MDM2 Overexpression Modulates the Angiogenesis-Related Gene Expression Profile of Prostate Cancer Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture and Nutlin-3 Treatment
2.2. Silencing of MDM2 in LNCaP-MSI Cells
2.3. RNA Extraction
2.4. Human Angiogenesis RT2 Profiler PCR Array
2.5. Quantitative Reverse Transcription Polymerase Chain Reaction
2.6. Reverse Transcription Polymerase Chain Reaction (RT-PCR) Analysis of Gene Expression
2.7. Protein Preparation and Western Blot Analysis
2.8. Cell Migration Assay
2.9. MMP Activity Assay
2.10. Enzyme-Linked Immunosorbent Assay (ELISA) for MMP9
2.11. Statistical Analysis
3. Results
3.1. Identification of Differentially Expressed Genes in LNCaP-MST Using Human Angiogenesis RT2 Profiler PCR Array
3.2. Gene Expression in LNCaP-MST Using qRT-PCR
3.3. Determination of Gene Expression in LNCaP-MST Using RT-PCR
3.4. Levels of Protein Expression in LNCaP, LNCaP-MST, LNCaP-MSI and Expression Modulated by Nutlin-3 Treatment
3.5. Role of MDM2 on HUVECs Migration and Anti-Migratory Effects of Nutlin-3 Treatment
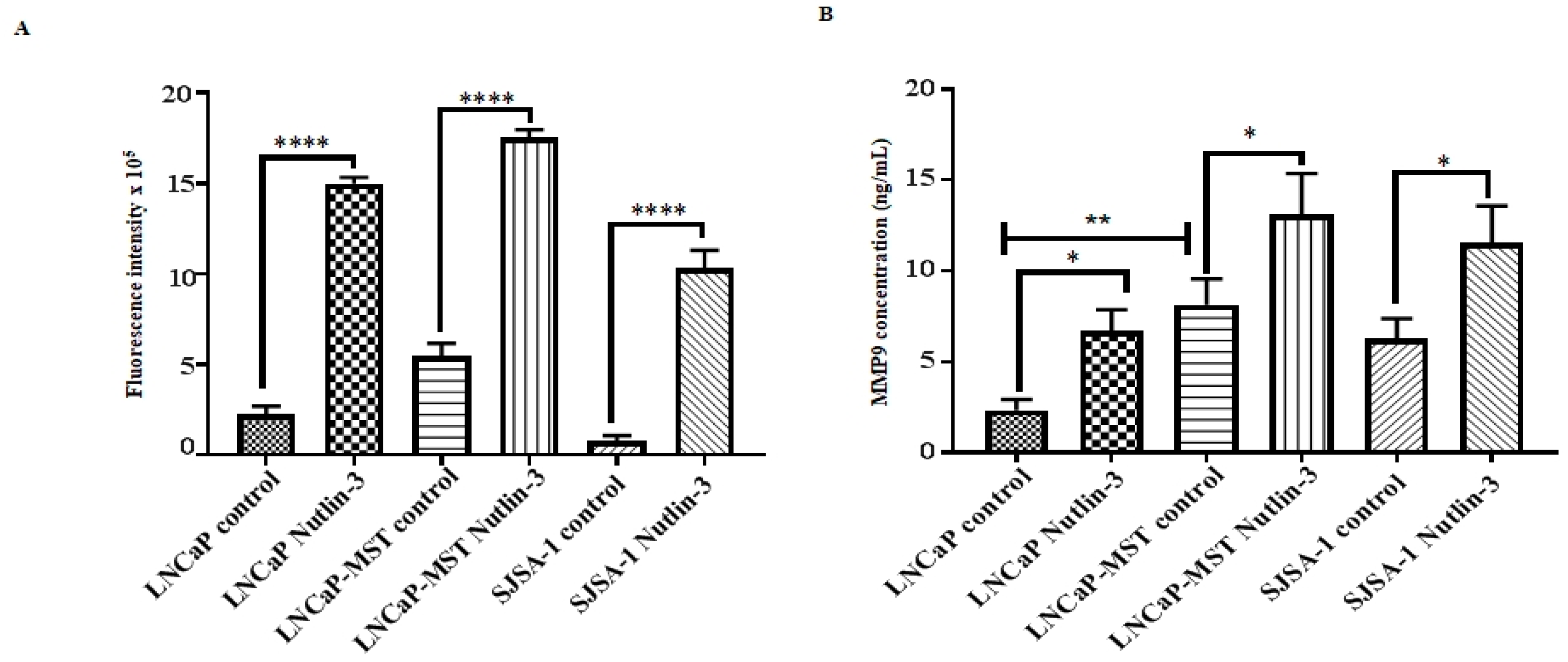
3.6. Impact of MDM2 and Nutlin-3 Treatment on MMP Activity
3.7. Effect of MDM2 and Nutlin-3 Treatment on MMP9 Expression
4. Discussion
5. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
References
- McCormick, F. Cancer therapy based on p53. Cancer J. 1999, 5, 139–144. [Google Scholar]
- Khor, L.Y.; Desilvio, M.; Al-Saleem, T.; Hammond, M.E.; Grignon, D.J.; Sause, W.; Pilepich, M.; Okunieff, P.; Sandler, H.; Pollack, A. MDM2 as a predictor of prostate carcinoma outcome: An analysis of Radiation Therapy Oncology Group Protocol 8610. Cancer 2005, 104, 962–967. [Google Scholar] [CrossRef] [PubMed]
- Marine, J.C.; Jochemsen, A.G. Mdmx as an essential regulator of p53 activity. Biochem. Biophys. Res. Commun. 2005, 331, 750–760. [Google Scholar] [CrossRef] [PubMed]
- Orit Karni, S.; Maria, L.; Carol, P. The Roles of MDM2 and MDMX in Cancer. Annu. Rev. Pathol. Mech. Dis. 2016, 11, 617–644. [Google Scholar] [CrossRef] [PubMed]
- Stad, R.; Little, N.A.; Xirodimas, D.P.; Frenk, R.; Vander, A.J.; Lane, D.P.; Saville, M.K.; Jochemsen, A.G. Mdmx stabilizes p53 and Mdm2 via two distinct mechanisms. EMBO Rep. 2001, 2, 1029–1034. [Google Scholar] [CrossRef] [PubMed]
- Sharp, D.A.; Kratowicz, S.A.; Sank, M.J.; George, D.L. Stabilization of the MDM2 oncoprotein by interaction with the structurally related MDMX protein. J. Biol. Chem. 1999, 274, 38189–38196. [Google Scholar] [CrossRef] [PubMed]
