Identification of the Ovine Keratin-Associated Protein 22-1 (KAP22-1) Gene and Its Effect on Wool Traits
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sheep Blood and Wool Samples
2.2. Search for an Ovine Homolog of the Human KRTAP22-1 Gene in the Sheep Genome Sequence
2.3. PCR Primers and Amplification of Sheep Genomic DNA
2.4. Screening for Variation in KRTAP22-1
2.5. Sequencing of Allelic Variants and Sequence Analysis
2.6. Statistical Analyses
3. Results
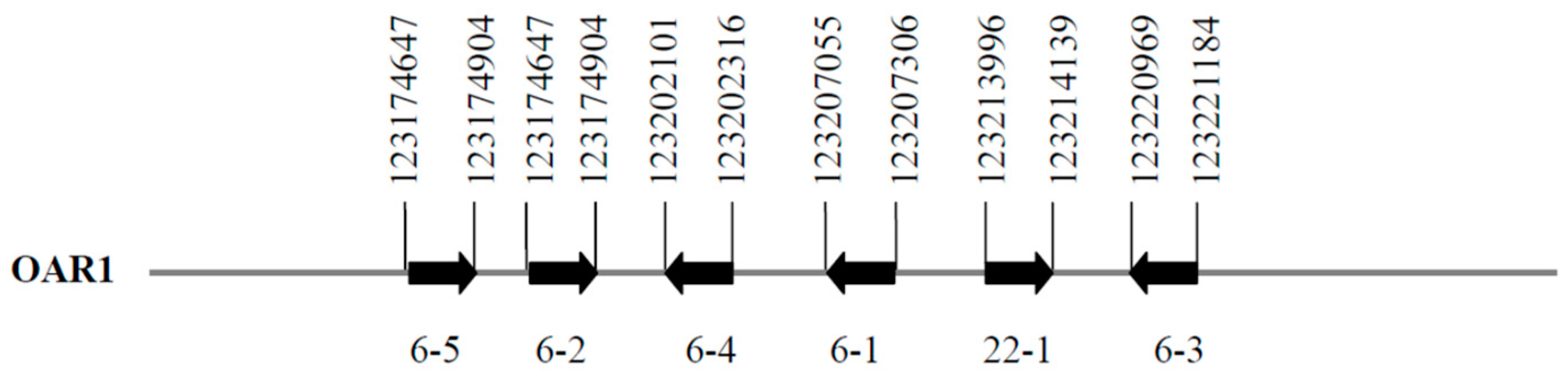
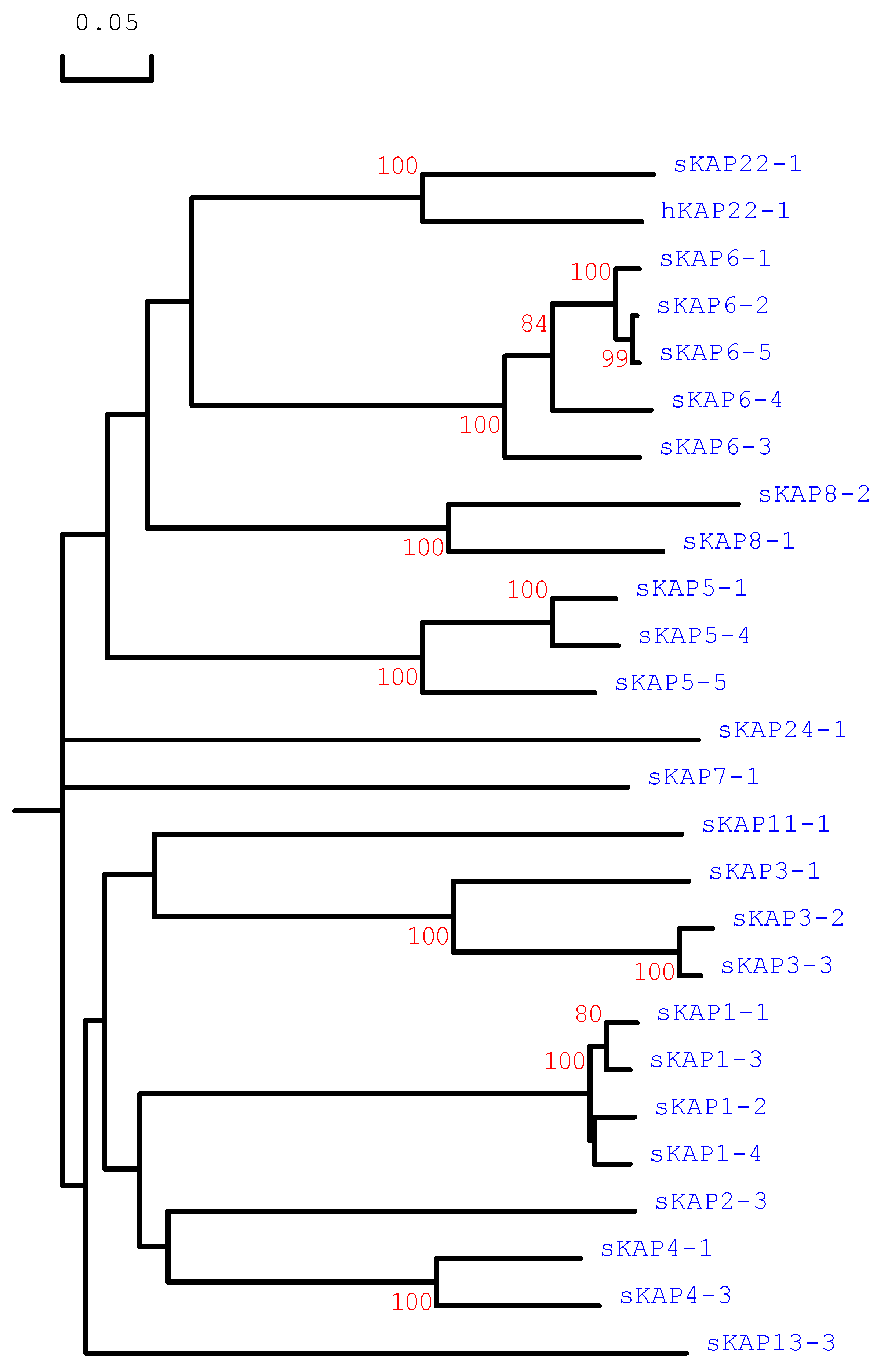
3.1. Identification of KRTAP22-1 in the Sheep Genome
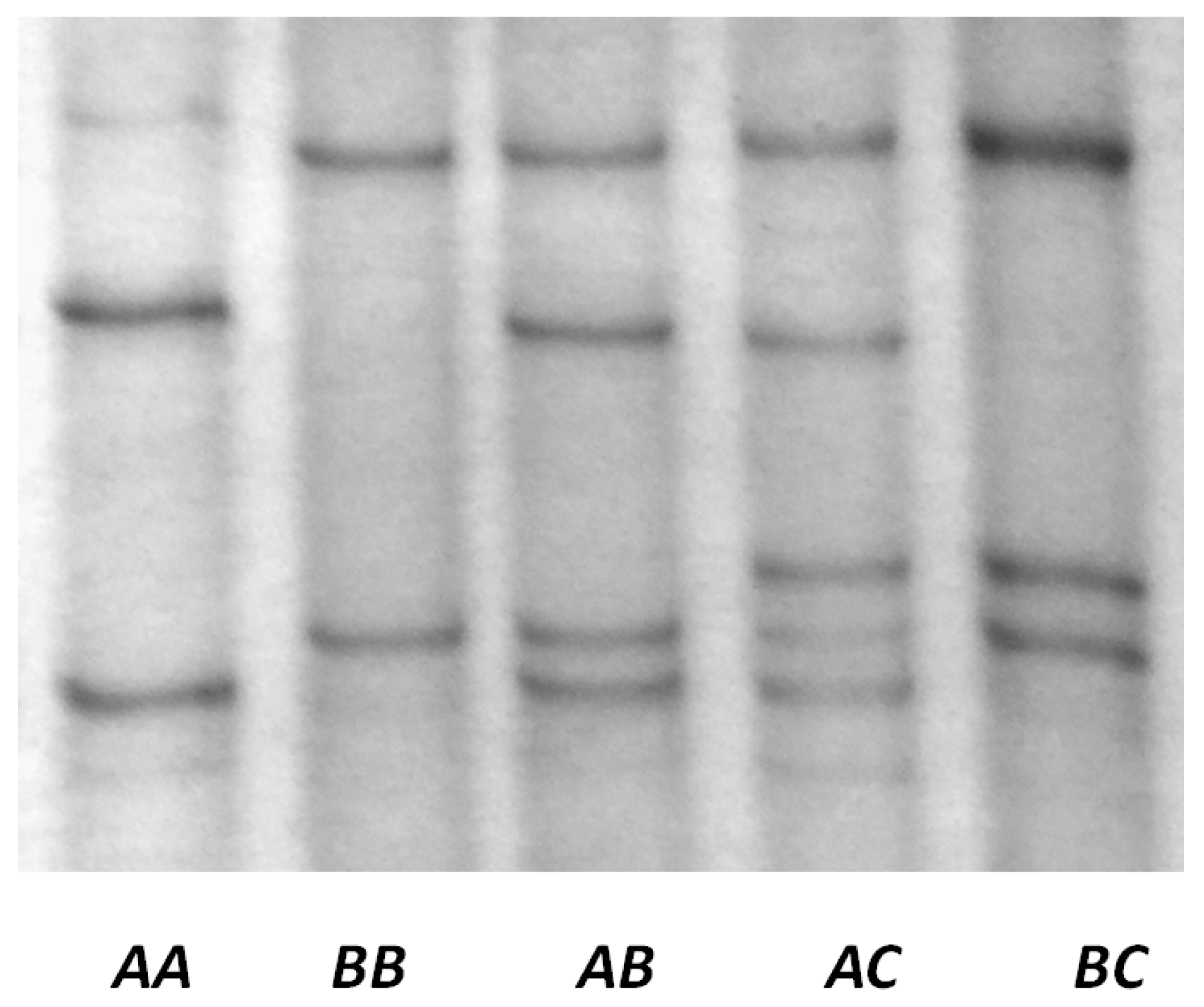
3.2. Detection of Variation in Ovine KRTAP22-1
3.3. Amino Acid Composition of Ovine KAP22-1
3.4. Genotypes and Allele Frequencies in NZ Romney and Merino-Cross Sheep
3.5. Effect of Variation in KRTAP22-1 on Wool Traits
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Powell, B.C.; Rogers, G.E. The role of keratin proteins and their genes in the growth, structure and properties of hair. In Formation and Structure of Human Hair; Jollès, P., Zahn, H., Höcker, H., Eds.; Birkhäuser Verlag: Basel, Switzerland, 1997; pp. 59–148. [Google Scholar]
- Gong, H.; Zhou, H.; Forrest, R.H.J.; Li, S.; Wang, J.; Dyer, J.M.; Luo, Y.; Hickford, J.G. Wool keratin-associated protein genes in Sheep—A Review. Genes 2016. [Google Scholar] [CrossRef] [PubMed]
- Gong, H.; Zhou, H.; McKenzie, G.W.; Yu, Z.D.; Clerens, S.; Dyer, J.M.; Plowman, J.E.; Wright, M.W.; Arora, R.; Bawden, C.S.; et al. An updated nomenclature for keratin-associated proteins (KAPs). Int. J. Bio. Sci. 2012, 8, 258–264. [Google Scholar] [CrossRef] [PubMed]
