 991–1001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joseph, J.; Liu, S.T.; Jablonski, S.A.; Yen, T.J.; Dasso, M. The RanGAP1-RanBP2 complex is essential for microtubule-kinetochore interactions in vivo. Curr. Biol. 2004, 14, 611–617. [Google Scholar] [CrossRef] [PubMed]
- Arnaoutov, A.; Azuma, Y.; Ribbeck, K.; Joseph, J.; Boyarchuk, Y.; Karpova, T.; McNally, J.; Dasso, M. CRM1 is a mitotic effector of Ran-GTP in somatic cells. Nat. Cell Biol. 2005, 7, 626–632. [Google Scholar] [CrossRef] [PubMed]
- Joseph, J.; Tan, S.H.; Karpova, T.S.; McNally, J.G.; Dasso, M. Sumo-1 targets RanGAP1 to kinetochores and mitotic spindles. J. Cell Biol. 2002, 156, 595–602. [Google Scholar] [CrossRef] [PubMed]
- Tulu, U.S.; Fagerstrom, C.; Ferenz, N.P.; Wadsworth, P. Molecular requirements for kinetochore-associated microtubule formation in mammalian cells. Curr. Biol. 2006, 16, 536–541. [Google Scholar] [CrossRef] [PubMed]
- Torosantucci, L.; de Luca, M.; Guarguaglini, G.; Lavia, P.; Degrassi, F. Localized rangtp accumulation promotes microtubule nucleation at kinetochores in somatic mammalian cells. Mol. Biol. Cell 2008, 19, 1873–1882. [Google Scholar] [CrossRef] [PubMed]
- Roscioli, E.; di Francesco, L.; Bolognesi, A.; Giubettini, M.; Orlando, S.; Harel, A.; Schinina, M.E.; Lavia, P. Importin-β negatively regulates multiple aspects of mitosis including RanGAP1 recruitment to kinetochores. J. Cell Biol. 2012, 196, 435–450. [Google Scholar] [CrossRef] [PubMed]
- Hashizume, C.; Kobayashi, A.; Wong, R.W. Down-modulation of nucleoporin RanBP2/Nup358 impaired chromosomal alignment and induced mitotic catastrophe. Cell Death Dis. 2013, 4, e854. [Google Scholar] [CrossRef] [PubMed]
- Dawlaty, M.M.; Malureanu, L.; Jeganathan, K.B.; Kao, E.; Sustmann, C.; Tahk, S.; Shuai, K.; Grosschedl, R.; van Deursen, J.M. Resolution of sister centromeres requires RanBP2-mediated sumoylation of topoisomerase IIα. Cell 2008, 133, 103–115. [Google Scholar] [CrossRef] [PubMed]
- Bhat, M.A.; Philp, A.V.; Glover, D.M.; Bellen, H.J. Chromatid segregation at anaphase requires the barren product, a novel chromosome-associated protein that interacts with topoisomerase II. Cell 1996, 87, 1103–1114. [Google Scholar] [CrossRef]
- Klein, U.R.; Haindl, M.; Nigg, E.A.; Muller, S. RanBP2 and SENP3 function in a mitotic SUMO2/3 conjugation-deconjugation cycle on borealin. Mol. Biol. Cell 2009, 20, 410–418. [Google Scholar] [CrossRef] [PubMed]
- Musacchio, A.; Salmon, E.D. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 2007, 8, 379–393. [Google Scholar] [CrossRef] [PubMed]
- Yu, H. Structural activation of Mad2 in the mitotic spindle checkpoint: The two-state Mad2 model versus the Mad2 template model. J. Cell Biol. 2006, 173, 153–157. [Google Scholar] [CrossRef] [PubMed]
- Fang, G. Checkpoint protein bubr1 acts synergistically with Mad2 to inhibit anaphase-promoting complex. Mol. Biol. Cell 2002, 13, 755–766. [Google Scholar] [CrossRef] [PubMed]
- Fang, G.; Yu, H.; Kirschner, M.W. Direct binding of CDC20 protein family members activates the anaphase-promoting complex in mitosis and G1. Mol. Cell 1998, 2, 163–171. [Google Scholar] [CrossRef]
- Peters, J.M. The anaphase promoting complex/cyclosome: A machine designed to destroy. Nat. Rev. Mol. Cell Biol. 2006, 7, 644–656. [Google Scholar] [CrossRef] [PubMed]
- Campbell, M.S.; Chan, G.K.; Yen, T.J. Mitotic checkpoint proteins HSMad1 and HSMad2 are associated with nuclear pore complexes in interphase. J. Cell Sci. 2001, 114, 953–963. [Google Scholar] [PubMed]
