Sulfolobus acidocaldarius UDG Can Remove dU from the RNA Backbone: Insight into the Specific Recognition of Uracil Linked with Deoxyribose
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
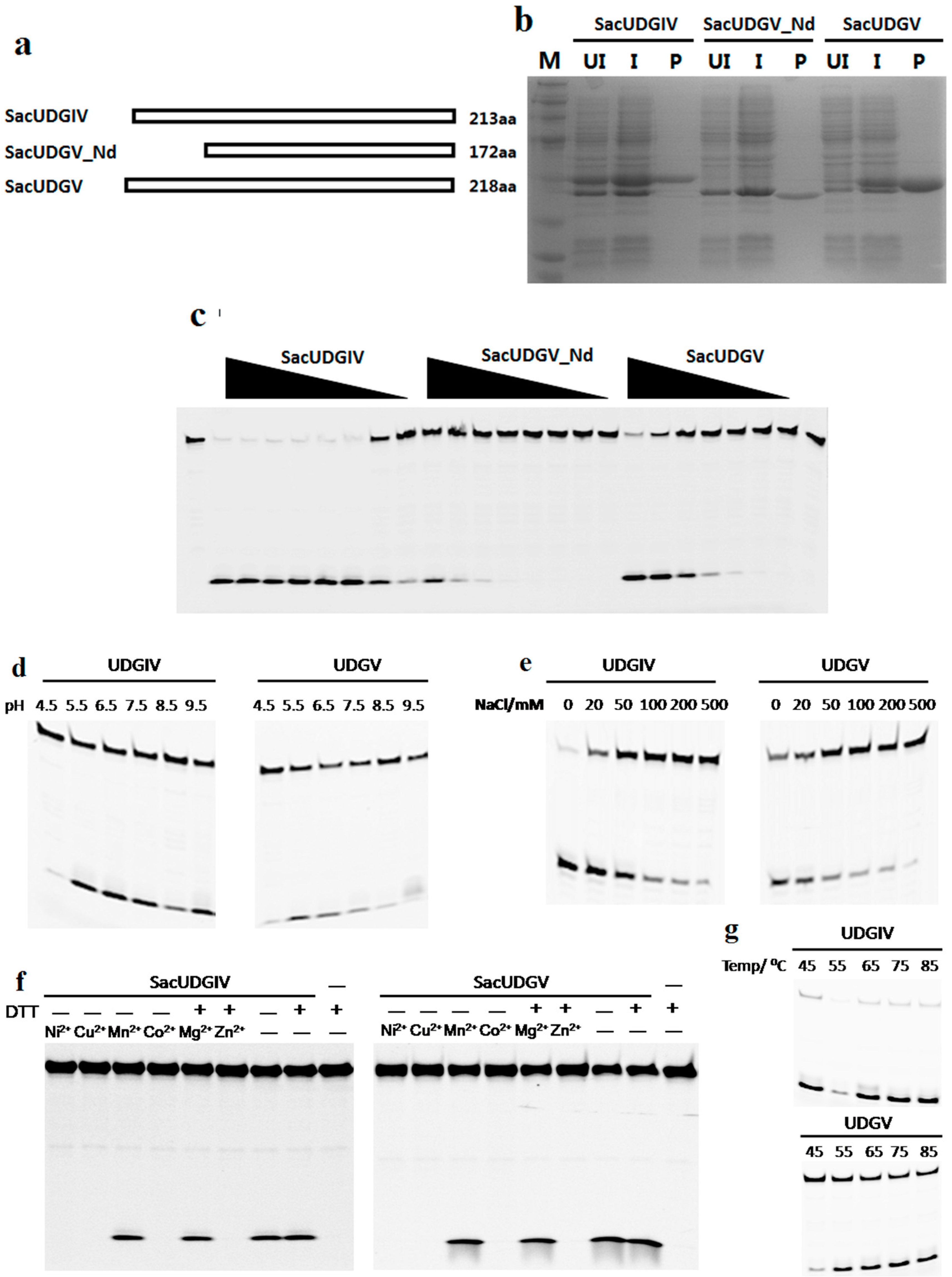
2.2. Expression and Purification of Recombinant UDGs
2.3. Biochemical Characterization of SacUDGs
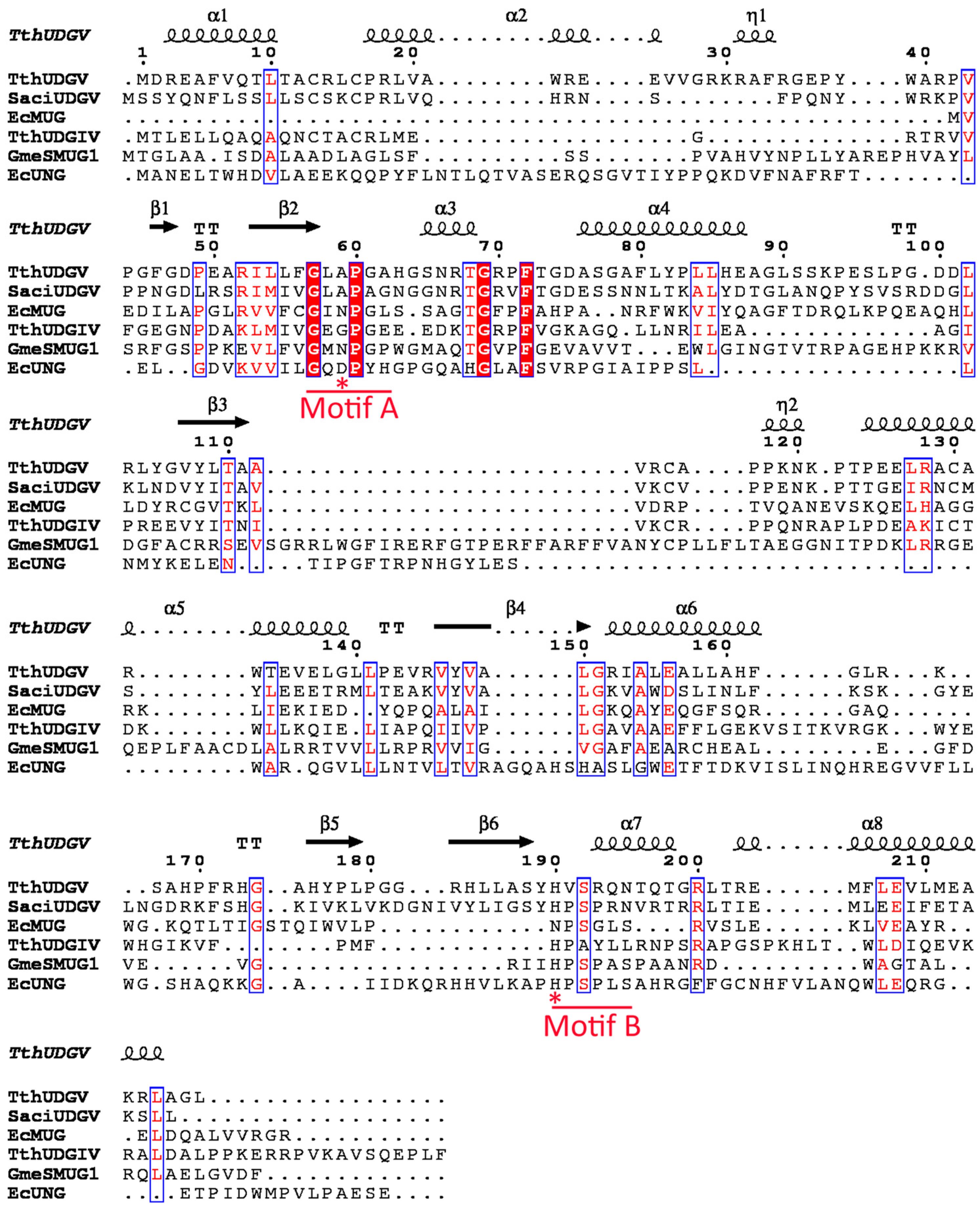
2.4. Multialignment and Phylogenetic Analysis
3. Results
3.1. Family 4 UDG Is More Widely Distributed than Family 5
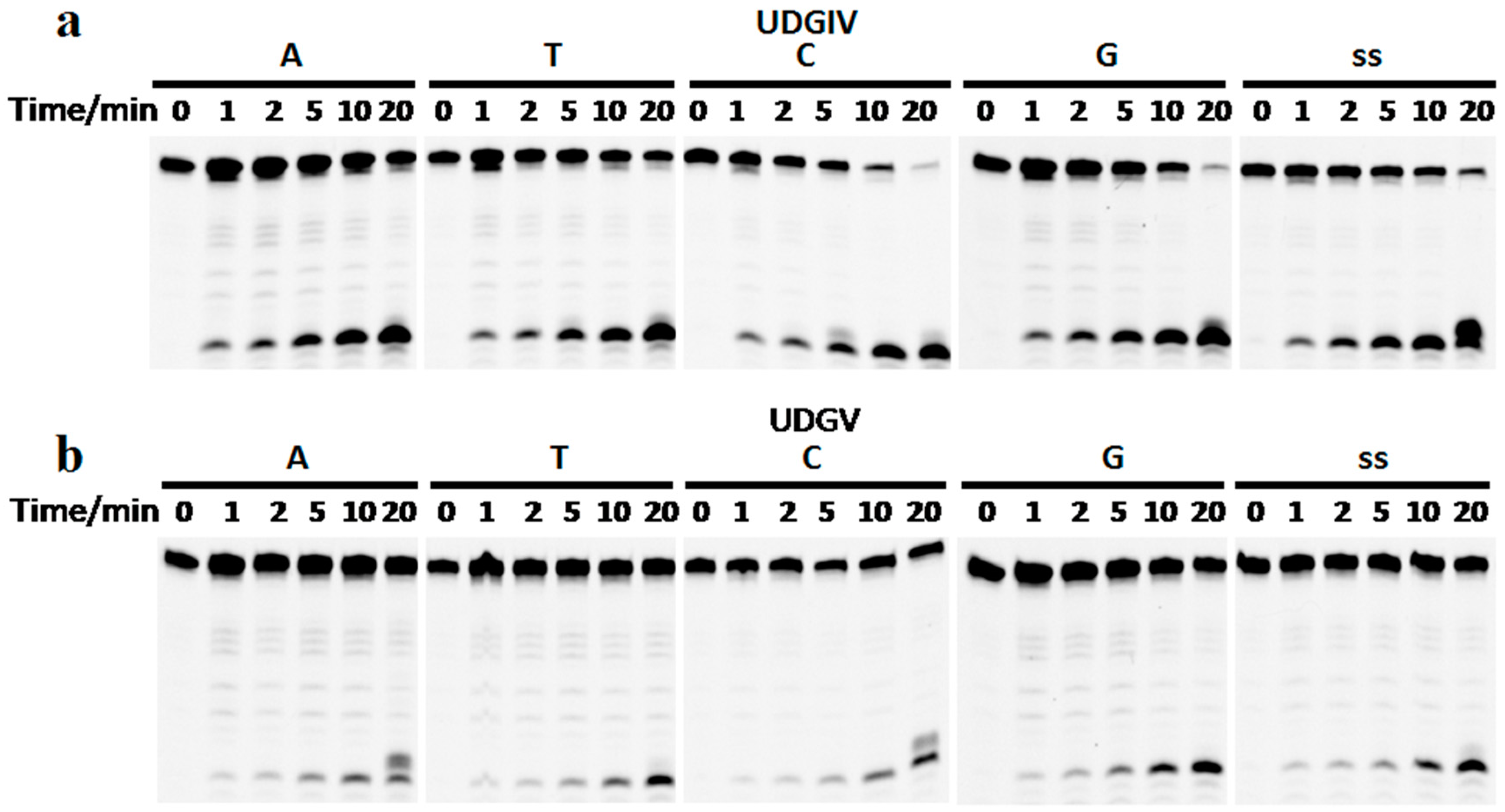
3.2. Both SacUDGs Are Uracil-DNA Glycosylases
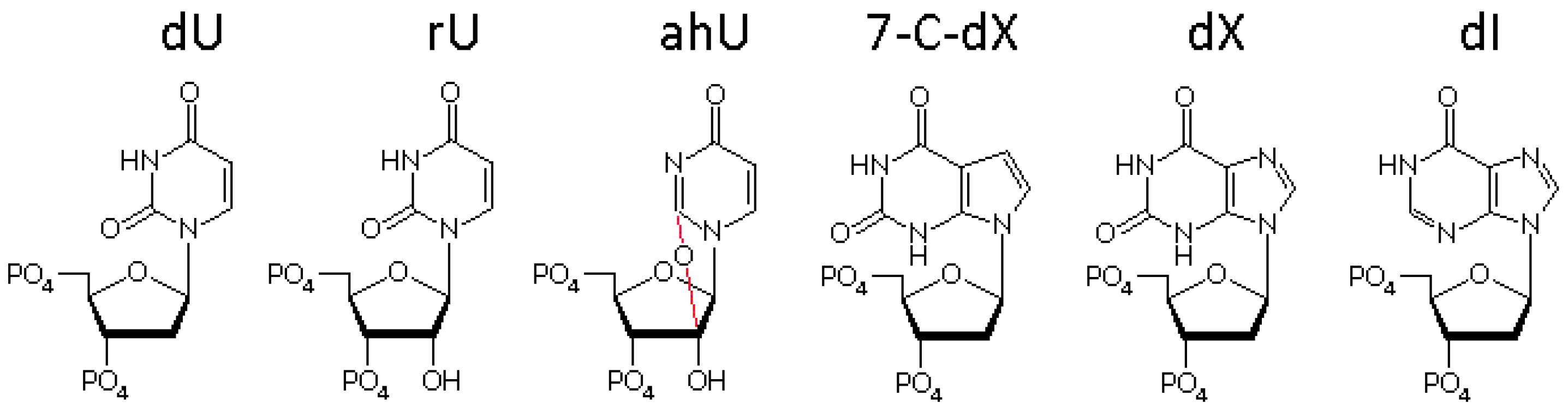
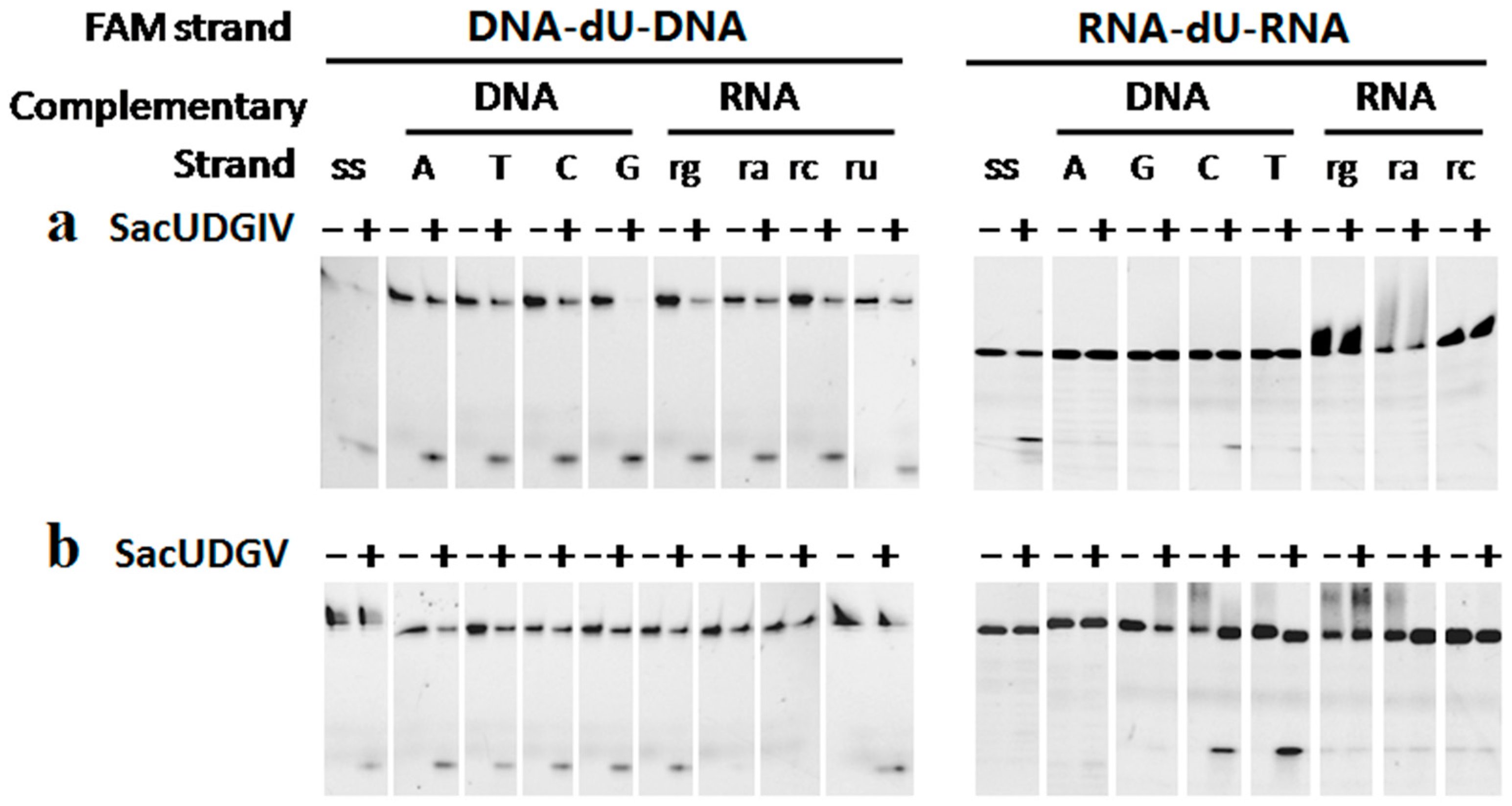
3.3. The Bases Opposite dU Have Little Effect on the Removal of dU
3.4. The Ribose Backbone Decreases dU Removal by UDGs
3.5. Effects of N-Terminal Sequence on UDGV’s Activity
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Suzuki, T.; Yamaoka, R.; Nishi, M.; Ide, H.; Makino, K. Isolation and characterization of a novel product, 2′-deoxyoxanosine, from 2′-deoxyguanosine, oligodeoxynucleotide and calf thymus DNA treated by nitrous-acid and nitric-oxide. J. Am. Chem. Soc. 1996, 118, 2515–2516. [Google Scholar] [CrossRef]
- Lindahl, T.; Nyberg, B. Heat-induced deamination of cytosine residues in deoxyribonucleic acid. Biochemistry 1974, 13, 3405–3410. [Google Scholar] [CrossRef] [PubMed]