- Rogers, M.A.; Langbein, L.; Praetzel-Wunder, S.; Winter, H.; Schweizer, J. Human hair keratin-associated proteins (KAPs). Int. Rev. Cytol. 2006, 251, 209–263. [Google Scholar] [PubMed]
- Gillespie, J.M. The proteins of hair and other hard α-keratins. In Cellular and Molecular Biology of Intermediate Filaments; Goldman, R.D.S., Peter, M., Eds.; Plenum press: New York, NY, USA, 1990; pp. 95–128. [Google Scholar]
- Gong, H.; Zhou, H.; Hickford, J.G.H. Diversity of the glycine/tyrosine-rich keratin-associated protein 6 gene (KAP6) family in sheep. Mol. Biol. Rep. 2011, 38, 31–35. [Google Scholar] [CrossRef] [PubMed]
- Gong, H.; Zhou, H.; Plowman, J.E.; Dyer, J.M.; Hickford, J.G.H. Search for variation in the ovine KAP7-1 and KAP8-1 genes using polymerase chain reaction–single-stranded conformational polymorphism screening. DNA Cell B 2012, 31, 367–370. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Gong, H.; Wang, J.Q.; Dyer, J.M.; Luo, Y.Z.; Hickford, J.G.H. Identification of four new gene members of the KAP6 gene family in sheep. Sci. Rep. 2016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rogers, M.A.; Langbein, L.; Winter, H.; Ehmann, C.; Praetzel, S.; Schweizer, J. Characterization of a first domain of human high glycine-tyrosine and high sulfur keratin-associated protein (KAP) genes on chromosome 21q22.1. J. Biol. Chem. 2002, 277, 48993–49002. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Hickford, J.G.H.; Fang, Q. A two-step procedure for extracting genomic DNA from dried blood spots on filter paper for polymerase chainreaction amplification. Anal. Biochem. 2006, 354, 159–161. [Google Scholar] [CrossRef] [PubMed]