- Lussi, Y.C.; Shumaker, D.K.; Shimi, T.; Fahrenkrog, B. The nucleoporin Nup153 affects spindle checkpoint activity due to an association with Mad1. Nucleus 2010, 1, 71–84. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Sterling, H.; Burlingame, A.; McCormick, F. Tpr directly binds to Mad1 and Mad2 and is important for the Mad1-Mad2-mediated mitotic spindle checkpoint. Genes Dev. 2008, 22, 2926–2931. [Google Scholar] [CrossRef] [PubMed]
- De Souza, C.P.; Hashmi, S.B.; Nayak, T.; Oakley, B.; Osmani, S.A. Mlp1 acts as a mitotic scaffold to spatially regulate spindle assembly checkpoint proteins in aspergillus nidulans. Mol. Biol. Cell 2009, 20, 2146–2159. [Google Scholar] [CrossRef] [PubMed]
- Ding, D.; Muthuswamy, S.; Meier, I. Functional interaction between the arabidopsis orthologs of spindle assembly checkpoint proteins Mad1 and Mad2 and the nucleoporin nua. Plant Mol. Biol. 2012, 79, 203–216. [Google Scholar] [CrossRef] [PubMed]
- Iouk, T.; Kerscher, O.; Scott, R.J.; Basrai, M.A.; Wozniak, R.W. The yeast nuclear pore complex functionally interacts with components of the spindle assembly checkpoint. J. Cell Biol. 2002, 159, 807–819. [Google Scholar] [CrossRef] [PubMed]
- Lince-Faria, M.; Maffini, S.; Orr, B.; Ding, Y.; Claudia, F.; Sunkel, C.E.; Tavares, A.; Johansen, J.; Johansen, K.M.; Maiato, H. Spatiotemporal control of mitosis by the conserved spindle matrix protein megator. J. Cell Biol. 2009, 184, 647–657. [Google Scholar] [CrossRef] [PubMed]
- Scott, R.J.; Lusk, C.P.; Dilworth, D.J.; Aitchison, J.D.; Wozniak, R.W. Interactions between Mad1p and the nuclear transport machinery in the yeast saccharomyces cerevisiae. Mol. Biol. Cell 2005, 16, 4362–4374. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Bravo, V.; Maciejowski, J.; Corona, J.; Buch, H.K.; Collin, P.; Kanemaki, M.T.; Shah, J.V.; Jallepalli, P.V. Nuclear pores protect genome integrity by assembling a premitotic and Mad1-dependent anaphase inhibitor. Cell 2014, 156, 1017–1031. [Google Scholar] [CrossRef] [PubMed]
- Schweizer, N.; Ferras, C.; Kern, D.M.; Logarinho, E.; Cheeseman, I.M.; Maiato, H. Spindle assembly checkpoint robustness requires Tpr-mediated regulation of Mad1/Mad2 proteostasis. J. Cell Biol. 2013, 203, 883–893. [Google Scholar] [CrossRef] [PubMed]
- Ibarra, A.; Hetzer, M.W. Nuclear pore proteins and the control of genome functions. Genes Dev. 2015, 29, 337–349. [Google Scholar] [CrossRef] [PubMed]
- Mackay, D.R.; Elgort, S.W.; Ullman, K.S. The nucleoporin Nup153 has separable roles in both early mitotic progression and the resolution of mitosis. Mol. Biol. Cell 2009, 20, 1652–1660. [Google Scholar] [CrossRef] [PubMed]
- Mackay, D.R.; Makise, M.; Ullman, K.S. Defects in nuclear pore assembly lead to activation of an aurora B-mediated abscission checkpoint. J. Cell Biol. 2010, 191, 923–931. [Google Scholar] [CrossRef] [PubMed]
- Rajanala, K.; Sarkar, A.; Jhingan, G.D.; Priyadarshini, R.; Jalan, M.; Sengupta, S.; Nandicoori, V.K. Phosphorylation of nucleoporin Tpr governs its differential localization and is required for its mitotic function. J. Cell Sci. 2014, 127, 3505–3520. [Google Scholar] [CrossRef] [PubMed]
- Peters, J.M. The anaphase-promoting complex: Proteolysis in mitosis and beyond. Mol. Cell 2002, 9, 931–943. [Google Scholar] [CrossRef]
- Visintin, R.; Prinz, S.; Amon, A. CDC20 and CDH1: A family of substrate-specific activators of APC-dependent proteolysis. Science 1997, 278, 460–463. [Google Scholar] [CrossRef] [PubMed]
- Pfleger, C.M.; Kirschner, M.W. The ken box: An APC recognition signal distinct from the D box targeted by CDH1. Genes Dev. 2000, 14, 655–665. [Google Scholar] [PubMed]
- Zur, A.; Brandeis, M. Securin degradation is mediated by Fzy and Fzr, and is required for complete chromatid separation but not for cytokinesis. EMBO J. 2001, 20, 792–801. [Google Scholar] [CrossRef] [PubMed]