- Vassilev, L.T. MDM2 Inhibitors for Cancer Therapy. Trends Mol. Med. 2007, 13, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Rousseau, R.F.; Middleton, S.A.; Nichols, G.L.; Newell, R.; Lunec, J.; Tweddle, D.A. Pre-Clinical Evaluation of the MDM2-p53 Antagonist RG7388 Alone and in Combination with Chemotherapy in Neuroblastoma. Oncotarget 2015, 6, 10207–10221. [Google Scholar] [CrossRef] [PubMed]
- Manfredi, J.J. The Mdm2-p53 Relationship Evolves: Mdm2 Swings Both Ways as an Oncogene and a Tumor Suppressor. Gene Dev. 2010, 24, 1580–1589. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zeng, S.X.; Lu, H. Targeting p53-MDM2-MDMX Loop for Cancer Therapy. Subcell. Biochem. 2014, 85, 281–319. [Google Scholar] [CrossRef] [PubMed]
- Rathinavelu, P.; Malave, A.; Raney, S.R.; Hurst, J.; Roberson, C.T.; Rathinavelu, A. Expression of mdm-2 oncoprotein in the primary and metastatic sites of mammary tumor (GI-101) implanted athymic nude mice. Cancer Biochem. Biophys. 1999, 17, 133–146. [Google Scholar] [PubMed]
- Madhusudhanan, N.; Rajiv, R.; Muthusamy, K.; Rathinavelu, A. Detection of HDM2 and VEGF co-expression in cancer cell lines: Novel effect of HDM2 antisense treatment on VEGF expression. Life Sci. 2007, 81, 1362–1372. [Google Scholar] [CrossRef]
- Madhusudhanan, N.; Rajiv, R.; Ramugounder, R.; Jason, A.Z.; Rathinavelu, A. Identification of HDM2 as a regulator of VEGF expression in cancer cells. Life Sci. 2008, 82, 1231–1241. [Google Scholar] [CrossRef]
- Alaseem, A.; Venkatesan, T.; Alhazzani, K.; Rathinavelu, A. Analysis of the regulation of angiogenesis pathway by inhibiting MDM2 function in LNCaP-MST prostate cancer cells using PCR array. In Proceedings of the 106th Annual Meeting of the American Association for Cancer Research, Philadelphia, PA, USA, 18–22 April 2015; AACR: Philadelphia, PA, USA, 2015. [Google Scholar] [CrossRef]
- Chen, X.; Qiu, J.; Yang, D.; Lu, J.; Yan, C.; Zha, X.; Yin, Y. MDM2 promotes invasion and metastasis in invasive ductal breast carcinoma by inducing matrix metalloproteinase-9. PLoS ONE 2013, 8, e78794. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.D.; Meng, Z.Q.; Chen, Z.; Lin, J.H.; Zhou, Z.H.; Liu, L.M. Identification of liver metastasis-related genes in a novel human pancreatic carcinoma cell model by microarray analysis. Cancer Lett. 2009, 283, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Meng, Z.; Chen, Z.; Hua, Y.; Gao, H.; Wang, P.; Lin, J.; Zhou, Z.; Luo, J.; Liu, L. RNA interference against MDM2 suppresses tumor growth and metastasis in pancreatic carcinoma SW1990HM cells. Mol. Cell. Biochem. 