- Byun, S.O.; Fang, Q.; Zhou, H.; Hickford, J.G.H. An effective method for silver-staining DNA in large numbers of polyacrylamide gels. Anal. Biochem. 2009, 385, 174–175. [Google Scholar] [CrossRef] [PubMed]
- Gong, H.; Zhou, H.; Dyer, J.M.; Hickford, J.G. Identification of the ovine KAP11-1 gene (KRTAP11-1) and genetic variation in its coding sequence. Mol. Biol. Rep. 2011, 38, 5429–5433. [Google Scholar] [CrossRef] [PubMed]
- Gong, H.; Zhou, H.; Hodge, S.; Dyer, J.M.; Hickford, J.G. Association of wool traits with variation in the ovine KAP1-2 gene in Merino cross lambs. Small Rum. Res. 2015, 124, 24–29. [Google Scholar] [CrossRef]
- Elliott, K.H.; Bigham, M.L.; Sumner, R.M.W.; Dalton, D.C. Wool production of yealing ewes of different breeds on hill country. N. Z. J. Agric. Res. 1978, 21, 179–186. [Google Scholar] [CrossRef]
- Nimbs, M.A.; Hygate, L.; Behrendt, R. The relationship between fibre curvature, crimp frequence and other wool traits. Anim. Prod. Aust. 1998, 22, 396. [Google Scholar]
- Sumner, R.M.W.; Young, S.R.; Upsdell, M.P. Wool yellowing and pH within Merino and Romney fleeces. Proc. N. Z. Soc. Anim. Prod. 2003, 63, 155–159. [Google Scholar]
- Li, S.W.; Ouyang, H.S.; Rogers, G.E.; Bawden, C.S. Characterization of the structural and molecular defectsin fibres and follicles of the merino felting lustre mutant. Exp. Dermatol. 2009, 18, 134–142. [Google Scholar] [CrossRef] [PubMed]
- Caldwell, J.P.; Mastronarde, D.N.; Woods, J.L.; Bryson, W.G. The three-dimensional arrangement of intermediate filaments in Romney wool cortical cells. J. Struct. Biol. 2005, 151, 298–305. [Google Scholar] [CrossRef] [PubMed]
