Divergent Expression Patterns in Two Vernicia Species Revealed the Potential Role of the Hub Gene VmAP2/ERF036 in Resistance to Fusarium oxysporum in Vernicia montana
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification of AP2/ERF Superfamily in Vernicia Species
2.2. Multiple Sequence Alignment of AP2/ERF Superfamily
2.3. Phylogenetic Analysis of the AP2/ERF Superfamily
2.4. Motif Recognition of the ERF Family
2.5. Evolution and Selection of the AP2/ERF Superfamily in Vernicia Species
2.6. Plant Materials and Pathogen Inoculation
2.7. RNA Isolation and Expression Analysis
2.8. Quantitative Real-Time PCR Assay
2.9. Transcriptome Analysis and Expression Patterns of the ERF Family during Infection
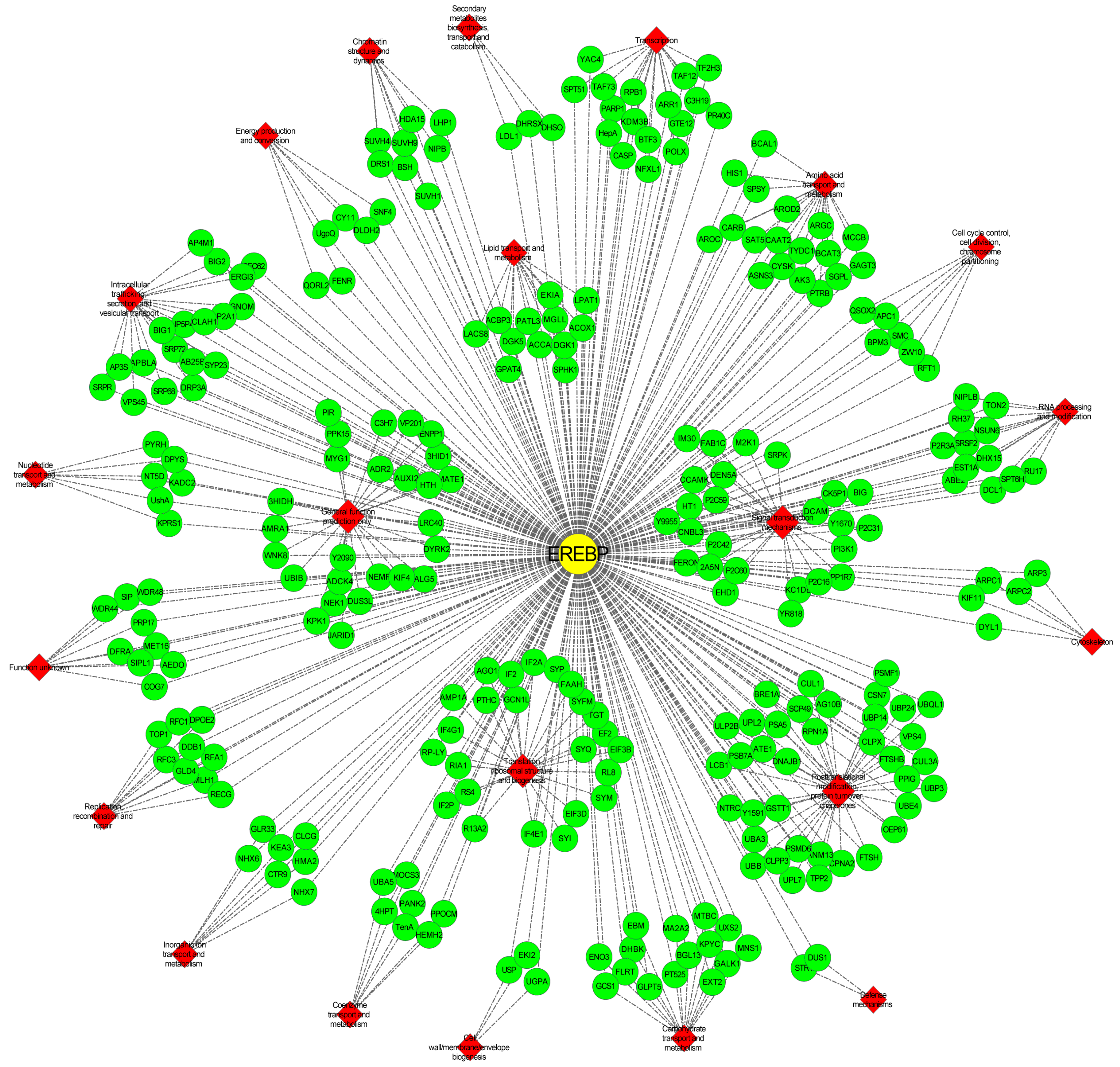
2.10. Network Analysis of Vf/VmAP2/ERF Superfamily Genes in Response to F. oxysporum
3. Results
3.1. Detection of AP2/ERF Transcription Factors in Two Vernicia Species
3.2. Structure and Group-Specific Residues of the AP2 Domains of Vf/VmERF Genes
3.3. Phylogenetic Relationships between AP2/ERF Superfamily Transcription Factors
3.4.Distribution of Conserved Motifs outside of the AP2/ERF Domain
3.5. Evolution and Selection of the Vf/VmAP2/ERF Superfamily in Vernicia Species
3.6. Expression Patterns of Vf/VmERF Genes in V. fordii and V. montana
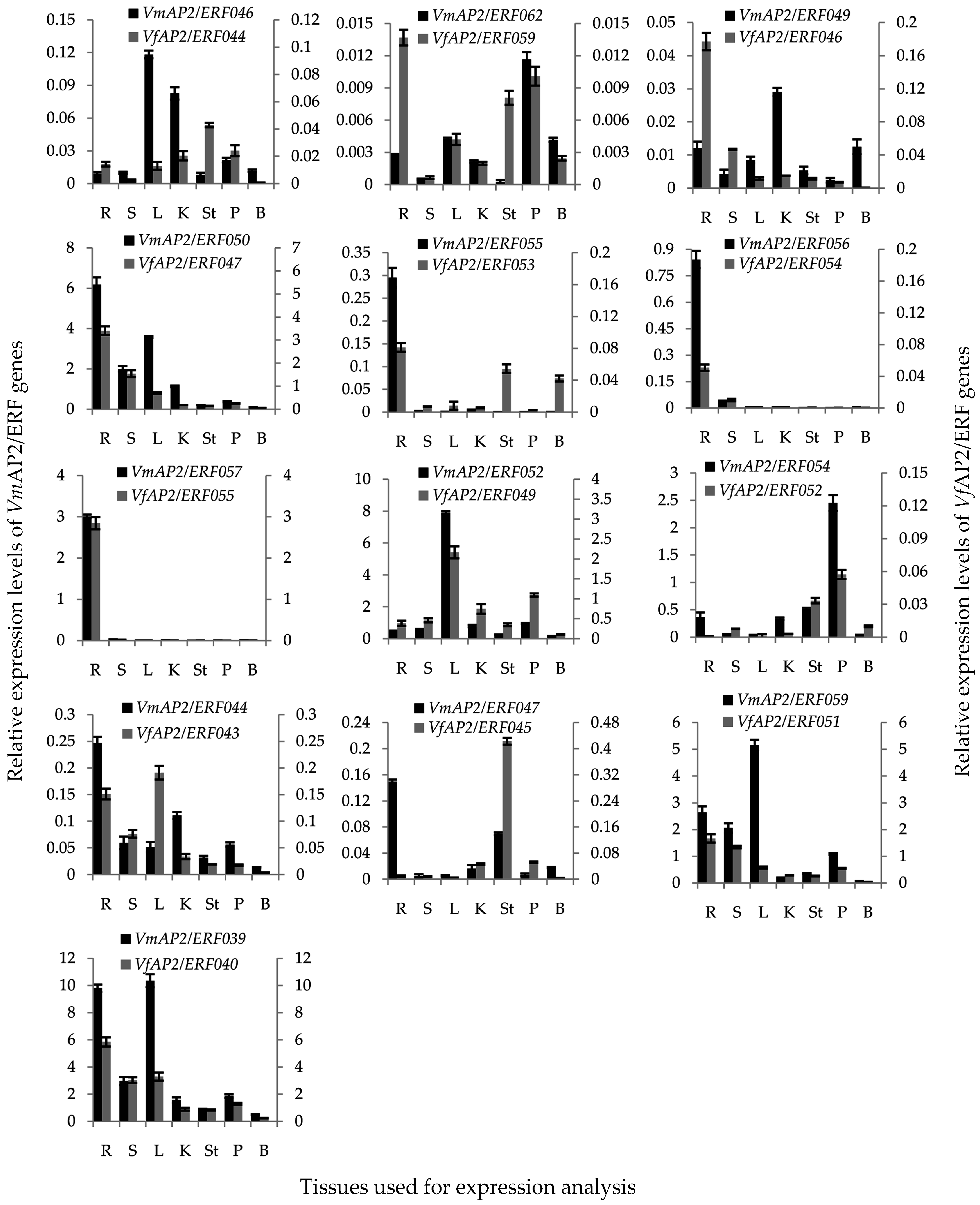
3.7. Validation of Vf/VmAP2/ERF Gene Expression by qRT-PCR
3.8. Tissue-Specific Expression Pattern of Vf/VmERF Genes
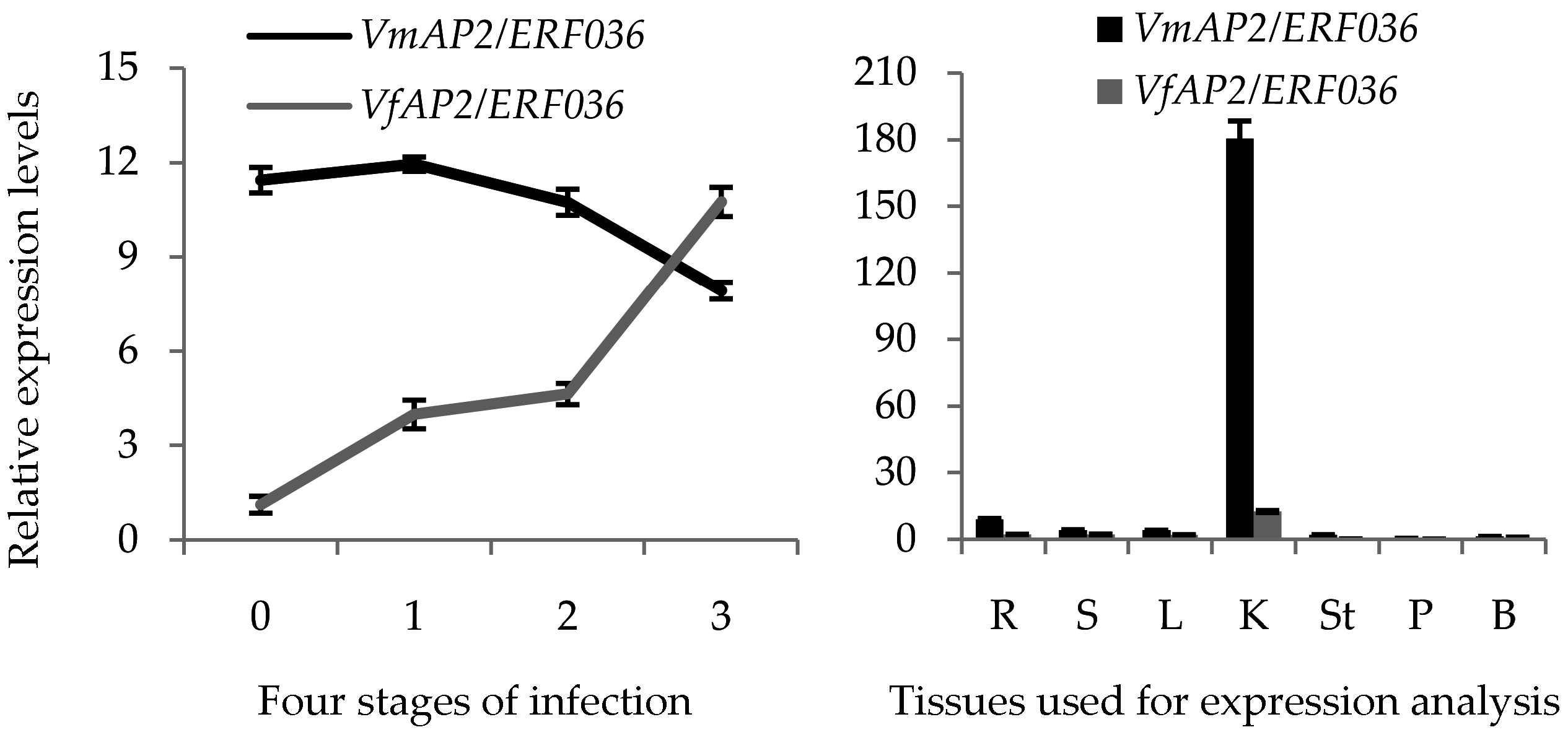
3.9. Hub Gene VmAP2/ERF036 Mediates Resistance to F. oxysporum in V. montana
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- McGrath, K.C.; Dombrecht, B.; Manners, J.M.; Schenk, P.M.; Edgar, C.I.; Maclean, D.J.; Scheible, W.R.; Udvardi, M.K.; Kazan, K. Repressor- and activator-type ethylene response factors functioning in jasmonate signaling and disease resistance identified via a genome-wide screen of Arabidopsis transcription factor gene expression. Plant Physiol. 2005, 139, 949–959. [Google Scholar] [CrossRef] [PubMed]
- Licausi, F.; Ohme-Takagi, M.; Perata, P. APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: Mediators of stress responses and developmental programs. New Phytol. 2013, 199, 639–649. [Google Scholar] [CrossRef] [PubMed]
- Gutterson, N.; Reuber, T.L. Regulation of disease resistance pathways by AP2/ERF transcription factors. Curr. Opin. Plant Biol. 2004, 7, 465–471. [Google Scholar] [CrossRef] [PubMed]
- Dey, S.; Corina Vlot, A. Ethylene responsive factors in the orchestration of stress responses in monocotyledonous plants. Front. Plant Sci. 2015, 6, 640. [Google Scholar] [CrossRef] [PubMed]
- Nakano, T.; Suzuki, K.; Fujimura, T.; Shinshi, H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 2006, 140, 411–432. [Google Scholar] [CrossRef] [PubMed]