2014, 387, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.H.; Zhang, L.Y.; Liu, D.J.; Yang, F.; Zhao, J.Z. Expression and significance of MMP-9 and MDM2 in the oncogenesis of lung cancer in rats. Asian Pac. J. Trop. Med. 2014, 7, 585–588. [Google Scholar] [CrossRef]
- Ling, W.; Yang, J.; Yke, J.A.; Ida, J.; Su, Q.; Gunhild, M.M.; Alexandr, K.; Bjorn, R.; Hokon, W.; Havard, E.W.; et al. STAMP1 Is Bota a Proliferative and Antiapoptotic Factor in Prostate Cancer. Cancer Res. 2010, 70, 5818–5828. [Google Scholar] [CrossRef]
- Datta, M.W.; Macri, E.; Signoretti, S.; Renshaw, A.A.; Loda, M. Transition from in situ to invasive testicular germ cell neoplasia is associated with the loss of p21 and gain of MDM2 expression. Mod. Pathol. 2001, 14, 437–442. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.D.; Chen, S.H.; Lin, C.L.; Tsai, S.H.; Liang, T.C. Inhibition of melanoma growth and metastasis by combination with epigallocatechin-3-gallate and dacarbazine in mice. J. Cell. Biochem. 2001, 83, 631–642. [Google Scholar] [CrossRef] [PubMed]
- Sanceau, J.; Truchet, S.; Bauvois, B. Matrix metalloproteinase 9 silencing by RNA interference triggers the migration-adhesive switch in Ewing sarcoma cells. J. Biochem. 2003, 278, 36537–36546. [Google Scholar] [CrossRef]
- Bergers, G.; Brekken, R.; McMahon, G.; Vu, T.H.; Itoh, T.; Tamaki, K.; Tanzawa, K.; Thorpe, P.; Itohara, S.; Werb, Z.; et al. Matrix metalloproteinase 9 triggers the angiogenic switch during carcinogenesis. Nat. Cell Biol. 2000, 2, 737–744. [Google Scholar] [CrossRef] [PubMed]
- Benezra, D. Angiogenesis-Critical Assessment of In vitro Assays and In vivo Models. In Angiogenesis: Models, Modulators, and Clinical Applications; Plenum Press: New York, NY, USA, 1998; Volume 298, pp. 85–89. [Google Scholar] [CrossRef]
- Zhang, J.; Ito, R.; Oue, N.; Zhu, X.; Kitadai, Y.; Yoshida, K.; Nakayama, H.; Yasui, W. Expression of thrombospondin 1 is correlated with microvessel density in gastric carcinoma. Virchows Arch. 2003, 442, 563–568. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.N.; Albo, D.; Tuszynski, G.P. Fibroblasts promote breast cancer cell invasion by upregulating tumor matrix metalloproteinase 9 production. Surgery 2002, 132, 220–225. [Google Scholar] [CrossRef] [PubMed]
