Entamoeba histolytica under Oxidative Stress: What Countermeasure Mechanisms Are in Place?
Abstract
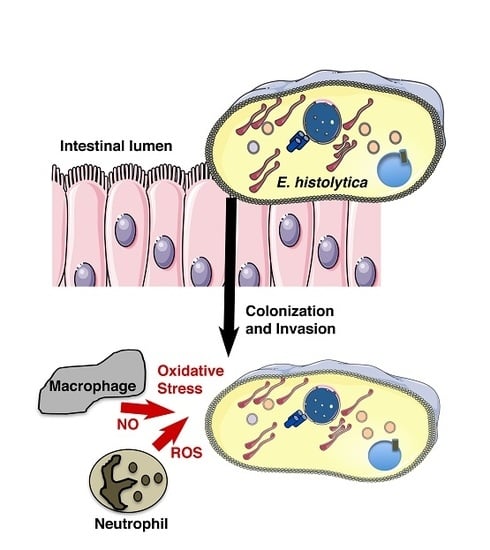
:1. Introduction
2. Amoeba Survival
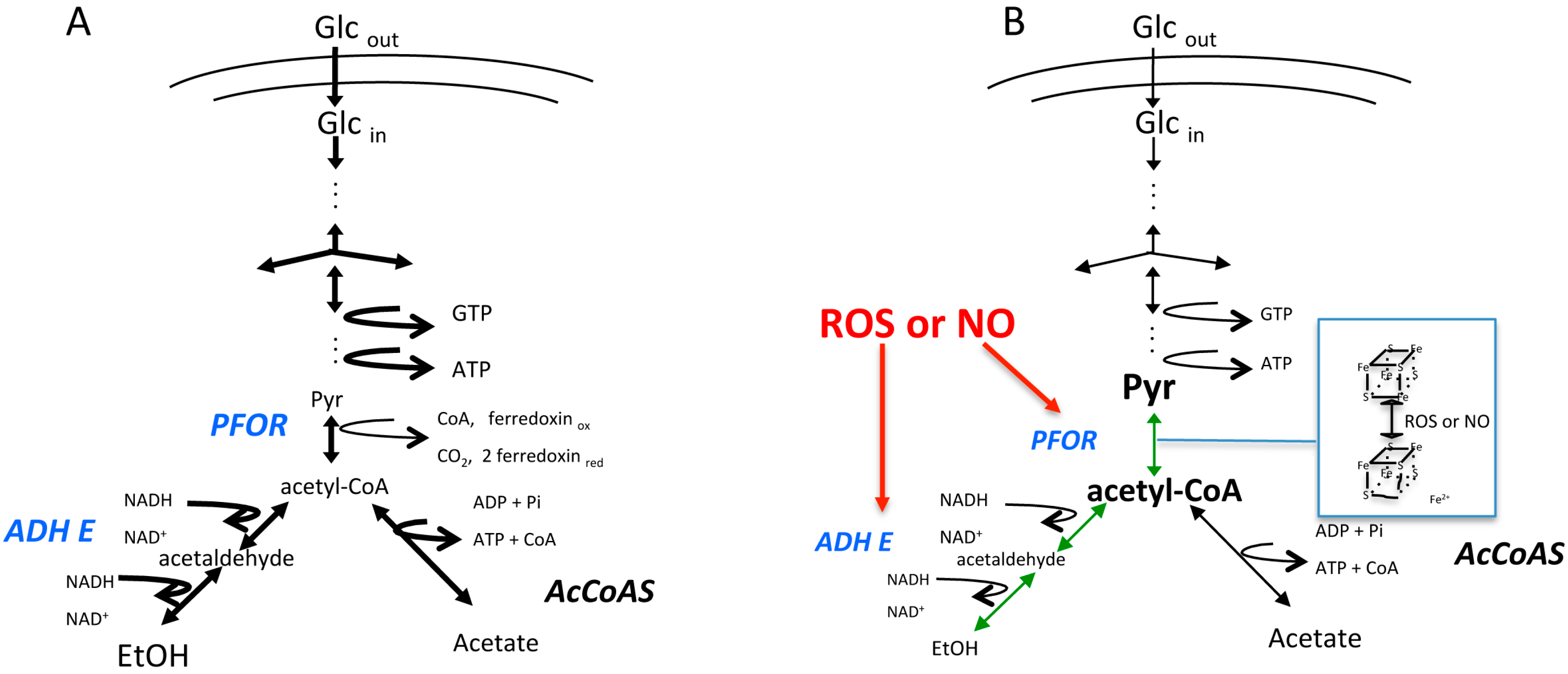
Survival Mode on: What Metabolic Pathways are Affected and to What Purpose?