- Lakhwani, D.; Pandey, A.; Dhar, Y.V.; Bag, S.K.; Trivedi, P.K.; Asif, M.H. Genome-wide analysis of the AP2/ERF family in Musa species reveals divergence and neofunctionalisation during evolution. Sci. Rep. 2016, 6, 18878. [Google Scholar] [CrossRef] [PubMed]
- Aukerman, M.J.; Sakai, H. Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 2003, 15, 2730–2741. [Google Scholar] [CrossRef] [PubMed]
- El Ouakfaoui, S.; Schnell, J.; Abdeen, A.; Colville, A.; Labbé, H.; Han, S.; Baum, B.; Laberge, S.; Miki, B. Control of somatic embryogenesis and embryo development by AP2 transcription factors. Plant Mol. Biol. 2010, 74, 313–326. [Google Scholar] [CrossRef] [PubMed]
- Imin, N.; Nizamidin, M.; Wu, T.; Rolfe, B.G. Factors involved in root formation in Medicago truncatula. J. Exp. Bot. 2007, 58, 439–451. [Google Scholar] [CrossRef] [PubMed]
- Jofuku, K.D.; Omidyar, P.K.; Gee, Z.; Okamuro, J.K. Control of seed mass and seed yield by the floral homeotic gene APETALA2. Proc. Natl. Acad. Sci. USA 2005, 102, 3117–3122. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Liu, S. Genome-wide identification and phylogenetic analysis of the ERF gene family in cucumbers. Genet. Mol. Biol. 2011, 34, 624–633. [Google Scholar] [CrossRef] [PubMed]
- Sohn, K.H.; Lee, S.C.; Jung, H.W.; Hong, J.K.; Hwang, B.K. Expression and functional roles of the pepper pathogen-induced transcription factor RAV1 in bacterial disease resistance, and drought and salt stress tolerance. Plant Mol. Biol. 2006, 61, 897–915. [Google Scholar] [CrossRef] [PubMed]
- Li, C.W.; Su, R.C.; Cheng, C.P.; Sanjaya; You, S.J.; Hsieh, T.H.; Chao, T.C.; Chan, M.T. Tomato RAV transcription factor is a pivotal modulator involved in the AP2/EREBP-mediated defense pathway. Plant Physiol. 2011, 156, 213–227. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.X.; Wang, Y.X.; Liu, X.F.; Li, J.Y. Arabidopsis RAV1 is down-regulated by brassinosteroid and may act as a negative regulator during plant development. Cell Res. 2004, 14, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Alonso, J.M.; Stepanova, A.N.; Leisse, T.J.; Kim, C.J.; Chen, H.; Shinn, P.; Stevenson, D.K.; Zimmerman, J.; Barajas, P.; Cheuk, R.; et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 2003, 301, 653–657. [Google Scholar] [CrossRef] [PubMed]
- Sakuma, Y.; Liu, Q.; Dubouzet, J.G.; Abe, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 2002, 290, 998–1009. [Google Scholar] [CrossRef] [PubMed]
- Ohme-Takagi, M.; Shinshi, H. Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 1995, 7, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Chen, M.; Li, L.; Xu, Z.; Chen, X.; Guo, J.; Ma, Y. Overexpression of the soybean GmERF3 gene, an AP2/ERF type transcription factor for increased tolerances to salt, drought, and diseases in transgenic tobacco. J. Exp. Bot. 2009, 60, 3781–3796. [Google Scholar] [CrossRef] [PubMed]