- Ren, B.; Yee, K.O.; Lawler, J.; Khosravi, R.F. Regulation of tumor angiogenesis by thrombospondin 1. Biochim. Biophys. Acta 2006, 1765, 178–188. [Google Scholar] [CrossRef] [PubMed]
- Westbrook, C.A.; Gasson, J.C.; Gerber, S.E.; Selsted, M.E.; Golde, D.W. Purification and characterization of human T-Lymphocyte-derived erythroid-potentiating activity. J. Biol. Chem. 1984, 259, 9992–9996. [Google Scholar] [PubMed]
- Brew, K.; Nagase, H. The tissue inhibitors of metalloproteinases (TIMPs): An ancient family with structural and functional diversity. Biochim. Biophys. Acta 2010, 1803, 55–71. [Google Scholar] [CrossRef] [PubMed]
- Henriet, P.; Blavier, L.; Declerck, Y.A. Tissue inhibitors of metalloproteinases (TIMP) in invasion and proliferation. APMIS 1999, 107, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Aggarwal, B.B. Signalling pathways of the TNF superfamily: A double-edged sword. Nat. Rev. Immunol. 2003, 3, 745–756. [Google Scholar] [CrossRef] [PubMed]
- Sethi, G.; Sung, B.; Aggarwal, B.B. TNF: A master switch for inflammation to cancer. Front. Biosci. 2008, 13, 5094–5107. [Google Scholar] [CrossRef] [PubMed]
- Paola, R.; Laura, L.; Francesco, A.; Mario, S.; Sergio, R. CXC chemokines: The regulatory link between inflammation and angiogenesis. Trends Immunol. 2004, 25, 201–209. [Google Scholar] [CrossRef]
- Worrall, C.; Suleymanova, N.; Crudden, C.; Trocoli Drakensjö, C.; Candrea, E.; Nedelcu, D.; Takahashi, S.I.; Girnita, L.; Girnita, A. Unbalancing p53/Mdm2/IGF-1R axis by Mdm2 activation restrains the IGF-1-dependent invasive phenotype of skin melanoma. Oncogene 2017, 36, 3274–3286. [Google Scholar] [CrossRef] [PubMed]
- Lowe, J.M.; Menendez, D.; Bushel, P.R.; Shatz, M.; Kirk, E.L.; Troester, M.A.; Garantziotis, S.; Fessler, M.B.; Resnick, M.A. p53 and NF-κB coregulate proinflammatory gene responses in human macrophages. Cancer Res. 2014, 74, 2182–2192. [Google Scholar] [CrossRef] [PubMed]
- Vandooren, J.; Van Den Steen, P.E.; Opdenakker, G. Biochemistry and molecular biology of gelatinase B or matrix metalloproteinase-9 (MMP-9): The next decade. Crit. Rev. Biochem. Mol. 2013, 48, 222–272. [Google Scholar] [CrossRef] [PubMed]
- Toyoda, H.; Komurasaki, T.; Uchida, D.; Takayama, Y.; Isobe, T.; Okuyama, T.; Hanada, K. Epiregulin: A novel epidermal growth factor with mitogenic activity for rat primary hepatocytes. J. Biol. Chem. 1995, 270, 7495–7500. [Google Scholar] [CrossRef] [PubMed]