3. Under Oxidative Stress, to Be or Not to Be Virulent, Matters
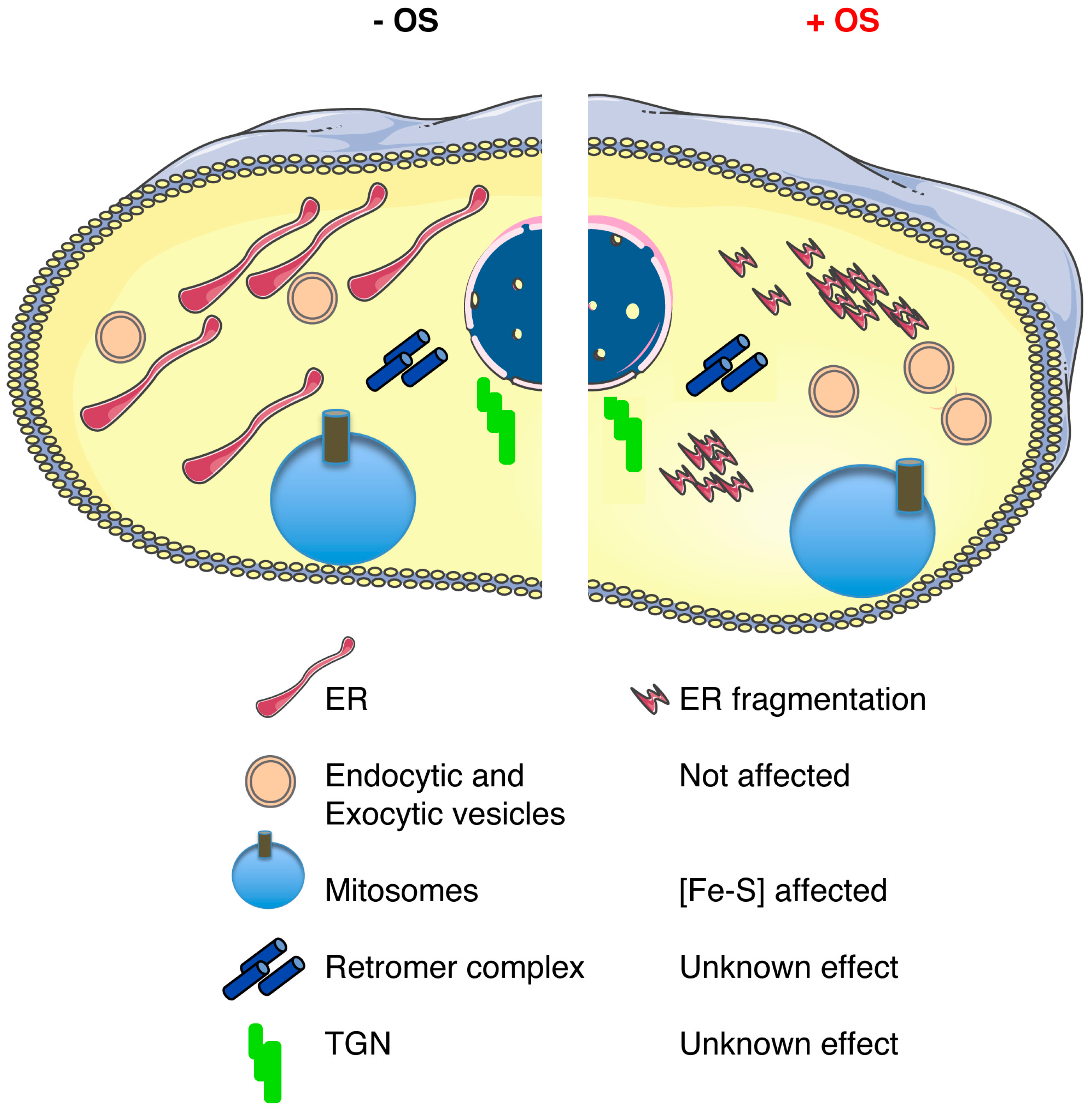
4. E. histolytica Endomembrane Network is a Puzzle of Vesicles
4.1. E. histolytica Endomembrane Network is Affected by Oxidative and Nitrosative Stresses
4.2. ER, Where Are You?
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| AcCoAS | Acetyl-CoA synthetase |
| ADHE | Aldehyde/alcohol dehydrogenase |
| ALA | Amebic liver abscess |
| ATP | Adenosine triphosphate |
| CRT | Calreticulin |
| EhCP-5 | Cysteine proteinase 5 |
| ALAH | Hamster amebic liver abscess |
| NO | Nitric oxide |
| NOS | Nitric oxide synthase |
| OS | Oxidative Stress |
| SRPR | Particle receptor |
| SNO | Protein S-nitrosothiols |
| PFOR | Pyruvate:ferredoxin oxidoreductase |
| ROS | Reactive oxygen species |
| SPC | Signal peptidases |
| NaNO3 | Sodium nitrate |
| NaNO2 | Sodium nitrite |
| SNP | Sodium nitroprusside |
| SOD | Superoxide dismutase |
| Eh29 | Thiol-dependent peroxidase |
| UPR | Unfolded Protein Response |
References
- WHO/PAHO/UNESCO report. A consultation with experts on amoebiasis. Mexico city, Mexico 28–29 January, 1997. Epidemiol. Bull. 1997, 18, 13–14.
