Regulation of DNA Replication in Early Embryonic Cleavages
Abstract
:1. Introduction
2. Onset of S-Phase in the Fertilized Egg
2.1. Developmental Regulation of DNA Replication Origin Usage
2.2. Assembly of Replication Forks in Early Embryos
2.3. Once-Per-Cell Cycle Regulation of DNA Replication in Early Embryos
3. Positive and Negative Regulation of Replication Initiation by S-CDKs and CHK1
4. DNA Replication-Dependent Inheritance of Epigenetic Marks: Methylation Program
5. Mechanisms Leading to S-Phase Lengthening at the Mid Blastula Transition
5.1. Similarities and Differences between Different Organisms
5.1.1. Drosophila melanogaster
5.1.2. Xenopus laevis
5.1.3. Zebrafish
5.1.4. Mammals
5.2. The Role of CDKs
5.3. The Role of the Replication Checkpoint
5.4. The Role of Zygotic Transcription Activation
6. Consequences of Fast Replication and Absence of Checkpoint Activation on Early Embryos Genome Integrity
7. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| 53BP | p53 binding protein |
| APC | anaphase-promoting complex |
| ATM | ataxia telangiectasia mutated |
| ATR | ataxia telangiectasia mutated- and Rad3-related |
| ATRIP | ATR-interacting protein |
| CCR4 | C-C chemokine receptor type 4 |
| CDC | cell cycle division |
| CDK | Cyclin Dependent Kinase |
| CDT | CDC10-dependent transcription |
| CHK | Checkpoint kinase |
| CMG | CDC45-MCM-GINS |
| DBF | dumbbell factor |
| DDB | DNA damage binding |
| DDK | DBF4-dependent kinase |
| DNA-PK | DNA-dependent protein kinase |
| DRF | dumbell-related factor |
| DUE | DNA unwinding element |
| Gy | Gray |
| kb | kilobase |
| MCM | mini chromosome maintenance |
| MMS | methyl methane sulphonate |
| mUb | monoubiquitination |
| NOT | Negative Regulator Of Transcription 1 |
| ORC | origin recognition complex |
| PCNA | proliferating cell nuclear antigen |
| POP | posterior pharynx defect protein |
| RPA | replication protein A |
| RF-C | replication factor C |
| Rad | Radiation sensitive |
| ssDNA | single stranded DNA |
| Sld | synthetic lethal with Dpb11 |
| Ticrr | TopBP1 interacting checkpoint and replication regulator |
| TopBP1 | Topoisomerase II binding protein 1 |
| 9-1-1 | Rad9-Rad1-Hus1 |
References
- Yasuda, G.K.; Schubiger, G. Temporal regulation in the early embryo: Is mbt too good to be true? Trends Genet. 1992, 8, 124–127. [Google Scholar] [CrossRef]
- Hormanseder, E.; Tischer, T.; Mayer, T.U. Modulation of cell cycle control during oocyte-to-embryo transitions. EMBO J. 2013, 32, 2191–2203. [Google Scholar] [CrossRef] [PubMed]
- Farrell, J.A.; O’Farrell, P.H. From egg to gastrula: How the cell cycle is remodeled during the Drosophila mid-blastula transition. Annu. Rev. Genet. 2014, 48, 269–294. [Google Scholar] [CrossRef] [PubMed]
- Graham, C.F.; Morgan, R.W. Changes in the cell cycle during early amphibian development. Dev. Biol. 1966, 14, 439–460. [Google Scholar] [CrossRef]
- Blumenthal, A.B.; Kriegstein, H.J.; Hogness, D.S. The units of DNA replication in Drosophila melanogaster chromosomes. Cold Spring Harb. Symp. Quant. Biol. 1974, 38, 205–223. [Google Scholar] [CrossRef] [PubMed]
- Longo, F.J.; Anderson, E. The fine structure of pronuclear development and fusion in the sea urchin, Arbacia punctulata. J. Cell Biol. 1968, 39, 339–368. [Google Scholar] [CrossRef] [PubMed]
- Strome, S.; Wood, W.B. Generation of asymmetry and segregation of germ-line granules in early C. elegans embryos. Cell 1983, 35, 15–25. [Google Scholar] [CrossRef]
- Ubbels, G.A.; Hara, K.; Koster, C.H.; Kirschner, M.W. Evidence for a functional role of the cytoskeleton in determination of the dorsoventral axis in Xenopus laevis eggs. J. Embryol. Exp. Morphol. 1983, 77, 15–37. [Google Scholar] [PubMed]
- Das, N.K.; Barker, C. Mitotic chromosome condensation in the sperm nucleus during postfertilization maturation division in urechis eggs. J. Cell Biol. 1976, 68, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Ciemerych, M.A.; Czolowska, R. Differential chromatin condensation of female and male pronuclei in mouse zygotes. Mol. Reprod. Dev. 1993, 34, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Mayer, W.; Smith, A.; Fundele, R.; Haaf, T. Spatial separation of parental genomes in preimplantation mouse embryos. J. Cell Biol. 2000, 148, 629–634. [Google Scholar] [CrossRef] [PubMed]
- Bomar, J.; Moreira, P.; Balise, J.J.; Collas, P. Differential regulation of maternal and paternal chromosome condensation in mitotic zygotes. J. Cell Sci. 2002, 115, 2931–2940. [Google Scholar] [PubMed]