- Toyoda, H.; Komurasaki, T.; Uchida, D.; Morimoto, S. Distribution of mRNA for human epiregulin, a differentially expressed member of the epidermal growth factor family. Biochem. J. 1997, 326, 69–75. [Google Scholar] [CrossRef] [PubMed]
- David, J.R.; Richard, L.C. Epiregulin: Roles in normal physiology and cancer. Semin. Cell Dev. Biol. 2014, 28, 49–56. [Google Scholar] [CrossRef]
- Pellikainen, J.; Kataja, V.; Ropponen, K.; Kellokoski, J.; Pietilainen, T.; Bohm, J.; Eskelinen, M.; Kosma, V.M. Reduced nuclear expression of transcription factor AP-2 associates with aggressive breast cancer. Clin. Cancer Res. 2002, 8, 3487–3495. [Google Scholar] [PubMed]
- Francesca, O.; Elisa, P.; Daniela, C.; Elena, A.; Federica, M.; Donatella, V.; Enrico, G.; Guido, S.; Piero, S.; Michele, D.B.; et al. AP-2α and AP-2γ regulate tumor progression via specific genetic programs. FASEB J. 2008, 22, 2702–2714. [Google Scholar] [CrossRef]






| Transcript | Forward and Reverse Primers |
|---|---|
| qRT-PCR Primers | |
| THBS1 | Forward: 5′-AGCGTCTTCACCAGAGACCT-3′ |
| Reverse: 5′-CATTCACCACGTTGTTGTCA-3′ | |
| TIMP1 | Forward: 5′-TACTTCCACAGGTCCCACAA-3′ |
| Reverse: 5′-ATTCCTCACAGCCAACAGTG-3′ | |
| CXCL3 | Forward: 5′-CCACACTCAAGAATGGGAAG-3′ |
| Reverse: 5′-CTGTCCCTAGAAAGCTGCTG-3′ | |
| MDM2 | Forward: 5′-CACCTCACAGATTCCAGCTT-3′ |
| Reverse: 5′-CGCCAAACAAATCTCCTAGA-3′ | |
| β-actin | Forward: 5′-GGACTTCGAGCAAGAGATGG-3′ |
| Reverse: 5′-AGCACTGTGTTGGCGTACAG-3′ | |
| RT-PCR Primers | |
| THBS1 | Forward: 5′-GACTAGGCGTCCTGTTCCTG-3′ |
| Reverse: 5′-ACCTGGCCAGAGTGGTCTTT-3′ | |
| TIMP1 | Forward: 5′-GGACACCAGAAGTCAACCAGACC-3′ |
| Reverse: 5′-CGTCCACAAGCAATGAGTGCC-3′ | |
| CXCL3 | Forward: 5′-GCAGGAGCGTCCGTGGTCAC-3′ |
| Reverse: 5′-GCTCTGGTAAGGGCAGGGACC-3′ | |
| MDM2 | Forward: 5′-CTGGGGAGTCTTGAGGGACC-3′ |
| Reverse: 5′-CAGGTTGTCTAAATTCCTAG-3′ | |
| β-actin | Forward: 5′-GTGGGGCGCCCCAGGCACCA-3′ |
| Reverse: 5′-CTCCTTAATGTCACGCACGATTTC-3′ | |
| Primers Used for Both qRT-PCR and RT-PCR | |
| TNF-α | Forward: 5′-TCCTTCAGACACCCTCAACC-3′ |
| Reverse: 5′-AGGCCCCAGTTTGAATTCTT-3′ | |
| MMP9 | Forward: 5′-CTCTGGAGGTTCGACGTG-3′ |
| Reverse: 5′-GTCCACCTGGTTCAACTCAC-3′ | |
| CXCL10 | Forward: 5′-GCTTAGACATATTCTGAGCCTAC-3′ |
| Reverse: 5′-AGCTGATTTGGTGACCATCATTG-3′ | |
| EREG | Forward: 5′- TCCATCTTCTACAGGCAGTCC-3′ |
| Reverse: 5′-CACGGTCAAAGCCACATACTC-3′ | |
| Gene | Accession Number | Description | Fold Change |
|---|---|---|---|
| Up-Regulated Genes | |||
| THBS1 | NM_003246 | Thrombospondin 1 | 155.3 |