- GBD 2013 Mortality and Causes of Death Collaborators. Global, regional, and national age-sex specific all-cause and cause-specific mortality for 240 causes of death, 1990–2013: A systematic analysis for the global burden of disease study 2013. Lancet 2015, 385, 117–171. [Google Scholar]
- Herricks, J.R.; Hotez, P.J.; Wanga, V.; Coffeng, L.E.; Haagsma, J.A.; Basanez, M.G.; Buckle, G.; Budke, C.M.; Carabin, H.; Fevre, E.M.; et al. The global burden of disease study 2013: What does it mean for the ntds? PLoS Negl. Trop. Dis. 2017, 11, e0005424. [Google Scholar] [CrossRef] [PubMed]
- de Lalla, F.; Rinaldi, E.; Santoro, D.; Nicolin, R.; Tramarin, A. Outbreak of Entamoeba histolytica and Giardia lamblia infections in travellers returning from the tropics. Infection 1992, 20, 78–82. [Google Scholar] [CrossRef] [PubMed]
- Nourse, C.B.; Robson, J.M.; Whitby, M.R.; Francis, J.R. First report of Entamoeba histolytica infection from timor-leste–acute amoebic colitis and concurrent late development of amoebic liver abscess in returned travellers to Australia. J. Travel Med. 2016, 23, tav027. [Google Scholar] [CrossRef] [PubMed]
- Welch, J.S.; Dobson, C.; Chopra, S. Immunodiagnosis of Entamoeba histolytica and Ascaris lumbricoides infections in caucasian and aboriginal Australians. Trans. R. Soc. Trop. Med. Hyg. 1986, 80, 240–247. [Google Scholar] [CrossRef]
- Petri, W.A., Jr.; Mondal, D.; Peterson, K.M.; Duggal, P.; Haque, R. Association of malnutrition with amebiasis. Nutr. Rev. 2009, 67 (Suppl. S2), S207–S215. [Google Scholar] [CrossRef] [PubMed]
- Nath, J.; Ghosh, S.K.; Singha, B.; Paul, J. Molecular epidemiology of amoebiasis: A cross-sectional study among north east Indian population. PLoS Negl. Trop. Dis. 2015, 9, e0004225. [Google Scholar] [CrossRef] [PubMed]
- Mondal, D.; Petri, W.A., Jr.; Sack, R.B.; Kirkpatrick, B.D.; Haque, R. Entamoeba histolytica-associated diarrheal illness is negatively associated with the growth of preschool children: Evidence from a prospective study. Trans. R. Soc. Trop. Med. Hyg. 2006, 100, 1032–1038. [Google Scholar] [CrossRef] [PubMed]
- Petri, W.A., Jr.; Smith, R.D.; Schlesinger, P.H.; Murphy, C.F.; Ravdin, J.I. Isolation of the galactose-binding lectin that mediates the in vitro adherence of Entamoeba histolytica. J. Clin. Investig. 1987, 80, 1238–1244. [Google Scholar] [CrossRef] [PubMed]
- Braga, L.L.; Ninomiya, H.; McCoy, J.J.; Eacker, S.; Wiedmer, T.; Pham, C.; Wood, S.; Sims, P.J.; Petri, W.A., Jr. Inhibition of the complement membrane attack complex by the galactose-specific adhesion of Entamoeba histolytica. J. Clin. Investig. 1992, 90, 1131–1137. [Google Scholar] [CrossRef] [PubMed]
- Perdomo, D.; Baron, B.; Rojo-Dominguez, A.; Raynal, B.; England, P.; Guillen, N. The alpha-helical regions of kerp1 are important in Entamoeba histolytica adherence to human cells. Sci. Rep. 2013, 3, 1171. [Google Scholar] [CrossRef] [PubMed]
- Moncada, D.; Keller, K.; Chadee, K. Entamoeba histolytica cysteine proteinases disrupt the polymeric structure of colonic mucin and alter its protective function. Infect. Immun. 2003, 71, 838–844. [Google Scholar] [CrossRef] [PubMed]
- Thibeaux, R.; Dufour, A.; Roux, P.; Bernier, M.; Baglin, A.C.; Frileux, P.; Olivo-Marin, J.C.; Guillen, N.; Labruyere, E. Newly visualized fibrillar collagen scaffolds dictate Entamoeba histolytica invasion route in the human colon. Cell. Microbiol. 2012, 14, 609–621. [Google Scholar] [CrossRef] [PubMed]
- Lauwaet, T.; Oliveira, M.J.; Callewaert, B.; De Bruyne, G.; Mareel, M.; Leroy, A. Proteinase inhibitors tpck and tlck prevent Entamoeba histolytica induced disturbance of tight junctions and microvilli in enteric cell layers in vitro. Int. J. Parasitol. 2004, 34, 785–794. [Google Scholar] [CrossRef] [PubMed]