- Hao, D.; Ohme-Takagi, M.; Sarai, A. Unique mode of GCC box recognition by the DNA-binding domain of ethylene responsive element-binding factor (ERF domain) in plants. J. Biol. Chem. 1998, 273, 26857–26861. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Yu, J.P.; Chen, F.; Zhao, T.J.; Fang, X.H.; Li, Y.Q.; Sui, S.F. TINY, a dehydration-responsive element (DRE)-binding protein-like transcription factor connecting the DRE and ethylene-responsive element-mediated signaling pathways in Arabidopsis. J. Biol. Chem. 2008, 283, 6261–6271. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Li, F.; Ling, L.; Liu, A. Genome-wide survey and expression profiles of the AP2/ERF family in castor bean (Ricinus communis L.). BMC Genom. 2013, 14, 785. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Li, Y.; Hou, X. Genome-wide analysis of the AP2/ERF transcription factor superfamily in Chinese cabbage (Brassica rapa ssp. pekinensis). BMC Genom. 2013, 14, 573. [Google Scholar] [CrossRef] [PubMed]
- Licausi, F.; Giorgi, F.M.; Zenoni, S.; Osti, F.; Pezzotti, M.; Perata, P. Genomic and transcriptomic analysis of the AP2/ERF superfamily in Vitis vinifera. BMC Genom. 2010, 11, 719. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, J.; Cai, B.; Peng, R.H.; Zhu, B.; Jin, X.F.; Xue, Y.; Gao, F.; Fu, X.Y.; Tian, Y.S.; Zhao, W.; et al. Genome-wide analysis of the AP2/ERF gene family in Populus trichocarpa. Biochem. Biophys. Res. Commun. 2008, 371, 468–474. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Qin, S.; Guo, Y.; Chen, Y.; Wu, P.; Chen, Y.; Li, M.; Jiang, H.; Wu, G. Genome-wide analysis of the AP2/ERF gene family in physic nut and overexpression of the JcERF011 gene in rice increased its sensitivity to salinity stress. PLoS ONE 2016, 11, e0150879. [Google Scholar] [CrossRef] [PubMed]
- Shu, Y.; Liu, Y.; Zhang, J.; Song, L.; Guo, C. Genome-Wide Analysis of the AP2/ERF Superfamily Genes and their Responses to Abiotic Stress in Medicago truncatula. Front. Plant Sci. 2016, 6, 1247. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Lv, H.; Li, L.; Liu, J.; Mu, S.; Li, X.; Gao, J. Genome-Wide Analysis of the AP2/ERF Transcription Factors Family and the Expression Patterns of DREB Genes in Moso Bamboo (Phyllostachys edulis). PLoS ONE 2015, 10, e0126657. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, G.; Garg, V.; Kudapa, H.; Doddamani, D.; Pazhamala, L.T.; Khan, A.W.; Thudi, M.; Lee, S.H.; Varshney, R.K. Genome-wide dissection of AP2/ERF and HSP90 gene families in five legumes and expression profiles in chickpea and pigeonpea. Plant Biotechnol. J. 2016, 14, 1562–1577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berrocal-Lobo, M.; Molina, A. Ethylene response factor 1 mediates Arabidopsis resistance to the soilborne fungus Fusarium oxysporum. Mol. Plant Microbe Interact. 2004, 17, 763–770. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.Y.; Catinot, J.; Zimmerli, L. Ethylene response factors in Arabidopsis immunity. J. Exp. Bot. 2016, 67, 1231–1241. [Google Scholar] [CrossRef] [PubMed]