| CXCL10 | NM_001565 | Chemokine (C-X-C motif) ligand 10 | 41.5 |
| TNF | NM_000594 | Tumor necrosis factor | 40.6 |
| MMP9 | NM_004994 | Matrix metallopeptidase 9 (gelatinase B, 92 kDa gelatinase, 92 kDa type IV collagenase) | 11.5 |
| Down-Regulated Genes | |||
| EREG | NM_001432 | Epiregulin | 0.0001 |
| TIMP1 | NM_003254 | TIMP metallopeptidase inhibitor 1 | 0.0007 |
| CXCL3 | NM_002090 | Chemokine (C-X-C motif) ligand 3 | 0.004 |
| CXCL1 | NM_001511 | Chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) | 0.009 |
| CXCL6 | NM_002993 | Chemokine (C-X-C motif) ligand 6 (granulocyte chemotactic protein 2) | 0.24 |
| Gene | Accession Number | Description | Fold Change |
|---|---|---|---|
| Down-Regulated Gene | |||
| THBS1 | NM_003246 | Thrombospondin 1 | 0.26 |
| Gene | ΔCt: LNCaP | ΔCt: LNCaP-MST | Fold Change |
|---|---|---|---|
| Up-Regulated Genes | |||
| THBS1 | 12.9 | 5.9 | 125.8 |
| TNF-α | 10.2 | 6.4 | 14.3 |
| MDM2 | 5.1 | 1.8 | 10.3 |
| CXCL10 | 22.6 | 20.2 | 5.2 |
| MMP9 | 12 | 11.5 | 1.4 |
| Down-Regulated Genes | |||
| EREG | 3.5 | 13.4 | 0.001 |
| TIMP1 | 2.9 | 10.3 | 0.006 |
| CXCL3 | 5.8 | 13.4 | 0.005 |
| Gene | ΔCt: LNCaP-MST | ΔCt: LNCaP-MST | Fold Change |
|---|---|---|---|
| Control | Nutlin-3 Treated | ||
| Up-Regulated Genes | |||
| EREG | 16.7 | 15.2 | 2.8 |
| CXCL3 | 15.9 | 14.2 | 3.4 |
| Down-Regulated Genes | |||
| THBS1 | 5 | 7.4 | 0.18 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Venkatesan, T.; Alaseem, A.; Chinnaiyan, A.; Dhandayuthapani, S.; Kanagasabai, T.; Alhazzani, K.; Dondapati, P.; Alobid, S.; Natarajan, U.; Schwartz, R.; et al. MDM2 Overexpression Modulates the Angiogenesis-Related Gene Expression Profile of Prostate Cancer Cells. Cells 2018, 7, 41. https://doi.org/10.3390/cells7050041
Venkatesan T, Alaseem A, Chinnaiyan A, Dhandayuthapani S, Kanagasabai T, Alhazzani K, Dondapati P, Alobid S, Natarajan U, Schwartz R, et al. MDM2 Overexpression Modulates the Angiogenesis-Related Gene Expression Profile of Prostate Cancer Cells. Cells. 2018; 7(5):41. https://doi.org/10.3390/cells7050041
Chicago/Turabian StyleVenkatesan, Thiagarajan, Ali Alaseem, Aiyavu Chinnaiyan, Sivanesan Dhandayuthapani, Thanigaivelan Kanagasabai, Khalid Alhazzani, Priya Dondapati, Saad Alobid, Umamaheswari Natarajan, Ruben Schwartz, and et al. 2018. "MDM2 Overexpression Modulates the Angiogenesis-Related Gene Expression Profile of Prostate Cancer Cells" Cells 7, no. 5: 41. https://doi.org/10.3390/cells7050041