- Tran, V.Q.; Herdman, D.S.; Torian, B.E.; Reed, S.L. The neutral cysteine proteinase of Entamoeba histolytica degrades igg and prevents its binding. J. Infect. Dis. 1998, 177, 508–511. [Google Scholar] [CrossRef] [PubMed]
- Reed, S.L.; Keene, W.E.; McKerrow, J.H.; Gigli, I. Cleavage of c3 by a neutral cysteine proteinase of Entamoeba histolytica. J. Immunol. 1989, 143, 189–195. [Google Scholar] [PubMed]
- Ralston, K.S.; Solga, M.D.; Mackey-Lawrence, N.M.; Somlata; Bhattacharya, A.; Petri, W.A., Jr. Trogocytosis by Entamoeba histolytica contributes to cell killing and tissue invasion. Nature 2014, 508, 526–530. [Google Scholar] [CrossRef] [PubMed]
- Orozco, E.; Guarneros, G.; Martinez-Palomo, A.; Sanchez, T. Entamoeba histolytica. Phagocytosis as a virulence factor. J. Exp. Med. 1983, 158, 1511–1521. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Palomo, A.; Gonzalez-Robles, A.; Chavez, B.; Orozco, E.; Fernandez-Castelo, S.; Cervantes, A. Structural bases of the cytolytic mechanisms of Entamoeba histolytica. J. Protozool. 1985, 32, 166–175. [Google Scholar] [CrossRef] [PubMed]
- Huston, C.D.; Boettner, D.R.; Miller-Sims, V.; Petri, W.A., Jr. Apoptotic killing and phagocytosis of host cells by the parasite Entamoeba histolytica. Infect. Immun. 2003, 71, 964–972. [Google Scholar] [CrossRef] [PubMed]
- Loftus, B.; Anderson, I.; Davies, R.; Alsmark, U.C.; Samuelson, J.; Amedeo, P.; Roncaglia, P.; Berriman, M.; Hirt, R.P.; Mann, B.J.; et al. The genome of the protist parasite Entamoeba histolytica. Nature 2005, 433, 865–868. [Google Scholar] [CrossRef] [PubMed]
- Bansal, D.; Ave, P.; Kerneis, S.; Frileux, P.; Boche, O.; Baglin, A.C.; Dubost, G.; Leguern, A.S.; Prevost, M.C.; Bracha, R.; et al. An ex-vivo human intestinal model to study Entamoeba histolytica pathogenesis. PLoS Negl. Trop. Dis. 2009, 3, e551. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.Y.; Chadee, K. Macrophage cytotoxicity against Entamoeba histolytica trophozoites is mediated by nitric oxide from l-arginine. J. Immunol. 1992, 148, 3999–4005. [Google Scholar] [PubMed]
- Santi-Rocca, J.; Smith, S.; Weber, C.; Pineda, E.; Hon, C.C.; Saavedra, E.; Olivos-Garcia, A.; Rousseau, S.; Dillies, M.A.; Coppee, J.Y.; et al. Endoplasmic reticulum stress-sensing mechanism is activated in Entamoeba histolytica upon treatment with nitric oxide. PLoS ONE 2012, 7, e31777. [Google Scholar] [CrossRef] [PubMed]
- Fahey, R.C.; Newton, G.L.; Arrick, B.; Overdank-Bogart, T.; Aley, S.B. Entamoeba histolytica: A eukaryote without glutathione metabolism. Science 1984, 224, 70–72. [Google Scholar] [CrossRef] [PubMed]
- Flohe, L.; Hecht, H.J.; Steinert, P. Glutathione and trypanothione in parasitic hydroperoxide metabolism. Free Radic. Biol. Med. 1999, 27, 966–984. [Google Scholar] [CrossRef]
- Jeelani, G.; Sato, D.; Husain, A.; Escueta-de Cadiz, A.; Sugimoto, M.; Soga, T.; Suematsu, M.; Nozaki, T. Metabolic profiling of the protozoan parasite Entamoeba invadens revealed activation of unpredicted pathway during encystation. PLoS ONE 2012, 7, e37740. [Google Scholar] [CrossRef] [PubMed]
- Leitsch, D.; Kolarich, D.; Wilson, I.B.; Altmann, F.; Duchene, M. Nitroimidazole action in Entamoeba histolytica: A central role for thioredoxin reductase. PLoS Biol. 2007, 5, e211. [Google Scholar] [CrossRef] [PubMed]
- Jeelani, G.; Sato, D.; Soga, T.; Watanabe, H.; Nozaki, T. Mass spectrometric analysis of l-cysteine metabolism: Physiological role and fate of l-cysteine in the enteric protozoan parasite Entamoeba histolytica. MBio 2014, 5, e01995. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.C.; Dean, D.R.; Smith, A.D.; Johnson, M.K. Structure, function, and formation of biological iron-sulfur clusters. Annu. Rev. Biochem. 2005, 74, 247–281. [Google Scholar] [CrossRef] [PubMed]
- Biederbick, A.; Stehling, O.; Rosser, R.; Niggemeyer, B.; Nakai, Y.; Elsasser, H.P.; Lill, R. Role of human mitochondrial nfs1 in cytosolic iron-sulfur protein biogenesis and iron regulation. Mol. Cell. Biol. 2006, 26, 5675–5687. [Google Scholar] [CrossRef] [PubMed]