- Jeganathan, K.B.; Malureanu, L.; van Deursen, J.M. The RAE1-Nup98 complex prevents aneuploidy by inhibiting securin degradation. Nature 2005, 438, 1036–1039. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, C.E.; Fornerod, M.; Kasper, L.H.; van Deursen, J.M. RAE1 is a shuttling mRNA export factor that binds to a GLEBs-like Nup98 motif at the nuclear pore complex through multiple domains. J. Cell Biol. 1999, 145, 237–254. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Babu, J.R.; Harden, J.M.; Jablonski, S.A.; Gazi, M.H.; Lingle, W.L.; de Groen, P.C.; Yen, T.J.; van Deursen, J.M. The mitotic checkpoint protein hBUB3 and the mRNA export factor hRAE1 interact with GLE2p-binding sequence (GLEBs)-containing proteins. J. Biol. Chem. 2001, 276, 26559–26567. [Google Scholar] [CrossRef] [PubMed]
- Gough, S.M.; Slape, C.I.; Aplan, P.D. Nup98 gene fusions and hematopoietic malignancies: Common themes and new biologic insights. Blood 2011, 118, 6247–6257. [Google Scholar] [CrossRef] [PubMed]
- Fahrenkrog, B. Nucleoporin gene fusions and hematopoietic malignancies. New J. Sci. 2014, 2014. [Google Scholar] [CrossRef]
- Salsi, V.; Ferrari, S.; Gorello, P.; Fantini, S.; Chiavolelli, F.; Mecucci, C.; Zappavigna, V. Nup98 fusion oncoproteins promote aneuploidy by attenuating the mitotic spindle checkpoint. Cancer Res. 2014, 74, 1079–1090. [Google Scholar] [CrossRef] [PubMed]
- Raza-Egilmez, S.Z.; Jani-Sait, S.N.; Grossi, M.; Higgins, M.J.; Shows, T.B.; Aplan, P.D. Nup98-HoxD13 gene fusion in therapy-related acute myelogenous leukemia. Cancer Res. 1998, 58, 4269–4273. [Google Scholar] [PubMed]
- Ahuja, H.G.; Felix, C.A.; Aplan, P.D. The t(11;20)(p15;q11) chromosomal translocation associated with therapy-related myelodysplastic syndrome results in an Nup98-Top1 fusion. Blood 1999, 94, 3258–3261. [Google Scholar] [PubMed]
- Jaju, R.J.; Fidler, C.; Haas, O.A.; Strickson, A.J.; Watkins, F.; Clark, K.; Cross, N.C.; Cheng, J.F.; Aplan, P.D.; Kearney, L.; et al. A novel gene, Nsd1, is fused to Nup98 in the t(5;11)(q35;p15.5) in de novo childhood acute myeloid leukemia. Blood 2001, 98, 1264–1267. [Google Scholar] [CrossRef] [PubMed]
- Taketani, T.; Taki, T.; Shibuya, N.; Kikuchi, A.; Hanada, R.; Hayashi, Y. Novel Nup98-HoxC11 fusion gene resulted from a chromosomal break within EXON 1 of HoxC11 in acute myeloid leukemia with t(11;12)(p15;q13). Cancer Res. 2002, 62, 4571–4574. [Google Scholar] [PubMed]
- Chen, R.H.; Shevchenko, A.; Mann, M.; Murray, A.W. Spindle checkpoint protein XMad1 recruits XMad2 to unattached kinetochores. J. Cell Biol. 1998, 143, 283–295. [Google Scholar] [CrossRef] [PubMed]
- Katsani, K.R.; Karess, R.E.; Dostatni, N.; Doye, V. In vivo dynamics of drosophila nuclear envelope components. Mol. Biol. Cell 2008, 19, 3652–3666. [Google Scholar] [CrossRef] [PubMed]
- Buffin, E.; Lefebvre, C.; Huang, J.; Gagou, M.E.; Karess, R.E. Recruitment of Mad2 to the kinetochore requires the Rod/ZW10 complex. Curr. Biol. 2005, 15, 856–861. [Google Scholar] [CrossRef] [PubMed]
- Ikui, A.E.; Furuya, K.; Yanagida, M.; Matsumoto, T. Control of localization of a spindle checkpoint protein, Mad2, in fission yeast. J. Cell Sci. 2002, 115, 1603–1610. [Google Scholar] [PubMed]
- Rodenas, E.; Gonzalez-Aguilera, C.; Ayuso, C.; Askjaer, P. Dissection of the Nup107 nuclear pore subcomplex reveals a novel interaction with spindle assembly checkpoint protein Mad1 in caenorhabditis elegans. Mol. Biol. Cell 2012, 23, 930–944. [Google Scholar] [CrossRef] [PubMed]
- De Souza, C.P.; Osmani, A.H.; Hashmi, S.B.; Osmani, S.A. Partial nuclear pore complex disassembly during closed mitosis in aspergillus nidulans. Curr. Biol. 2004, 14, 1973–1984. [Google Scholar] [CrossRef] [PubMed]