- Oñate-Sánchez, L.; Anderson, J.P.; Young, J.; Singh, K.B. AtERF14, a member of the ERF family of transcription factors, plays a nonredundant role in plant defense. Plant Physiol. 2007, 143, 400–409. [Google Scholar] [CrossRef] [PubMed]
- Moffat, C.S.; Ingle, R.A.; Wathugala, D.L.; Saunders, N.J.; Knight, H.; Knight, M.R. ERF5 and ERF6 play redundant roles as positive regulators of JA/Et-mediated defense against Botrytis cinereal in Arabidopsis. PLoS ONE 2012, 7, e35995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, X.; Xu, J.; He, Y.; Yang, K.Y.; Mordorski, B.; Liu, Y.; Zhang, S. Phosphorylation of an ERF transcription factor by Arabidopsis MPK3/MPK6 regulates plant defense gene induction and fungal resistance. Plant Cell 2013, 25, 1126–1142. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Huang, L.; Dai, Y.; Liu, S.; Hong, Y.; Tian, L.; Huang, L.; Cao, Z.; Li, D.; Song, F. Arabidopsis AtERF15 positively regulates immunity against Pseudomonas syringae pv. tomato DC3000 and Botrytis cinerea. Front. Plant Sci. 2015, 6, 686. [Google Scholar] [PubMed]
- Pré, M.; Atallah, M.; Champion, A.; De Vos, M.; Pieterse, C.M.; Memelink, J. The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol. 2008, 147, 1347–1357. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.H.; Chen, J.H.; Chang, C.Y.; Chang, C.C. Biodiesel production from tung Vernicia montana oil and its blending properties in different fatty acid compositions. Bioresour. Technol. 2010, 101, 9521–9526. [Google Scholar] [CrossRef] [PubMed]
- PlantTFDB: Plant Transcription Factor Database. Available online: http://planttfdb.cbi.pku.edu.cn (accessed on 9 November 2016).
- Simple Modular Architecture Research Tool (SMART). Available online: http://smart.embl-heidelberg.de/ (accessed on 9 November 2016).
- Letunic, I.; Doerks, T.; Bork, P. SMART 7: Recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012, 40, D302–D305. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Lu, S.; Anderson, J.B.; Chitsaz, F.; Derbyshire, M.K.; DeWeese-Scott, C.; Fong, J.H.; Geer, L.Y.; Geer, R.C.; Gonzales, N.R.; et al. CDD: A conserved domain database for the functional annotation of proteins. Nucleic Acids Res. 2011, 39, D225–D229. [Google Scholar] [CrossRef] [PubMed]
- Hall, T. BioEdit: An important software for molecular biology. GERF Bull. Biosci. 2011, 2, 60–61. [Google Scholar]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, J.; Zhao, X.Q.; Wang, J.; Wong, G.K.; Yu, J. KaKs_Calculator: Calculating Ka and Ks through model selection and model averaging. Genom. Proteom. Bioinform. 2006, 4, 259–263. [Google Scholar] [CrossRef]
- Trinity. Available online: http://trinityrnaseq.sourceforge.net/ (accessed on 9 November 2016).