- Hogrefe, H.H.; Hansen, C.J.; Scott, B.R.; Nielson, K.B. Archaeal dUTPase enhances PCR amplifications with archaeal DNA polymerases by preventing dUTP incorporation. Proc. Natl. Acad. Sci. USA 2002, 99, 596–601. [Google Scholar] [CrossRef] [PubMed]
- Björnberg, O.; Neuhard, J.; Nyman, P.O. A bifunctional dCTP deaminase-dUTP nucleotidohydrolase from the hyperthermophilic archaeon Methanocaldococcus jannaschii. J. Biol. Chem. 2003, 278, 20667–20672. [Google Scholar] [CrossRef] [PubMed]
- Pearl, L.H. Structure and function in the uracil-DNA glycosylase superfamily. Mutat. Res. 2000, 460, 165–181. [Google Scholar] [CrossRef]
- Sartori, A.A.; Fitz-Gibbon, S.; Yang, H.; Miller, J.H.; Jiricny, J. A novel uracil-DNA glycosylase with broad substrate specificity and an unusual active site. EMBO J. 2002, 21, 3182–3191. [Google Scholar] [CrossRef] [PubMed]
- Kuraoka, I. Diversity of endonuclease V: From DNA repair to RNA editing. Biomolecules 2015, 5, 2194–2206. [Google Scholar] [CrossRef] [PubMed]
- Georg, J.; Schomacher, L.; Chong, J.P.; Majerník, A.I.; Raabe, M.; Urlaub, H.; Müller, S.; Ciirdaeva, E.; Kramer, W.; Fritz, H.J. The Methanothermobacter thermautotrophicus ExoIII homologue Mth212 is a DNA uridine endonuclease. Nucleic Acids Res. 2006, 34, 5325–5336. [Google Scholar] [CrossRef] [PubMed]
- Greagg, M.A.; Fogg, M.J.; Panayotou, G.; Evans, S.J.; Connolly, B.A.; Pearl, L.H. A read-ahead function in archaeal DNA polymerases detects promutagenic template-strand uracil. Proc. Natl. Acad. Sci. USA 1999, 96, 9045–9050. [Google Scholar] [CrossRef] [PubMed]