- Ali, V.; Nozaki, T. Iron-sulphur clusters, their biosynthesis, and biological functions in protozoan parasites. Adv. Parasitol. 2013, 83, 1–92. [Google Scholar] [PubMed]
- Imlay, J.A. Iron-sulphur clusters and the problem with oxygen. Mol. Microbiol. 2006, 59, 1073–1082. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.; Imlay, J.A. Micromolar intracellular hydrogen peroxide disrupts metabolism by damaging iron-sulfur enzymes. J. Biol. Chem. 2007, 282, 929–937. [Google Scholar] [CrossRef] [PubMed]
- Reeves, R.E. Metabolism of Entamoeba histolytica schaudinn, 1903. Adv. Parasitol. 1984, 23, 105–142. [Google Scholar] [PubMed]
- Wassmann, C.; Hellberg, A.; Tannich, E.; Bruchhaus, I. Metronidazole resistance in the protozoan parasite Entamoeba histolytica is associated with increased expression of iron-containing superoxide dismutase and peroxiredoxin and decreased expression of ferredoxin 1 and flavin reductase. J. Biol. Chem. 1999, 274, 26051–26056. [Google Scholar] [CrossRef] [PubMed]
- Pan, N.; Imlay, J.A. How does oxygen inhibit central metabolism in the obligate anaerobe bacteroides thetaiotaomicron. Mol. Microbiol. 2001, 39, 1562–1571. [Google Scholar] [CrossRef] [PubMed]
- Pieulle, L.; Guigliarelli, B.; Asso, M.; Dole, F.; Bernadac, A.; Hatchikian, E.C. Isolation and characterization of the pyruvate-ferredoxin oxidoreductase from the sulfate-reducing bacterium Desulfovibrio africanus. Biochim. Biophys. Acta 1995, 1250, 49–59. [Google Scholar] [CrossRef]
- Ramos-Martinez, E.; Olivos-Garcia, A.; Saavedra, E.; Nequiz, M.; Sanchez, E.C.; Tello, E.; El-Hafidi, M.; Saralegui, A.; Pineda, E.; Delgado, J.; et al. Entamoeba histolytica: Oxygen resistance and virulence. Int. J. Parasitol. 2009, 39, 693–702. [Google Scholar] [CrossRef] [PubMed]
- Pineda, E.; Encalada, R.; Rodriguez-Zavala, J.S.; Olivos-Garcia, A.; Moreno-Sanchez, R.; Saavedra, E. Pyruvate:Ferredoxin oxidoreductase and bifunctional aldehyde-alcohol dehydrogenase are essential for energy metabolism under oxidative stress in Entamoeba histolytica. FEBS J. 2010, 277, 3382–3395. [Google Scholar] [CrossRef] [PubMed]
- Lo, H.S.; Reeves, R.E. Pyruvate-to-ethanol pathway in Entamoeba histolytica. Biochem. J. 1978, 171, 225–230. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Li, E.; Kairong, T.; Stanley, S.L., Jr. Entamoeba histolytica has an alcohol dehydrogenase homologous to the multifunctional adhe gene product of Escherichia coli. Mol. Biochem. Parasitol. 1994, 64, 253–260. [Google Scholar] [CrossRef]
- Espinosa, A.; Yan, L.; Zhang, Z.; Foster, L.; Clark, D.; Li, E.; Stanley, S.L., Jr. The bifunctional Entamoeba histolytica alcohol dehydrogenase 2 (ehadh2) protein is necessary for amebic growth and survival and requires an intact c-terminal domain for both alcohol dahydrogenase and acetaldehyde dehydrogenase activity. J. Biol. Chem. 2001, 276, 20136–20143. [Google Scholar] [CrossRef] [PubMed]
- Echave, P.; Tamarit, J.; Cabiscol, E.; Ros, J. Novel antioxidant role of alcohol dehydrogenase e from Escherichia coli. J. Biol. Chem. 2003, 278, 30193–30198. [Google Scholar] [CrossRef] [PubMed]
- Pineda, E.; Encalada, R.; Vazquez, C.; Nequiz, M.; Olivos-Garcia, A.; Moreno-Sanchez, R.; Saavedra, E. In vivo identification of the steps that control energy metabolism and survival of Entamoeba histolytica. FEBS J. 2015, 282, 318–331. [Google Scholar] [CrossRef] [PubMed]
- Pineda, E.; Encalada, R.; Olivos-Garcia, A.; Nequiz, M.; Moreno-Sanchez, R.; Saavedra, E. The bifunctional aldehyde-alcohol dehydrogenase controls ethanol and acetate production in Entamoeba histolytica under aerobic conditions. FEBS Lett. 2013, 587, 178–184. [Google Scholar] [CrossRef] [PubMed]