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Saeed, A.I.; Sharov, V.; White, J.; Li, J.; Liang, W.; Bhagabati, N.; Braisted, J.; Klapa, M.; Currier, T.; Thiagarajan, M.; et al. TM4: A free, open-source system for microarray data management and analysis. Biotechniques 2003, 34, 374–378. [Google Scholar] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Allen, M.D.; Yamasaki, K.; Ohme-Takagi, M.; Tateno, M.; Suzuki, M. A novel mode of DNA recognition by a β-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA. EMBO J. 1998, 17, 5484–5496. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.F.; Li, J.; Chen, F.; Li, Y.Q.; Zhou, H.M.; Liu, Q. Effect of two conserved amino acid residues on DREB1A function. Biochemistry 2001, 66, 623–627. [Google Scholar] [PubMed]
- Liu, L.; White, M.J.; MacRae, T.H. Transcription factors and their genes in higher plants functional domains, evolution and regulation. Eur. J. Biochem. 1999, 262, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Reyes, J.C.; Muro-Pastor, M.I.; Florencio, F.J. The GATA family of transcription factors in Arabidopsis and rice. Plant Physiol. 2004, 134, 1718–1732. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.C.; Wong, C.L.; Muzzi, F.; Vlaardingerbroek, I.; Kidd, B.N.; Schenk, P.M. Root defense analysis against Fusarium oxysporum reveals new regulators to confer resistance. Sci. Rep. 2014, 4, 5584. [Google Scholar] [CrossRef] [PubMed]
- Duan, C.; Argout, X.; Gébelin, V.; Summo, M.; Dufayard, J.F.; Leclercq, J.; Piyatrakul, P.; Pirrello, J.; Rio, M.; Champion, A.; et al. Identification of the Hevea brasiliensis AP2/ERF superfamily by RNA sequencing. BMC Genom. 2013, 14, 30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohta, M.; Matsui, K.; Hiratsu, K.; Shinshi, H.; Ohme-Takagi, M. Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 2001, 13, 1959–1968. [Google Scholar] [CrossRef] [PubMed]
