Hints of tRNA-Derived Small RNAs Role in RNA Silencing Mechanisms
Abstract
:1. Introduction
2. Where They Live and What They Do
| Species | Cleavage specificity | Nuclease | Function | RNAi relation | Name |
|---|---|---|---|---|---|
| E. coli [1,15,16] | tRNA Tyr, His, Asn, Asp, Lys, Arg | Prrc, ColicinE5 and ColicinD | Phage T4 infection | N/D | N/D |
| T. thermophila [9] | All tRNAs | N/D | Reduction of uncharged tRNA. Cell cycle progression Essential amino acids starvation | N/D | N/D |
| S. coelicor [10] | All tRNAs | Cell differentiation | N/D | N/D | |
| A. fumigatus [11] | All tRNAs | N/D | Conidiogenesis Down-regulation of protein synthesis | N/D | tRNA halves |
| S. cerevisiae [2,13] | All tRNAs | Rny1 | Oxidative stress | N/D | N/D |
| G. Lamblia [6] | All tRNAs | N/D | Encystation and adverse environments Down-regulation of protein synthesis | N/D | tRNA halvessitRNAs |
| Mammalian cells [5,8,13,17,18,19] | All tRNAs | ELAC2 DICER Angiogenin tRNaseZ Other N/D | Some stress conditions (oxidative, heat shock, UV hyperosmolarity) Down-regulation of protein synthesis | Associate to Ago2, 3 and 4. Possible miRNA/siRNA function. | tRNA halves 5' tRFs 3' CCA tRFs 3' U tRFs |
| T. cruzi [14,20] | tRNA Glu, Asp, Tyr, Val, Arg, His, Thr | N/D | Nutritional stress | N/D | tRNA halves |
| Drosophila [21] | All tRNAs | PIWI | Some stress conditions | Associated to PIWI | N/D |
| A. Thaliana [13] | Al tRNAs | N/D | Nutritional and oxidative stress conditions Down-regulation of protein synthesis | ND | N/D |
| C. maxima [22] | All tRNAs | N/D | Translational Inhibition | N/D | N/D |
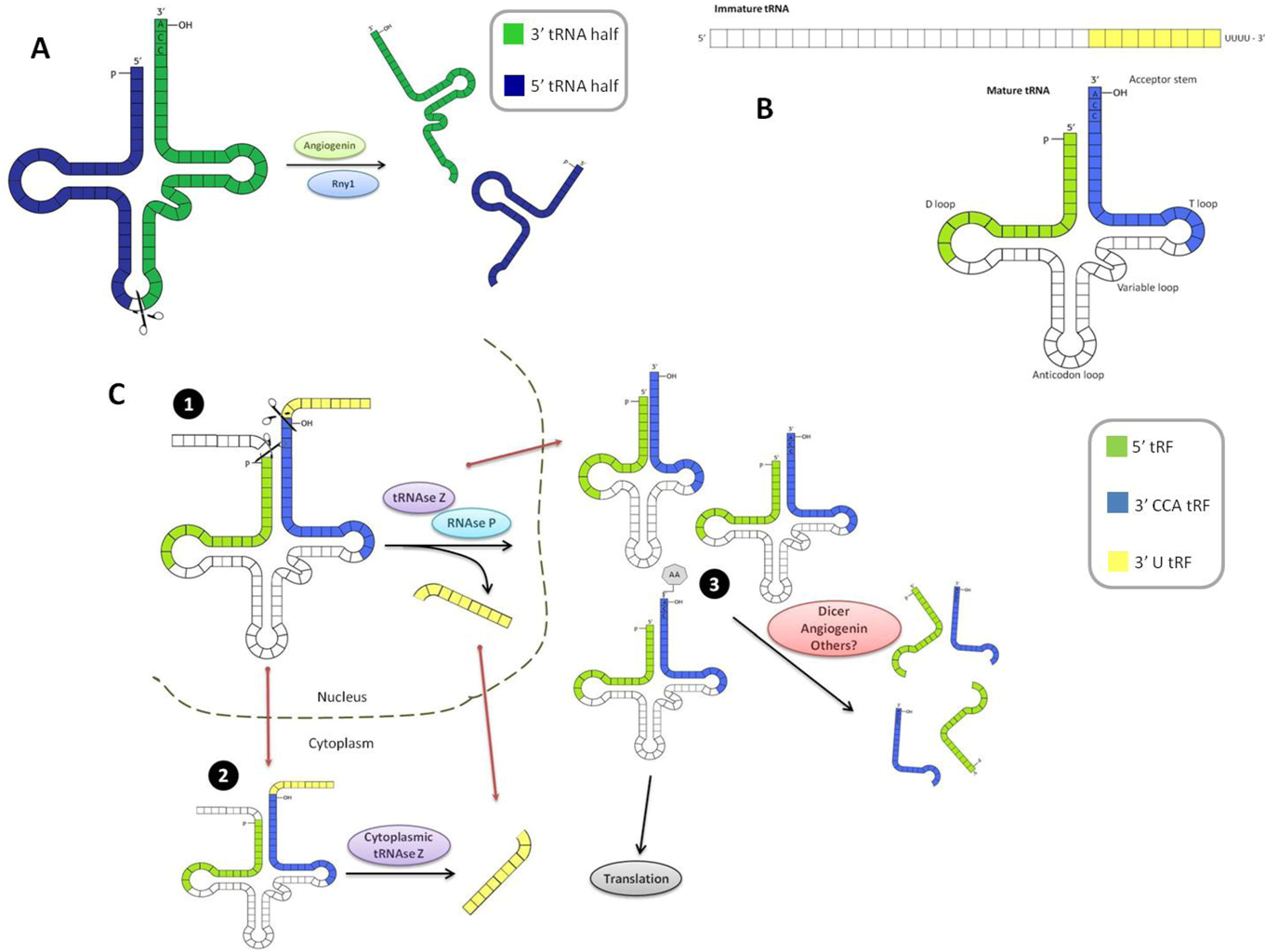
3. Classification and Biogenesis
3.1. tRNA Halves
3.2. Small tRFs
3.3. Biogenesis of Small tRFs
4. Evidence for tRFs as Small Interfering RNAs
5. Conclusions
References and Notes
- Ogawa, T.; Tomita, K.; Ueda, T.; Watanabe, K.; Uozumi, T.; Masaki, H. A cytotoxic ribonuclease targeting specific transfer rna anticodons. Science 1999, 283, 2097–2100. [Google Scholar]
- Thompson, D.M.; Parker, R. Stressing out over trna cleavage. Cell 2009, 138, 215–219. [Google Scholar] [CrossRef]
- Fu, H.; Feng, J.; Liu, Q.; Sun, F.; Tie, Y.; Zhu, J.; Xing, R.; Sun, Z.; Zheng, X. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett. 2009, 583, 437–442. [Google Scholar] [CrossRef]
- Lee, Y.S.; Shibata, Y.; Malhotra, A.; Dutta, A. A novel class of small rnas: tRNA-derived RNA fragments (tRFs). Genes Dev. 2009, 23, 2639–2649. [Google Scholar] [CrossRef]
- Haussecker, D.; Huang, Y.; Lau, A.; Parameswaran, P.; Fire, A.Z.; Kay, M.A. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA 2010, 16, 673–695. [Google Scholar] [CrossRef]
- Li, Y.; Luo, J.; Zhou, H.; Liao, J.Y.; Ma, L.M.; Chen, Y.Q.; Qu, L.H. Stress-induced tRNA-derived rRNA: A novel class of small rRNA in the primitive eukaryote giardia lamblia. Nucleic Acids Res. 2008, 36, 6048–6055. [Google Scholar] [CrossRef]