- Powell, B.C.; Rogers, G.E. Hard keratin IF and associatedproteins. In Cellular and Molecular Biology of Intermediate Filaments; Goldman, R.D., Steinert, P.M., Eds.; Plenum Press: New York, NY, USA, 1990; pp. 267–300. [Google Scholar]
- Hurst, L.D. Molecular genetics: The sound of silence. Nature 2011, 471, 582–583. [Google Scholar] [CrossRef] [PubMed]
| SNP | Allele | Amino Acid Change | ||
|---|---|---|---|---|
| A | B | C | ||
| C.-100C/T | C | C | T | 5′UTR |
| C.45T/C | T | C | C | No change |
| Trait 1 | Variant | n | Mean ± SE 2 | p 3 | ||
|---|---|---|---|---|---|---|
| Absent | Present | Absent | Present | |||
| GFW (kg) | A | 79 | 311 | 2.30 ± 0.07 | 2.33 ± 0.06 | 0.628 |
| B | 99 | 291 | 2.37 ± 0.07 | 2.31 ± 0.06 | 0.191 | |
| CFW (kg) | A | 79 | 311 | 1.69 ± 0.06 | 1.69 ± 0.05 | 0.931 |
| B | 99 | 291 | 1.70 ± 0.05 | 1.69 ± 0.05 | 0.844 | |
| Yield (%) | A | 79 | 311 | 72.9 ± 0.99 | 72.0 ± 0.79 | 0.244 |
| B | 99 | 291 | 70.8 ± 0.91 | 72.6 ± 0.79 | 0.008 | |
| MFD (µm) | A | 79 | 311 | 19.6 ± 0.31 | 19.5 ± 0.25 | 0.705 |
| B | 99 | 291 | 19.4 ± 0.30 | 19.6 ± 0.25 | 0.547 | |
| FDSD (µm) | A | 79 | 311 | 4.28 ± 0.11 | 4.16 ± 0.09 | 0.139 |
| B | 99 | 291 | 4.17 ± 0.10 | 4.19 ± 0.30 | 0.828 | |
| CVFD (%) | A | 79 | 311 | 22.0 ± 0.36 | 21.7 ± 0.30 | 0.281 |
| B | 99 | 291 | 21.9 ± 0.35 | 21.7 ± 0.30 | 0.503 | |
| MSL (mm) | A | 79 | 311 | 84.2 ± 1.99 | 84.6 ± 1.63 | 0.796 |
| B | 99 | 291 | 85.5 ± 1.93 | 84.3 ± 1.62 | 0.368 | |
| MSS (N/ktex) | A | 79 | 311 | 21.1 ± 1.25 | 23.4 ± 1.02 | 0.192 |
| B | 99 | 291 | 21.7 ± 1.21 | 22.2 ± 1.02 | 0.526 | |
| MFC (o/mm) | A | 79 | 311 | 86.5 ± 2.43 | 89.4 ± 1.98 | 0.128 |
| B | 99 | 291 | 91.6 ± 2.34 | 87.9 ± 1.97 | 0.032 | |