- Kagale, S.; Rozwadowski, K. EAR motif-mediated transcriptional repression in plants: An underlying mechanism for epigenetic regulation of gene expression. Epigenetics 2011, 6, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Rashid, M.; Guangyuan, H.; Guangxiao, Y.; Hussain, J.; Xu, Y. AP2/ERF transcription factor in rice: Genome-wide canvas and syntenic relationships between monocots and eudicots. Evol. Bioinform. Online 2012, 8, 321–355. [Google Scholar] [CrossRef] [PubMed]
- Çevik, V.; Kidd, B.N.; Zhang, P.; Hill, C.; Kiddle, S.; Denby, K.J.; Holub, E.B.; Cahill, D.M.; Manners, J.M.; Schenk, P.M.; et al. MEDIATOR25 acts as an integrative hub for the regulation of jasmonate-responsive gene expression in Arabidopsis. Plant Physiol. 2012, 160, 541–555. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, Y.; Jia, H.X.; Li, J.B.; Huang, J.; Lu, M.Z.; Hu, J.J. The heat shock factor gene family in Salix suchowensis: A genome-wide survey and expression profiling during development and abiotic stresses. Front. Plant Sci. 2015, 6, 748. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, Y.; Yamoto, N.; Suzuki, Y.; Chiba, Y.; Yamazaki, K.; Sato, T.; Yamaguchi, J. The Arabidopsis transcriptional repressor ERF9 participates in resistance against necrotrophic fungi. Plant Sci. 2013, 213, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Shi, J.; Xu, W.; Li, H.; He, M.; Xu, Y.; Xu, T.; Yang, Y.; Cao, J.; Wang, Y. Three ERF transcription factors from Chinese wild grapevine Vitis pseudoreticulata participate in different biotic and abiotic stress-responsive pathways. J. Plant Physiol. 2013, 170, 923–933. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Chen, X.; Liu, J.; Ye, J.; Guo, Z. The rice ERF transcription factor OsERF922 negatively regulates resistance to Magnaporthe oryzae and salt tolerance. J. Exp. Bot. 2012, 63, 3899–3911. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Qi, L.; Liu, X.; Cai, S.; Xu, H.; Huang, R.; Li, J.; Wei, X.; Zhang, Z. The wheat ethylene response factor transcription factor pathogen induced ERF1 mediates host responses to both the necrotrophic pathogen Rhizoctonia cerealis and freezing stresses. Plant Physiol. 2014, 164, 1499–1514. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Guo, Z. Tobacco OPBP1 enhances salt tolerance and disease resistance of transgenic rice. Int. J. Mol. Sci. 2008, 9, 2601–2613. [Google Scholar] [CrossRef] [PubMed]
- De Wit, P.J. How plants recognize pathogens and defend themselves. Cell. Mol. Life Sci. 2007, 64, 2726–2732. [Google Scholar] [CrossRef] [PubMed]
- Li, H.Y.; Xiao, S.; Chye, M.L. Ethylene- and pathogen-inducible Arabidopsis acyl-CoA-binding protein 4 interacts with an ethylene-responsive element binding protein. J. Exp. Bot. 2008, 59, 3997–4006. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Chen, J.; Zhang, H.; Song, F. Overexpression of a rice diacylglycerol kinase gene OsBIDK1 enhances disease resistance in transgenic tobacco. Mol. Cell 2008, 26, 258–264. [Google Scholar]
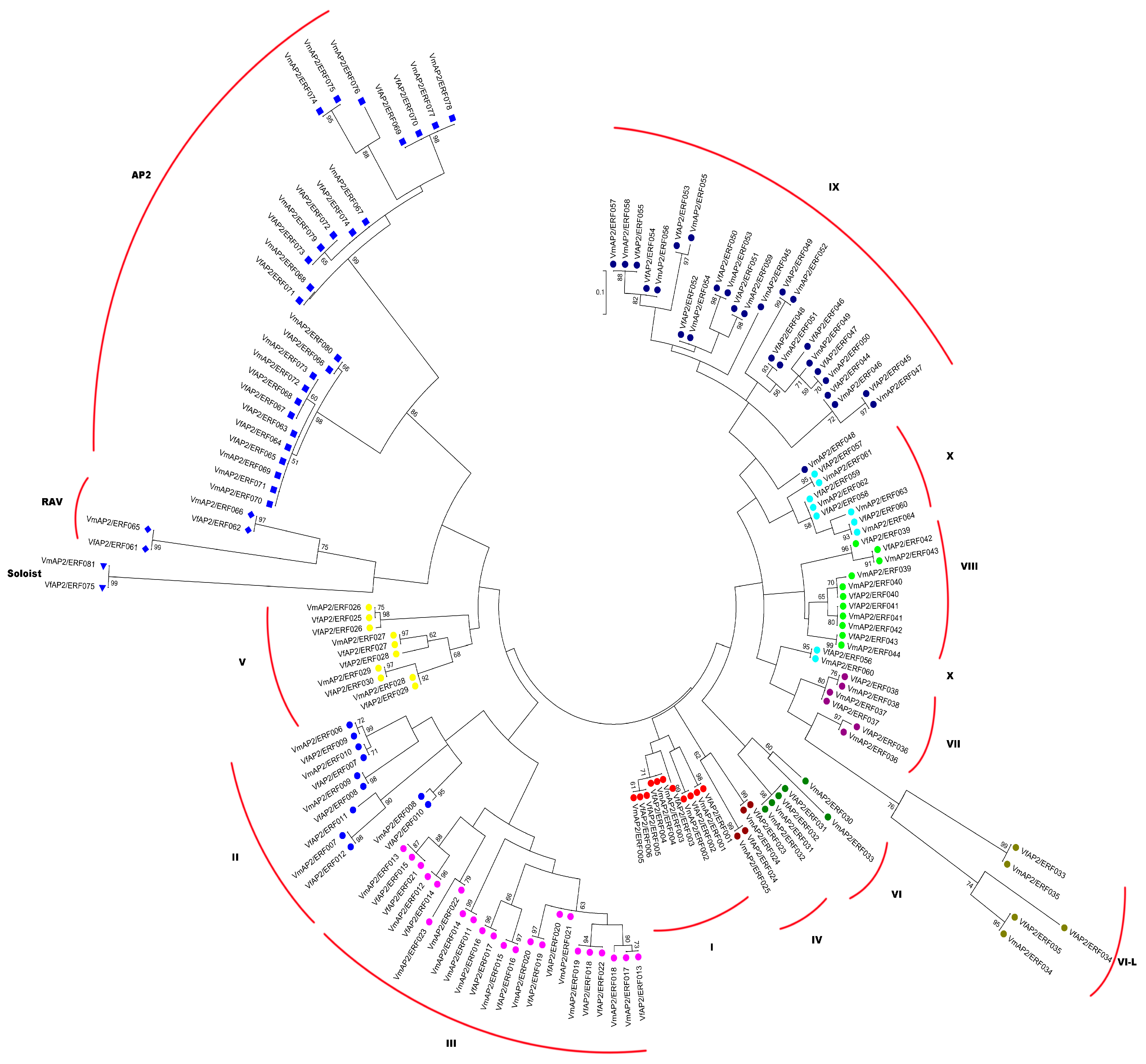
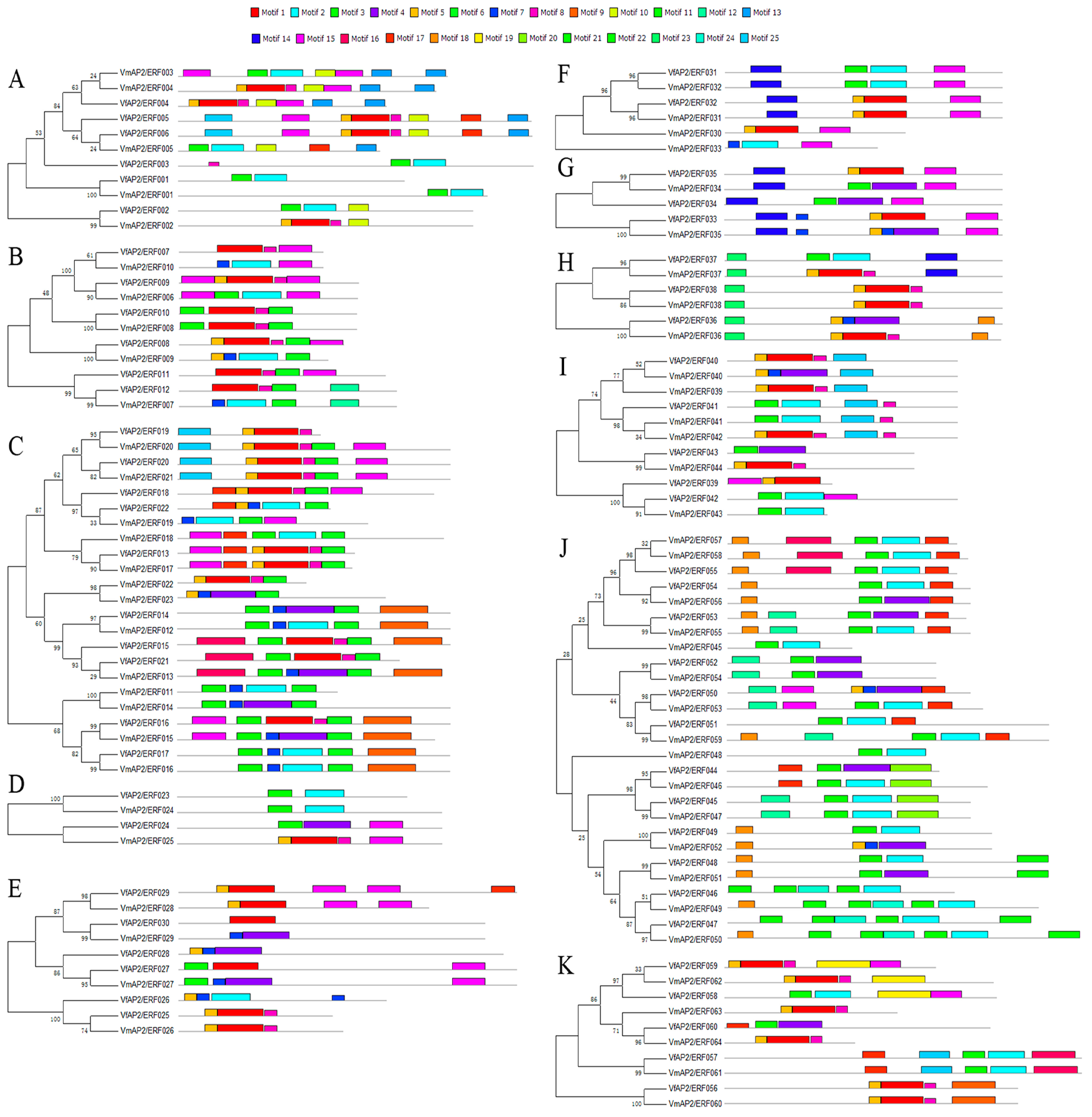
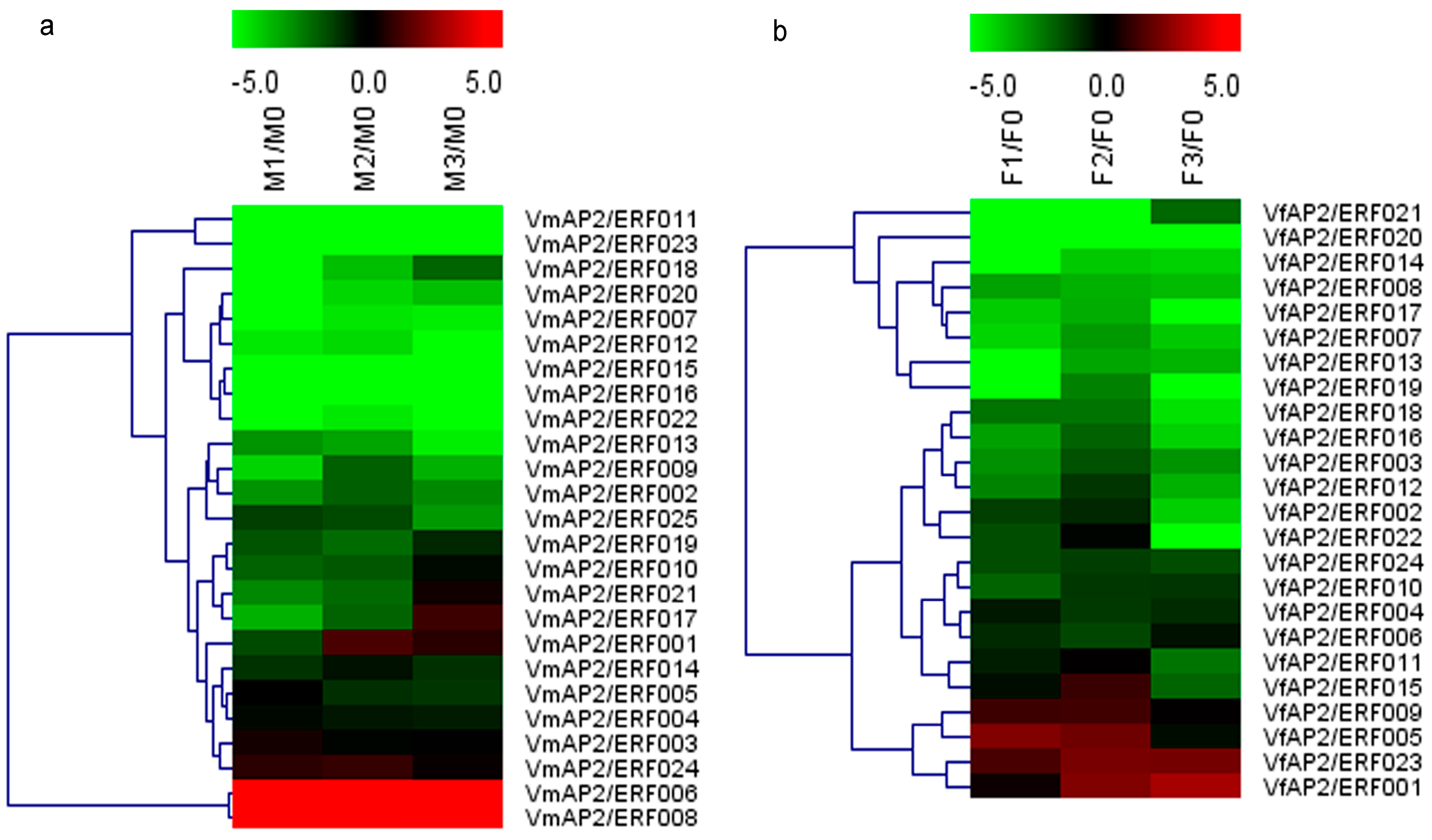

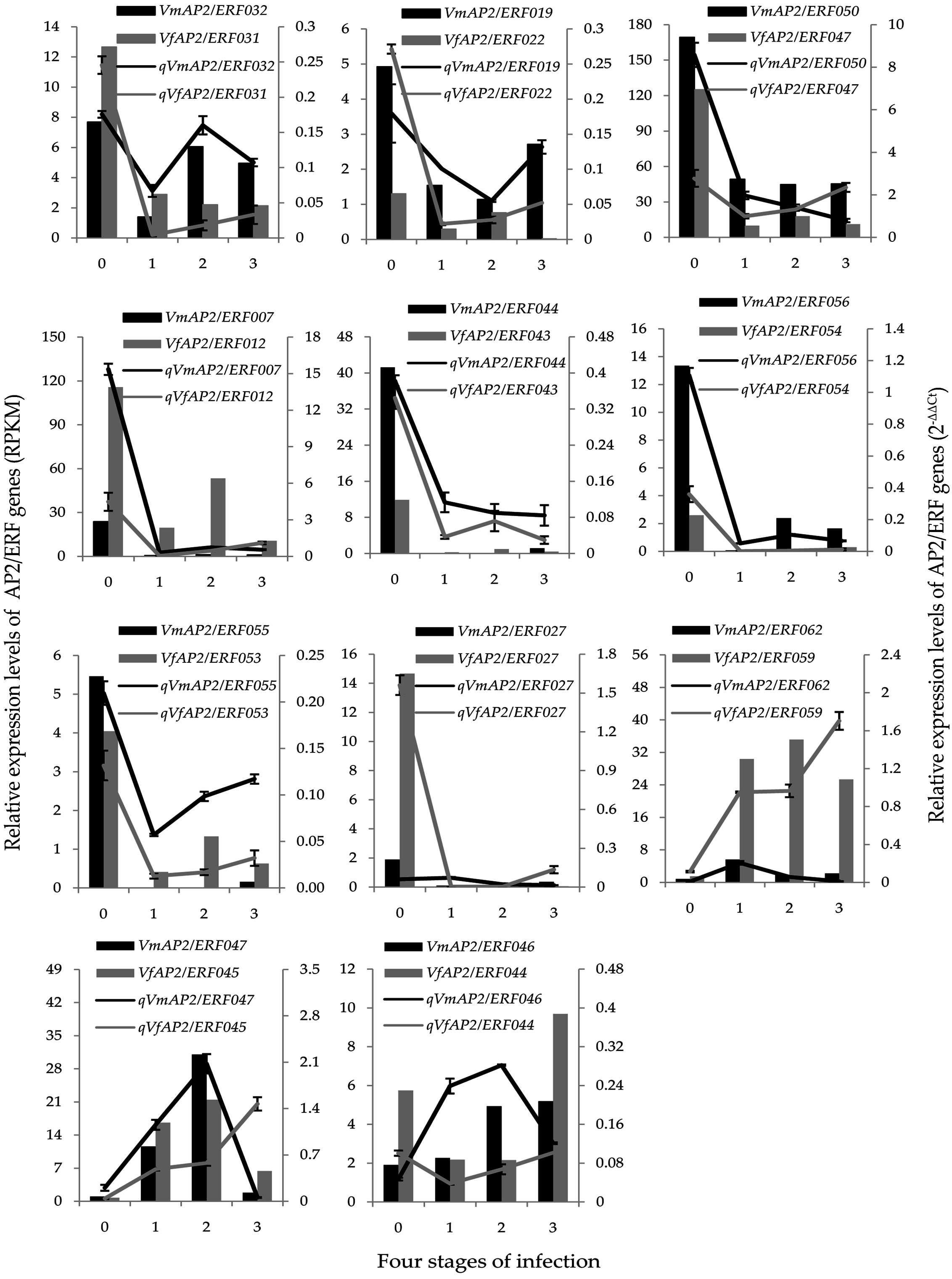
| Family | Subfamily | Group | Vernicia | Arabidopsis thaliana | Jatropha curcas | |
|---|---|---|---|---|---|---|
| V. montana | V. fordii | |||||
| AP2 | Total | 14 | 12 | 18 | 16 | |
| ERF | Total | 64 | 60 | 122 | 98 | |
| DREB | Total | 25 | 24 | 57 | 35 | |
| I | 5 | 6 | 10 | 5 | ||
| II | 5 | 6 | 15 | 8 | ||
| III | 13 | 10 | 23 | 16 | ||
| IV | 2 | 2 | 9 | 6 | ||
| ERF | Total | 39 | 36 | 65 | 63 | |
| V | 4 | 6 | 5 | 12 | ||
| VI | 4 | 2 | 8 | 4 | ||
| VII | 3 | 3 | 5 | 3 | ||
| VIII | 6 | 5 | 15 | 12 | ||
| IX | 15 | 12 | 17 | 21 | ||
| X | 5 | 5 | 8 | 8 | ||
| VI-L | 2 | 3 | 4 | 3 | ||
| Xb-L | 0 | 0 | 3 | 0 | ||
| RAV | 2 | 2 | 6 | 4 | ||
| Soloist | 1 | 1 | 1 | 1 | ||
| Total | 81 | 75 | 147 | 119 | ||