- Lucas-Lledó, J.I.; Maddamsetti, R.; Lynch, M. Phylogenomic analysis of the uracil-DNA glycosylase superfamily. Mol. Biol. Evol. 2011, 28, 1307–1317. [Google Scholar] [CrossRef] [PubMed]
- Sang, P.B.; Srinath, T.; Patil, A.G.; Woo, E.J.; Varshney, U. A unique uracil-DNA binding protein of the uracil DNA glycosylase superfamily. Nucleic Acids Res. 2015, 43, 8452–8463. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.W.; Dominy, B.N.; Cao, W. New family of deamination repair enzymes in uracil-DNA glycosylase superfamily. J. Biol. Chem. 2011, 286, 31282–31287. [Google Scholar] [CrossRef] [PubMed]
- Stivers, J.T.; Pankiewicz, K.W.; Watanabe, K.A. Kinetic mechanism of damage site recognition and uracil flipping by Escherichia coli uracil DNA glycosylase. Biochemistry 1999, 38, 952–963. [Google Scholar] [CrossRef] [PubMed]
- Gallinari, P.; Jiricny, J. A new class of uracil-DNA glycosylases related to human thymine-DNA glycosylase. Nature 1996, 383, 735–738. [Google Scholar] [CrossRef] [PubMed]
- Mi, R.; Dong, L.; Kaulgud, T.; Hackett, K.W.; Dominy, B.N.; Cao, W. Insights from xanthine and uracil DNA glycosylase activities of bacterial and human SMUG1: Switching SMUG1 to UDG. J. Mol. Biol. 2009, 385, 761–778. [Google Scholar] [CrossRef] [PubMed]