- Husain, A.; Sato, D.; Jeelani, G.; Soga, T.; Nozaki, T. Dramatic increase in glycerol biosynthesis upon oxidative stress in the anaerobic protozoan parasite Entamoeba histolytica. PLoS Negl. Trop. Dis. 2012, 6, e1831. [Google Scholar] [CrossRef] [PubMed]
- Santi-Rocca, J.; Rigothier, M.C.; Guillen, N. Host-microbe interactions and defense mechanisms in the development of amoebic liver abscesses. Clin. Microbiol. Rev. 2009, 22, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Tsutsumi, V.; Mena-Lopez, R.; Anaya-Velazquez, F.; Martinez-Palomo, A. Cellular bases of experimental amebic liver abscess formation. Am. J. Pathol. 1984, 117, 81–91. [Google Scholar] [PubMed]
- Thibeaux, R.; Weber, C.; Hon, C.C.; Dillies, M.A.; Ave, P.; Coppee, J.Y.; Labruyere, E.; Guillen, N. Identification of the virulence landscape essential for Entamoeba histolytica invasion of the human colon. PLoS Pathog. 2013, 9, e1003824. [Google Scholar] [CrossRef] [PubMed]
- MacFarlane, R.C.; Singh, U. Identification of differentially expressed genes in virulent and nonvirulent Entamoeba species: Potential implications for amebic pathogenesis. Infect. Immun. 2006, 74, 340–351. [Google Scholar] [CrossRef] [PubMed]
- Weber, C.; Koutero, M.; Dillies, M.A.; Varet, H.; Lopez-Camarillo, C.; Coppee, J.Y.; Hon, C.C.; Guillen, N. Extensive transcriptome analysis correlates the plasticity of Entamoeba histolytica pathogenesis to rapid phenotype changes depending on the environment. Sci. Rep. 2016, 6, 35852. [Google Scholar] [CrossRef] [PubMed]
- Ward, T.H.; Polishchuk, R.S.; Caplan, S.; Hirschberg, K.; Lippincott-Schwartz, J. Maintenance of Golgi structure and function depends on the integrity of er export. J. Cell Biol. 2001, 155, 557–570. [Google Scholar] [CrossRef] [PubMed]
- Klumperman, J. The growing golgi: In search of its independence. Nat. Cell Biol. 2000, 2, E217–E219. [Google Scholar] [CrossRef] [PubMed]
- Perdomo, D.; Ait-Ammar, N.; Syan, S.; Sachse, M.; Jhingan, G.D.; Guillen, N. Cellular and proteomics analysis of the endomembrane system from the unicellular Entamoeba histolytica. J. Proteom. 2015, 112, 125–140. [Google Scholar] [CrossRef] [PubMed]
- Akbar, M.A.; Chatterjee, N.S.; Sen, P.; Debnath, A.; Pal, A.; Bera, T.; Das, P. Genes induced by a high-oxygen environment in Entamoeba histolytica. Mol. Biochem. Parasitol. 2004, 133, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Vicente, J.B.; Ehrenkaufer, G.M.; Saraiva, L.M.; Teixeira, M.; Singh, U. Entamoeba histolytica modulates a complex repertoire of novel genes in response to oxidative and nitrosative stresses: Implications for amebic pathogenesis. Cell. Microbiol. 2009, 11, 51–69. [Google Scholar] [CrossRef] [PubMed]
- Rastew, E.; Vicente, J.B.; Singh, U. Oxidative stress resistance genes contribute to the pathogenic potential of the anaerobic protozoan parasite, Entamoeba histolytica. Int. J. Parasitol. 2012, 42, 1007–1015. [Google Scholar] [CrossRef] [PubMed]
- Ramos, E.; Olivos-Garcia, A.; Nequiz, M.; Saavedra, E.; Tello, E.; Saralegui, A.; Montfort, I.; Perez Tamayo, R. Entamoeba histolytica: Apoptosis induced in vitro by nitric oxide species. Exp. Parasitol. 2007, 116, 257–265. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, A.; Comatas, K.E.; Liu, L.; Stamler, J.S. Screening for nitric oxide-dependent protein-protein interactions. Science 2003, 301, 657–661. [Google Scholar] [CrossRef] [PubMed]
- Illi, B.; Colussi, C.; Grasselli, A.; Farsetti, A.; Capogrossi, M.C.; Gaetano, C. No sparks off chromatin: Tales of a multifaceted epigenetic regulator. Pharmacol. Ther. 2009, 123, 344–352. [Google Scholar] [CrossRef] [PubMed]
- Montagna, C.; Rizza, S.; Maiani, E.; Piredda, L.; Filomeni, G.; Cecconi, F. To eat, or not to eat: S-nitrosylation signaling in autophagy. FEBS J. 2016, 283, 3857–3869. [Google Scholar] [CrossRef] [PubMed]