- Elbarbary, R.A.; Takaku, H.; Uchiumi, N.; Tamiya, H.; Abe, M.; Takahashi, M.; Nishida, H.; Nashimoto, M. Modulation of gene expression by human cytosolic trnase z(l) through 5'-half-tRNA. PLoS One 2009, 4, e5908. [Google Scholar]
- Yamasaki, S.; Ivanov, P.; Hu, G.F.; Anderson, P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J. Cell Biol. 2009, 185, 35–42. [Google Scholar] [CrossRef]
- Lee, S.R.; Collins, K. Starvation-induced cleavage of the tRNA anticodon loop in tetrahymena thermophila. J. Biol. Chem. 2005, 280, 42744–42749. [Google Scholar]
- Haiser, H.J.; Karginov, F.V.; Hannon, G.J.; Elliot, M.A. Developmentally regulated cleavage of trnas in the bacterium streptomyces coelicolor. Nucleic Acids Res. 2008, 36, 732–741. [Google Scholar]
- Jochl, C.; Rederstorff, M.; Hertel, J.; Stadler, P.F.; Hofacker, I.L.; Schrettl, M.; Haas, H.; Huttenhofer, A. Small ncRNA transcriptome analysis from aspergillus fumigatus suggests a novel mechanism for regulation of protein synthesis. Nucleic Acids Res. 2008, 36, 2677–2689. [Google Scholar] [CrossRef]
- Kawaji, H.; Nakamura, M.; Takahashi, Y.; Sandelin, A.; Katayama, S.; Fukuda, S.; Daub, C.O.; Kai, C.; Kawai, J.; Yasuda, J.; et al. Hidden layers of human small RNAs. BMC Genomics 2008, 9, 157. [Google Scholar] [CrossRef]
- Thompson, D.M.; Lu, C.; Green, P.J.; Parker, R. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA 2008, 14, 2095–2103. [Google Scholar] [CrossRef]
- Garcia-Silva, M.R.; Frugier, M.; Tosar, J.P.; Correa-Dominguez, A.; Ronalte-Alves, L.; Parodi-Talice, A.; Rovira, C.; Robello, C.; Goldenberg, S.; Cayota, A. A population of tRNA-derived small RNAs is actively produced in trypanosoma cruzi and recruited to specific cytoplasmic granules. Mol. Biochem. Parasitol. 2011, 171, 64–73. [Google Scholar]
- Levitz, R.; Chapman, D.; Amitsur, M.; Green, R.; Snyder, L.; Kaufmann, G. The optional E. Coli prr locus encodes a latent form of phage t4-induced anticodon nuclease. EMBO J. 1990, 9, 1383–1389. [Google Scholar]
- Tomita, K.; Ogawa, T.; Uozumi, T.; Watanabe, K.; Masaki, H. A cytotoxic ribonuclease which specifically cleaves four isoaccepting arginine trnas at their anticodon loops. Proc. Natl. Acad. Sci. USA 2000, 97, 8278–8283. [Google Scholar]
- Emara, M.M.; Ivanov, P.; Hickman, T.; Dawra, N.; Tisdale, S.; Kedersha, N.; Hu, G.F.; Anderson, P. Angiogenin-induced tRNA-derived stress-induced rnas promote stress-induced stress granule assembly. J. Biol. Chem. 2010, 285, 10959–10968. [Google Scholar]
- Ivanov, P.; Emara, M.M.; Villen, J.; Gygi, S.P.; Anderson, P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol. Cell 2011, 43, 613–623. [Google Scholar] [CrossRef]