- Hinks, J.A.; Evans, M.C.; De Miguel, Y.; Sartori, A.A.; Jiricny, J.; Pearl, L.H. An iron-sulfur cluster in the family 4 uracil-DNA glycosylases. J. Biol. Chem. 2002, 277, 16936–16940. [Google Scholar] [CrossRef] [PubMed]
- Aravind, L.; Koonin, E.V. The alpha/beta fold uracil DNA glycosylases: A common origin with diverse fates. Genome Biol. 2000. [Google Scholar] [CrossRef] [PubMed]
- Slupphaug, G.; Mol, C.D.; Kavli, B.; Arvai, A.S.; Krokan, H.E.; Tainer, J.A. A nucleotide-flipping mechanism from the structure of human uracil-DNA glycosylase bound to DNA. Nature 1996, 384, 87–92. [Google Scholar] [CrossRef] [PubMed]
- Mol, D.C.; Arvai, A.S.; Begley, T.J.; Cunningham, R.P.; Tainer, J.A. Structure and activity of a thermostable thymine-DNA glycosylase: Evidence for base twisting to remove mismatched normal DNA bases. J. Mol. Biol. 2002, 315, 373–384. [Google Scholar] [CrossRef] [PubMed]
- Thayer, M.M.; Ahern, H.; Xing, D.; Cunningham, R.P.; Tainer, J.A. Novel DNA binding motifs in the DNA repair enzyme endonuclease III crystal. EMBO J. 1995, 14, 4108–4120. [Google Scholar] [PubMed]
- Trasviña-Arenas, C.H.; Lopez-Castillo, L.M.; Sanchez-Sandoval, E.; Brieba, L.G. Dispensability of the [4Fe-4S] cluster in novel homologues of adenine glycosylase MutY. FEBS J. 2016, 283, 521–540. [Google Scholar] [CrossRef] [PubMed]