- Forrester, M.T.; Thompson, J.W.; Foster, M.W.; Nogueira, L.; Moseley, M.A.; Stamler, J.S. Proteomic analysis of S-nitrosylation and denitrosylation by resin-assisted capture. Nat. Biotechnol. 2009, 27, 557–559. [Google Scholar] [CrossRef] [PubMed]
- Shahi, P.; Trebicz-Geffen, M.; Nagaraja, S.; Alterzon-Baumel, S.; Hertz, R.; Methling, K.; Lalk, M.; Ankri, S. Proteomic identification of oxidized proteins in Entamoeba histolytica by resin-assisted capture: Insights into the role of arginase in resistance to oxidative stress. PLoS Negl. Trop. Dis. 2016, 10, e0004340. [Google Scholar] [CrossRef] [PubMed]
- Shahi, P.; Trebicz-Geffen, M.; Nagaraja, S.; Hertz, R.; Baumel-Alterzon, S.; Methling, K.; Lalk, M.; Mazumder, M.; Samudrala, G.; Ankri, S. Corrigendum: N-acetyl ornithine deacetylase is a moonlighting protein and is involved in the adaptation of Entamoeba histolytica to nitrosative stress. Sci. Rep. 2017, 7, 45802. [Google Scholar] [CrossRef] [PubMed]
- Arhets, P.; Gounon, P.; Sansonetti, P.; Guillen, N. Myosin II is involved in capping and uroid formation in the human pathogen Entamoeba histolytica. Infect. Immun. 1995, 63, 4358–4367. [Google Scholar] [PubMed]
- Coudrier, E.; Amblard, F.; Zimmer, C.; Roux, P.; Olivo-Marin, J.C.; Rigothier, M.C.; Guillen, N. Myosin II and the gal-galnac lectin play a crucial role in tissue invasion by Entamoeba histolytica. Cell. Microbiol. 2005, 7, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Perdomo, D.; Manich, M.; Syan, S.; Olivo-Marin, J.C.; Dufour, A.C.; Guillen, N. Intracellular traffic of the lysine and glutamic acid rich protein kerp1 reveals features of endomembrane organization in Entamoeba histolytica. Cell. Microbiol. 2016, 18, 1134–1152. [Google Scholar] [CrossRef] [PubMed]
- Trebicz-Geffen, M.; Shahi, P.; Nagaraja, S.; Vanunu, S.; Manor, S.; Avrahami, A.; Ankri, S. Identification of s-nitrosylated (sno) proteins in Entamoeba histolytica adapted to nitrosative stress: Insights into the role of sno actin and in vitro virulence. Front. Cell. Infect. Microbiol. 2017, 7, 192. [Google Scholar] [CrossRef] [PubMed]
- Malhotra, J.D.; Miao, H.; Zhang, K.; Wolfson, A.; Pennathur, S.; Pipe, S.W.; Kaufman, R.J. Antioxidants reduce endoplasmic reticulum stress and improve protein secretion. Proc. Natl. Acad. Sci. USA 2008, 105, 18525–18530. [Google Scholar] [CrossRef] [PubMed]
- Grootjans, J.; Kaser, A.; Kaufman, R.J.; Blumberg, R.S. The unfolded protein response in immunity and inflammation. Nat. Rev. Immunol. 2016, 16, 469–484. [Google Scholar] [CrossRef] [PubMed]
- McGuckin, M.A.; Eri, R.D.; Das, I.; Lourie, R.; Florin, T.H. Er stress and the unfolded protein response in intestinal inflammation. Am. J. Physiol. Gastrointest. Liver Physiol. 2010, 298, G820–G832. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, J.E.; Huston, C.D. Evidence of a continuous endoplasmic reticulum in the protozoan parasite Entamoeba histolytica. Eukaryot. Cell 2008, 7, 1222–1226. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, E.; de Leon Mdel, C.; Meza, I.; Ocadiz-Delgado, R.; Gariglio, P.; Silva-Olivares, A.; Galindo-Gomez, S.; Shibayama, M.; Moran, P.; Valadez, A.; et al. Entamoeba histolytica calreticulin: An endoplasmic reticulum protein expressed by trophozoites into experimentally induced amoebic liver abscesses. Parasitol. Res. 2011, 108, 439–449. [Google Scholar] [CrossRef] [PubMed]
- Rajesh, K.; Krishnamoorthy, J.; Kazimierczak, U.; Tenkerian, C.; Papadakis, A.I.; Wang, S.; Huang, S.; Koromilas, A.E. Phosphorylation of the translation initiation factor eif2alpha at serine 51 determines the cell fate decisions of akt in response to oxidative stress. Cell Death Dis. 2015, 6, e1591. [Google Scholar] [CrossRef] [PubMed]
- Back, S.H.; Scheuner, D.; Han, J.; Song, B.; Ribick, M.; Wang, J.; Gildersleeve, R.D.; Pennathur, S.; Kaufman, R.J. Translation attenuation through eif2alpha phosphorylation prevents oxidative stress and maintains the differentiated state in beta cells. Cell Metab. 2009, 10, 13–26. [Google Scholar] [CrossRef] [PubMed]