- Lee, Y.S.; Pressman, S.; Andress, A.P.; Kim, K.; White, J.L.; Cassidy, J.J.; Li, X.; Lubell, K.; Lim do, H.; Cho, I.S.; et al. Silencing by small rnas is linked to endosomal trafficking. Nat. Cell Biol. 2009, 11, 1150–1156. [Google Scholar] [CrossRef]
- Franzen, O.; Arner, E.; Ferella, M.; Nilsson, D.; Respuela, P.; Carninci, P.; Hayashizaki, Y.; Aslund, L.; Andersson, B.; Daub, C.O. The short non-coding transcriptome of the protozoan parasite trypanosoma cruzi. PLoS Negl. Trop. Dis. 2011, 5, e1283. [Google Scholar] [CrossRef]
- Schaefer, M.; Pollex, T.; Hanna, K.; Tuorto, F.; Meusburger, M.; Helm, M.; Lyko, F. RNA methylation by dnmt2 protects transfer rnas against stress-induced cleavage. Genes Dev. 2010, 24, 1590–1595. [Google Scholar] [CrossRef]
- Zhang, S.; Sun, L.; Kragler, F. The phloem-delivered RNA pool contains small noncoding RNAs and interferes with translation. Plant Physiol. 2009, 150, 378–387. [Google Scholar] [CrossRef]
- Garcia Silva, M.R.; Tosar, J.P.; Frugier, M.; Pantano, S.; Bonilla, B.; Esteban, L.; Serra, E.; Rovira, C.; Robello, C.; Cayota, A. Cloning, characterization and subcellular localization of a trypanosoma cruzi argonaute protein defining a new subfamily distinctive of trypanosomatids. Gene 2010, 466, 26–35. [Google Scholar] [CrossRef]
- Brennecke, J.; Aravin, A.A.; Stark, A.; Dus, M.; Kellis, M.; Sachidanandam, R.; Hannon, G.J. Discrete small RNA-generating loci as master regulators of transposon activity in drosophila. Cell 2007, 128, 1089–1103. [Google Scholar] [CrossRef]
- Babiarz, J.E.; Ruby, J.G.; Wang, Y.; Bartel, D.P.; Blelloch, R. Mouse es cells express endogenous shrnas, siRNAs, and other microprocessor-independent, dicer-dependent small rnas. Genes Dev. 2008, 22, 2773–2785. [Google Scholar] [CrossRef]
- Cole, C.; Sobala, A.; Lu, C.; Thatcher, S.R.; Bowman, A.; Brown, J.W.; Green, P.J.; Barton, G.J.; Hutvagner, G. Filtering of deep sequencing data reveals the existence of abundant dicer-dependent small RNAs derived from tRNAs. RNA 2009, 15, 2147–2160. [Google Scholar] [CrossRef]
- Pederson, T. Regulatory RNAs derived from transfer RNA? RNA 2010, 16, 1865–1869. [Google Scholar] [CrossRef]
- Sobala, A.; Hutvagner, G. Transfer RNA-derived fragments: Origins, processing, and function. Wiley Interdiscip. Rev. 2011, 2, 853–862. [Google Scholar] [CrossRef]
- Liao, J.Y.; Ma, L.M.; Guo, Y.H.; Zhang, Y.C.; Zhou, H.; Shao, P.; Chen, Y.Q.; Qu, L.H. Deep sequencing of human nuclear and cytoplasmic small RNAs reveals an unexpectedly complex subcellular distribution of mirnas and tRNA 3' trailers. PLoS One 2010, 5, e10563. [Google Scholar]
- Li, Z.; Ender, C.; Meister, G.; Moore, P.S.; Chang, Y.; John, B. Extensive terminal and asymmetric processing of small RNAs from rrnas, snornas, snrnas, and trnas. Nucleic Acids Res. 2012, 40, 6787–6799. [Google Scholar]
- Jinek, M.; Doudna, J.A. A three-dimensional view of the molecular machinery of RNA interference. Nature 2009, 457, 405–412. [Google Scholar] [CrossRef]