- Constantinescu-Aruxandei, D.; Petrovic-Stojanovska, B.; Penedo, J.C.; White, M.F.; Naismith, J.H. Mechanism of DNA loading by the DNA repair helicase XPD. Nucleic Acids Res. 2016, 44, 2806–2815. [Google Scholar] [CrossRef] [PubMed]
- Kiyonari, S.; Uchimura, M.; Shirai, T.; Ishino, Y. Physical and functional interactions between uracil-DNA glycosylase and proliferating cell nuclear antigen from the euryarchaeon Pyrococcus furiosus. J. Biol. Chem. 2008, 283, 24185–24193. [Google Scholar] [CrossRef] [PubMed]
- Engstrom, L.M.; Partington, O.A.; David, S.S. An iron-sulfur cluster loop motif in the Archaeoglobus fulgidus uracil-DNA glycosylase mediates efficient uracil recognition and removal. Biochemistry 2012, 51, 5187–5197. [Google Scholar] [CrossRef] [PubMed]
- Hoseki, J.; Okamoto, A.; Masui, R.; Shibata, T.; Inoue, Y.; Yokoyama, S.; Kuramitsu, S. Crystal structure of a family 4 uracil-DNA glycosylase from Thermus thermophilus HB8. J. Mol. Biol. 2003, 333, 515–526. [Google Scholar] [CrossRef] [PubMed]
- Rogers, P.A.; Eide, L.; Klungland, A.; Ding, H. Reversible inactivation of E. coli endonuclease III via modification of its [4Fe-4S] cluster by nitric oxide. DNA Repair 2003, 2, 809–817. [Google Scholar] [CrossRef]
- Malshetty, V.S.; Jain, R.; Srinath, T.; Kurthkoti, K.; Varshney, U. Synergistic effects of UdgB and Ung in mutation prevention and protection against commonly encountered DNA damaging agents in Mycobacterium smegmatis. Microbiology 2010, 156, 940–949. [Google Scholar] [CrossRef] [PubMed]