| Candidate Gene | Group | Regulation | Source Plant | Fungal Pathogen | References |
|---|---|---|---|---|---|
| ERF1 | IX | activator | A. thaliana | F. oxysporum | [29] |
| AtERF2 | IX | activator | A. thaliana | F. oxysporum | [1] |
| AtERF4 | VIII | repressor | A. thaliana | F. oxysporum | [1] |
| AtERF14 | IX | activator | A. thaliana | F. oxysporum | [31] |
| ERF5 | IX | activator | A. thaliana | Botrytis cinerea | [32] |
| ERF6 | IX | activator | A. thaliana | B. cinerea | [2,32] |
| ERF9 | VIII | repressor | A. thaliana | B. cinerea | [60] |
| AtERF15 | IX | activator | A. thaliana | B. cinerea | [34] |
| ORA59 | IX | activator | A. thaliana | B. cinerea | [35] |
| VpERF3 | IX | activator | Vitis pseudoreticulata | Phytophtora parasitica | [61] |
| OsERF922 | IX | repressor | Oryza sativa | Magnaporthe oryzae | [62] |
| TaPIE1 | IX | activator | Triticum aestivum | Rhizoctonia cerealis | [63] |
| GmERF3 | IV | activator | Glycine max | Alternaria alternate | [18] |
| OPBP1 | IX | activator | Nicotiana tabacum | Rhizoctonia solani | [64] |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Q.; Gao, M.; Wu, L.; Wang, Y.; Chen, Y. Divergent Expression Patterns in Two Vernicia Species Revealed the Potential Role of the Hub Gene VmAP2/ERF036 in Resistance to Fusarium oxysporum in Vernicia montana. Genes 2016, 7, 109. https://doi.org/10.3390/genes7120109
Zhang Q, Gao M, Wu L, Wang Y, Chen Y. Divergent Expression Patterns in Two Vernicia Species Revealed the Potential Role of the Hub Gene VmAP2/ERF036 in Resistance to Fusarium oxysporum in Vernicia montana. Genes. 2016; 7(12):109. https://doi.org/10.3390/genes7120109
Chicago/Turabian StyleZhang, Qiyan, Ming Gao, Liwen Wu, Yangdong Wang, and Yicun Chen. 2016. "Divergent Expression Patterns in Two Vernicia Species Revealed the Potential Role of the Hub Gene VmAP2/ERF036 in Resistance to Fusarium oxysporum in Vernicia montana" Genes 7, no. 12: 109. https://doi.org/10.3390/genes7120109