| PF (%) | A | 79 | 311 | 2.12 ± 0.36 | 2.05 ± 0.29 | 0.816 |
| B | 99 | 291 | 2.13 ± 0.35 | 2.05 ± 0.28 | 0.762 | |
| Trait 1 | Mean ± SE 2 | p | ||
|---|---|---|---|---|
| AA (n = 93) | AB (n = 212) | BB (n = 77) | ||
| GFW (kg) | 2.38 ± 0.07 | 2.31 ± 0.06 | 2.29 ± 0.07 | 0.341 |
| CFW (kg) | 1.71 ± 0.06 | 1.69 ± 0.05 | 1.70 ± 0.06 | 0.918 |
| Yield (%) | 70.9 ± 0.97 b | 72.6 ± 0.83 a | 73.0 ± 1.00 a | 0.044 |
| MFD (µm) | 19.5 ± 0.31 | 19.6 ± 0.27 | 19.6 ± 0.32 | 0.838 |
| FDSD (µm) | 4.20 ± 0.10 | 4.16 ± 0.09 | 4.27 ± 0.11 | 0.385 |
| CVFD (%) | 21.9 ± 0.36 | 21.6 ± 0.31 | 21.9 ± 0.37 | 0.321 |
| MSL (mm) | 85.9 ± 1.98 | 84.1 ± 1.71 | 84.1 ± 2.03 | 0.488 |
| MSS (N/ktex) | 21.8 ± 1.24 | 22.7 ± 1.08 | 21.2 ± 1.28 | 0.306 |
| MFC (o/mm) | 91.7 ± 2.45 | 89.0 ± 2.12 | 86.8 ± 2.51 | 0.120 |
| PF (%) | 2.46 ± 0.53 | 2.45 ± 0.46 | 3.04 ± 0.55 | 0.387 |
© 2017 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, S.; Zhou, H.; Gong, H.; Zhao, F.; Wang, J.; Liu, X.; Luo, Y.; Hickford, J.G.H. Identification of the Ovine Keratin-Associated Protein 22-1 (KAP22-1) Gene and Its Effect on Wool Traits. Genes 2017, 8, 27. https://doi.org/10.3390/genes8010027
Li S, Zhou H, Gong H, Zhao F, Wang J, Liu X, Luo Y, Hickford JGH. Identification of the Ovine Keratin-Associated Protein 22-1 (KAP22-1) Gene and Its Effect on Wool Traits. Genes. 2017; 8(1):27. https://doi.org/10.3390/genes8010027
Chicago/Turabian StyleLi, Shaobin, Huitong Zhou, Hua Gong, Fangfang Zhao, Jiqing Wang, Xiu Liu, Yuzhu Luo, and Jon G. H. Hickford. 2017. "Identification of the Ovine Keratin-Associated Protein 22-1 (KAP22-1) Gene and Its Effect on Wool Traits" Genes 8, no. 1: 27. https://doi.org/10.3390/genes8010027