- Lari, S.U.; Chen, C.Y.; Vertéssy, B.G.; Morré, J.; Bennett, S.E. Quantitative determination of uracil residues in Escherichia coli DNA: Contribution of ung, dug, and dut genes to uracil avoidance. DNA Repair 2006, 5, 1407–1420. [Google Scholar] [CrossRef] [PubMed]
- Sakai, T.; Tokishita, S.; Mochizuki, K.; Motomiya, A.; Yamagata, H.; Ohta, T. Mutagenesis of uracil-DNA glycosylase deficient mutants of the extremely thermophilic eubacterium Thermus thermophilus. DNA Repair 2008, 7, 663–669. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Brügger, K.; Skovgaard, M.; Redder, P.; She, Q.; Torarinsson, E.; Greve, B.; Awayez, M.; Zibat, A.; Klenk, H.P.; et al. The Genome of Sulfolobus acidocaldarius, a Model Organism of the Crenarchaeota. J. Bacteriol. 2005, 187, 4992–4999. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.P.; Liu, J.H. The terminal 5′ phosphate and proximate phosphorothioate promote ligation-independent cloning. Protein Sci. 2010, 19, 967–973. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Liu, X.P.; Han, Z.; Allers, T.; Hou, J.L.; Liu, J.H. RecJ-like protein from Pyrococcus furiosus has 3′-5′ exonuclease activity on RNA: Implication of its proofreading capacity on 3′-mismatched RNA primer in DNA replication. Nucleic Acids Res. 2013, 41, 5817–5826. [Google Scholar] [CrossRef] [PubMed]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef] [PubMed]