- Hollien, J. Evolution of the unfolded protein response. Biochim. Biophys. Acta 2013, 1833, 2458–2463. [Google Scholar] [CrossRef] [PubMed]
- Hendrick, H.M.; Welter, B.H.; Hapstack, M.A.; Sykes, S.E.; Sullivan, W.J., Jr.; Temesvari, L.A. Phosphorylation of eukaryotic initiation factor-2alpha during stress and encystation in Entamoeba species. PLoS Pathog. 2016, 12, e1006085. [Google Scholar] [CrossRef] [PubMed]
- Doutheil, J.; Althausen, S.; Treiman, M.; Paschen, W. Effect of nitric oxide on endoplasmic reticulum calcium homeostasis, protein synthesis and energy metabolism. Cell Calcium 2000, 27, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Lopez, R.; Siminovich, B.; Alagon, A. Entamoeba histolytica codes for a protein homologue of the sec61 alpha subunit, a component of the endoplasmic reticulum translocon. Arch. Med. Res. 2000, 31, S168–S170. [Google Scholar] [CrossRef]
- Sanchez, R.; Saralegui, A.; Olivos-Garcia, A.; Scapolla, C.; Damonte, G.; Sanchez-Lopez, R.; Alagon, A.; Stock, R.P. Entamoeba histolytica: Intracellular distribution of the sec61alpha subunit of the secretory pathway and down-regulation by antisense peptide nucleic acids. Exp. Parasitol. 2005, 109, 241–251. [Google Scholar] [CrossRef] [PubMed]
- Biller, L.; Matthiesen, J.; Kuhne, V.; Lotter, H.; Handal, G.; Nozaki, T.; Saito-Nakano, Y.; Schumann, M.; Roeder, T.; Tannich, E.; et al. The cell surface proteome of Entamoeba histolytica. Mol. Cell. Proteom. 2014, 13, 132–144. [Google Scholar] [CrossRef] [PubMed]
- Schlenstedt, G.; Gudmundsson, G.H.; Boman, H.G.; Zimmermann, R. A large presecretory protein translocates both cotranslationally, using signal recognition particle and ribosome, and post-translationally, without these ribonucleoparticles, when synthesized in the presence of mammalian microsomes. J. Biol. Chem. 1990, 265, 13960–13968. [Google Scholar] [PubMed]
- Field, J.; Van Dellen, K.; Ghosh, S.K.; Samuelson, J. Responses of Entamoeba invadens to heat shock and encystation are related. J. Eukaryot. Microbiol. 2000, 47, 511–514. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Sharma, S.; Bhattacharya, A.; Tatu, U. Heat shock protein 90 regulates encystation in Entamoeba. Front. Microbiol. 2015, 6, 1125. [Google Scholar] [CrossRef] [PubMed]
- Manning-Cela, R.; Marquez, C.; Franco, E.; Talamas-Rohana, P.; Meza, I. BFA-sensitive and insensitive exocytic pathways in Entamoeba histolytica trophozoites: Their relationship to pathogenesis. Cell. Microbiol. 2003, 5, 921–932. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.S.; Zimmermann, E.M.; Chuang, B.M.; Song, B.; Nwokoye, A.; Wilkinson, J.E.; Eaton, K.A.; Kaufman, R.J. The unfolded protein response and chemical chaperones reduce protein misfolding and colitis in mice. Gastroenterology 2013, 144, 989–1000. [Google Scholar] [CrossRef] [PubMed]
- Bogaert, S.; De Vos, M.; Olievier, K.; Peeters, H.; Elewaut, D.; Lambrecht, B.; Pouliot, P.; Laukens, D. Involvement of endoplasmic reticulum stress in inflammatory bowel disease: A different implication for colonic and ileal disease? PLoS ONE 2011, 6, e25589. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pineda, E.; Perdomo, D. Entamoeba histolytica under Oxidative Stress: What Countermeasure Mechanisms Are in Place? Cells 2017, 6, 44. https://doi.org/10.3390/cells6040044
Pineda E, Perdomo D. Entamoeba histolytica under Oxidative Stress: What Countermeasure Mechanisms Are in Place? Cells. 2017; 6(4):44. https://doi.org/10.3390/cells6040044
Chicago/Turabian StylePineda, Erika, and Doranda Perdomo. 2017. "Entamoeba histolytica under Oxidative Stress: What Countermeasure Mechanisms Are in Place?" Cells 6, no. 4: 44. https://doi.org/10.3390/cells6040044