- Miyoshi, K.; Miyoshi, T.; Siomi, H. Many ways to generate microrna-like small RNAs: Non-canonical pathways for microrna production. Mol. Genet. Genomics 2010, 284, 95–103. [Google Scholar] [CrossRef]
- Tuck, A.C.; Tollervey, D. RNA in pieces. Trends Genet. 2011, 27, 422–432. [Google Scholar] [CrossRef]
- Kim, V.N. Small RNAs: Classification, biogenesis, and function. Mol. Cells 2005, 19, 1–15. [Google Scholar] [CrossRef]
- Hutvagner, G.; Simard, M.J. Argonaute proteins: Key players in RNA silencing. Nat. Rev. Mol. Cell Biol. 2008, 9, 22–32. [Google Scholar] [CrossRef]
- Burroughs, A.M.; Ando, Y.; de Hoon, M.J.; Tomaru, Y.; Suzuki, H.; Hayashizaki, Y.; Daub, C.O. Deep-sequencing of human argonaute-associated small RNAs provides insight into miRNA sorting and reveals argonaute association with RNA fragments of diverse origin. RNA Biol. 2011, 8, 158–177. [Google Scholar] [CrossRef]
- Ender, C.; Krek, A.; Friedlander, M.R.; Beitzinger, M.; Weinmann, L.; Chen, W.; Pfeffer, S.; Rajewsky, N.; Meister, G. A human snorna with microRNA-like functions. Mol. Cell 2008, 32, 519–528. [Google Scholar] [CrossRef]
- Yeung, M.L.; Bennasser, Y.; Watashi, K.; Le, S.Y.; Houzet, L.; Jeang, K.T. Pyrosequencing of small non-coding RNAs in HIV-1 infected cells: Evidence for the processing of a viral-cellular double-stranded RNA hybrid. Nucleic Acids Res. 2009, 37, 6575–6586. [Google Scholar] [CrossRef]
- Tamura, M.; Nashimoto, C.; Miyake, N.; Daikuhara, Y.; Ochi, K.; Nashimoto, M. Intracellular mrna cleavage by 3' trnase under the direction of 2'-o-methyl RNA heptamers. Nucleic Acids Res. 2003, 31, 4354–4360. [Google Scholar] [CrossRef]
- Nakashima, A.; Takaku, H.; Shibata, H.S.; Negishi, Y.; Takagi, M.; Tamura, M.; Nashimoto, M. Gene silencing by the tRNA maturase trnase zl under the direction of small-guide RNA. Gene Ther. 2007, 14, 78–85. [Google Scholar] [CrossRef]
- Takahashi, M.; Elbarbary, R.A.; Abe, M.; Sato, M.; Yoshida, T.; Yamada, Y.; Tamura, M.; Nashimoto, M. Elimination of specific mirnas by naked 14-nt sgRNAs. PLoS One 2012, 7, e38496. [Google Scholar]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. Elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Garcia-Silva, M.R.; Cabrera-Cabrera, F.; Güida, M.C.; Cayota, A. Hints of tRNA-Derived Small RNAs Role in RNA Silencing Mechanisms. Genes 2012, 3, 603-614. https://doi.org/10.3390/genes3040603
Garcia-Silva MR, Cabrera-Cabrera F, Güida MC, Cayota A. Hints of tRNA-Derived Small RNAs Role in RNA Silencing Mechanisms. Genes. 2012; 3(4):603-614. https://doi.org/10.3390/genes3040603
Chicago/Turabian StyleGarcia-Silva, Maria Rosa, Florencia Cabrera-Cabrera, Maria Catalina Güida, and Alfonso Cayota. 2012. "Hints of tRNA-Derived Small RNAs Role in RNA Silencing Mechanisms" Genes 3, no. 4: 603-614. https://doi.org/10.3390/genes3040603