- Kosaka, H.; Hoseki, J.; Nakagawa, N.; Kuramitsu, S.; Masui, R. Crystal structure of family 5 uracil-DNA glycosylase bound to DNA. J. Mol. Biol. 2007, 373, 839–850. [Google Scholar] [CrossRef] [PubMed]
- Parikh, S.S.; Walcher, G.; Jones, G.D.; Slupphaug, G.; Krokan, H.E.; Blackburn, G.M.; Tainer, J.A. Uracil-DNA glycosylase-DNA substrate and product structures: Conformational strain promotes catalytic efficiency by coupled stereoelectronic effects. Proc. Natl. Acad. Sci. USA 2000, 97, 5083–5088. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.W.; Brice, A.R.; Wright, C.B.; Dominy, B.N.; Cao, W. Identification of Escherichia coli mismatch-specific uracil DNA glycosylase as a robust xanthine DNA glycosylase. J. Biol. Chem. 2010, 285, 41483–41490. [Google Scholar] [CrossRef] [PubMed]
- Lindahl, T. Instability and decay of the primary structure of DNA. Nature 1993, 362, 709–715. [Google Scholar] [CrossRef] [PubMed]
- Kamiya, H. Mutagenic potentials of damaged nucleic acids produced by reactive oxygen/nitrogen species: Approaches using synthetic oligonucleotides and nucleotides: Survey and summary. Nucleic Acids Res. 2003, 31, 517–531. [Google Scholar] [CrossRef] [PubMed]
- Malik, S.S.; Coey, C.T.; Varney, K.M.; Pozharski, E.; Drohat, A.C. Thymine DNA glycosylase exhibits negligible affinity for nucleobases that it removes from DNA. Nucleic Acids Res. 2015, 43, 9541–9552. [Google Scholar] [CrossRef] [PubMed]