- Osmani, A.H.; Davies, J.; Liu, H.L.; Nile, A.; Osmani, S.A. Systematic deletion and mitotic localization of the nuclear pore complex proteins of aspergillus nidulans. Mol. Biol. Cell 2006, 17, 4946–4961. [Google Scholar] [CrossRef] [PubMed]
- De Souza, C.P.; Osmani, S.A. Mitosis, not just open or closed. Eukaryot. Cell 2007, 6, 1521–1527. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.K.; Gruenbaum, Y.; Spann, P.; Liu, J.; Wilson, K.L.C. Elegans nuclear envelope proteins emerin, man1, lamin, and nucleoporins reveal unique timing of nuclear envelope breakdown during mitosis. Mol. Biol. Cell 2000, 11, 3089–3099. [Google Scholar] [CrossRef] [PubMed]
- Quimby, B.B.; Arnaoutov, A.; Dasso, M. Ran GTpase regulates Mad2 localization to the nuclear pore complex. Eukaryot. Cell 2005, 4, 274–280. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Hara-Nishimura, I. The molecular architecture of the plant nuclear pore complex. J. Exp. Bot. 2013, 64, 823–832. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Meier, I. Identification and characterization of the arabidopsis FG-repeat nucleoporin Nup62. Plant Signal. Behav. 2011, 6, 330–334. [Google Scholar] [CrossRef] [PubMed]
- Holden, J.M.; Koreny, L.; Obado, S.; Ratushny, A.V.; Chen, W.M.; Chiang, J.H.; Kelly, S.; Chait, B.T.; Aitchison, J.D.; Rout, M.P.; et al. Nuclear pore complex evolution: A trypanosome Mlp analogue functions in chromosomal segregation but lacks transcriptional barrier activity. Mol. Biol. Cell 2014, 25, 1421–1436. [Google Scholar] [CrossRef] [PubMed]
- Morelle, C.; Sterkers, Y.; Crobu, L.; MBang-Benet, D.E.; Kuk, N.; Portales, P.; Bastien, P.; Pages, M.; Lachaud, L. The nucleoporin Mlp2 is involved in chromosomal distribution during mitosis in trypanosomatids. Nucleic Acids Res. 2015, 43, 4013–4027. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Li, Z. Comparative analysis of chromosome segregation in human, yeasts and trypanosome. Front. Biol. (Beijing) 2014, 9, 472–480. [Google Scholar] [CrossRef] [PubMed]
- Markossian, S.; Suresh, S.; Osmani, A.H.; Osmani, S.A. Nup2 requires a highly divergent partner, nupa, to fulfill functions at nuclear pore complexes and the mitotic chromatin region. Mol. Biol. Cell 2015, 26, 605–621. [Google Scholar] [CrossRef] [PubMed]
- Sudakin, V.; Chan, G.K.; Yen, T.J. Checkpoint inhibition of the APC/C in hela cells is mediated by a complex of BUBR1, BUB3, CDC20, and Mad2. J. Cell Biol. 2001, 154, 925–936. [Google Scholar] [CrossRef] [PubMed]
- Malureanu, L.A.; Jeganathan, K.B.; Hamada, M.; Wasilewski, L.; Davenport, J.; van Deursen, J.M. Bubr1 N-terminus acts as a soluble inhibitor of cyclin b degradation by APC/C(CDC20) in interphase. Dev. Cell 2009, 16, 118–131. [Google Scholar] [CrossRef] [PubMed]
- Cairo, L.V.; Ptak, C.; Wozniak, R.W. Mitosis-specific regulation of nuclear transport by the spindle assembly checkpoint protein Mad1p. Mol. Cell 2013, 49, 109–120. [Google Scholar] [CrossRef] [PubMed]
- Cairo, L.V.; Ptak, C.; Wozniak, R.W. Dual personality of Mad1: Regulation of nuclear import by a spindle assembly checkpoint protein. Nucleus 2013, 4, 367–373. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mossaid, I.; Fahrenkrog, B. Complex Commingling: Nucleoporins and the Spindle Assembly Checkpoint. Cells 2015, 4, 706-725. https://doi.org/10.3390/cells4040706
Mossaid I, Fahrenkrog B. Complex Commingling: Nucleoporins and the Spindle Assembly Checkpoint. Cells. 2015; 4(4):706-725. https://doi.org/10.3390/cells4040706
Chicago/Turabian StyleMossaid, Ikram, and Birthe Fahrenkrog. 2015. "Complex Commingling: Nucleoporins and the Spindle Assembly Checkpoint" Cells 4, no. 4: 706-725. https://doi.org/10.3390/cells4040706