- Shiraishi, M.; Ishino, S.; Yamagami, T.; Egashira, Y.; Kiyonari, S.; Ishino, Y. A novel endonuclease that may be responsible for damaged DNA base repair in Pyrococcus furiosus. Nucleic Acids Res. 2015, 43, 2853–2863. [Google Scholar] [CrossRef] [PubMed]
- Kavli, B.; Otterlei, M.; Slupphaug, G.; Krokan, H.E. Uracil in DNA—general mutagen, but normal intermediate in acquired immunity. DNA Repair 2007, 6, 505–516. [Google Scholar] [CrossRef] [PubMed]

© 2017 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yi, G.-S.; Wang, W.-W.; Cao, W.-G.; Wang, F.-P.; Liu, X.-P. Sulfolobus acidocaldarius UDG Can Remove dU from the RNA Backbone: Insight into the Specific Recognition of Uracil Linked with Deoxyribose. Genes 2017, 8, 38. https://doi.org/10.3390/genes8010038
Yi G-S, Wang W-W, Cao W-G, Wang F-P, Liu X-P. Sulfolobus acidocaldarius UDG Can Remove dU from the RNA Backbone: Insight into the Specific Recognition of Uracil Linked with Deoxyribose. Genes. 2017; 8(1):38. https://doi.org/10.3390/genes8010038
Chicago/Turabian StyleYi, Gang-Shun, Wei-Wei Wang, Wei-Guo Cao, Feng-Ping Wang, and Xi-Peng Liu. 2017. "Sulfolobus acidocaldarius UDG Can Remove dU from the RNA Backbone: Insight into the Specific Recognition of Uracil Linked with Deoxyribose" Genes 8, no. 1: 38. https://doi.org/10.3390/genes8010038