- Blow, J.J.; Laskey, R.A. Xenopus cell-free extracts and their contribution to the study of DNA replication and other complex biological processes. Int. J. Dev. Biol. 2016, 60, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Anderson, K.V.; Lengyel, J.A. Rates of synthesis of major classes of RNA in Drosophila embryos. Dev. Biol. 1979, 70, 217–231. [Google Scholar] [CrossRef]
- Newport, J.; Kirschner, M. A major developmental transition in early Xenopus embryos: I. Characterization and timing of cellular changes at the midblastula stage. Cell 1982, 30, 675–686. [Google Scholar] [CrossRef]
- Anderson, K.V.; Lengyel, J.A. Changing rates of histone mrna synthesis and turnover in Drosophila embryos. Cell 1980, 21, 717–727. [Google Scholar] [CrossRef]
- Adamson, E.D.; Woodland, H.R. Histone synthesis in early amphibian development: Histone and DNA syntheses are not co-ordinated. J. Mol. Biol. 1974, 88, 263–285. [Google Scholar] [CrossRef]
- Sansam, C.G.; Goins, D.; Siefert, J.C.; Clowdus, E.A.; Sansam, C.L. Cyclin-dependent kinase regulates the length of s phase through TICRR/TRESLIN phosphorylation. Genes Dev. 2015, 29, 555–566. [Google Scholar] [CrossRef] [PubMed]
- Fragkos, M.; Ganier, O.; Coulombe, P.; Mechali, M. DNA replication origin activation in space and time. Nat. Rev. Mol. Cell Biol. 2015, 16, 360–374. [Google Scholar] [CrossRef] [PubMed]
- Bell, S.P.; Labib, K. Chromosome duplication in saccharomyces cerevisiae. Genetics 2016, 203, 1027–1067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hyrien, O.; Maric, C.; Mechali, M. Transition in specification of embryonic metazoan DNA replication origins. Science 1995, 270, 994–997. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, T.; Sawado, T.; Yamaguchi, M.; Shinomiya, T. Specification of regions of DNA replication initiation during embryogenesis in the 65-kilobase DNApolalpha-dE2F locus of Drosophila melanogaster. Mol. Cell. Biol. 1999, 19, 547–555. [Google Scholar] [CrossRef] [PubMed]
- Laskey, R.A. Chromosome replication in early development of Xenopus laevis. J. Embryol. Exp. Morphol. 1985, 89, 285–296. [Google Scholar] [PubMed]
- Mahbubani, H.M.; Paull, T.; Elder, J.K.; Blow, J.J. DNA replication initiates at multiple sites on plasmid DNA in Xenopus egg extracts. Nucleic Acids Res. 1992, 20, 1457–1462. [Google Scholar] [CrossRef] [PubMed]
- Hyrien, O.; Mechali, M. Chromosomal replication initiates and terminates at random sequences but at regular intervals in the ribosomal DNA of Xenopus early embryos. EMBO J. 1993, 12, 4511–4520. [Google Scholar] [PubMed]
- Walter, J.; Newport, J.W. Regulation of replicon size in Xenopus egg extracts. Science 1997, 275, 993–995. [Google Scholar] [CrossRef] [PubMed]
- Cayrou, C.; Coulombe, P.; Vigneron, A.; Stanojcic, S.; Ganier, O.; Peiffer, I.; Rivals, E.; Puy, A.; Laurent-Chabalier, S.; Desprat, R.; et al. Genome-scale analysis of metazoan replication origins reveals their organization in specific but flexible sites defined by conserved features. Genome Res. 2011, 21, 1438–1449. [Google Scholar] [CrossRef] [PubMed]
- Hartwell, L.H.; Weinert, T.A. Checkpoints: Controls that ensure the order of cell cycle events. Science 1989, 246, 629–634. [Google Scholar] [CrossRef] [PubMed]
- Newport, J.; Kirschner, M. A major developmental transition in early Xenopus embryos: II. Control of the onset of transcription. Cell 1982, 30, 687–696. [Google Scholar] [CrossRef]
- Raff, J.W.; Glover, D.M. Nuclear and cytoplasmic mitotic cycles continue in Drosophila embryos in which DNA synthesis is inhibited with aphidicolin. J. Cell Biol. 1988, 107, 2009–2019. [Google Scholar] [CrossRef] [PubMed]
- Marheineke, K.; Hyrien, O. Control of replication origin density and firing time in Xenopus egg extracts: Role of a caffeine-sensitive, atr-dependent checkpoint. J. Biol. Chem. 2004, 279, 28071–28081. [Google Scholar] [CrossRef] [PubMed]
- Ahuja, A.K.; Jodkowska, K.; Teloni, F.; Bizard, A.H.; Zellweger, R.; Herrador, R.; Ortega, S.; Hickson, I.D.; Altmeyer, M.; Mendez, J.; et al. A short G1 phase imposes constitutive replication stress and fork remodelling in mouse embryonic stem cells. Nat. Commun. 2016. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, T.S.; Walter, J.C. CDC7-DRF1 is a developmentally regulated protein kinase required for the initiation of vertebrate DNA replication. Genes Dev. 2005, 19, 2295–2300. [Google Scholar] [CrossRef] [PubMed]
- Collart, C.; Allen, G.E.; Bradshaw, C.R.; Smith, J.C.; Zegerman, P. Titration of four replication factors is essential for the Xenopus laevis midblastula transition. Science 2013, 341, 893–896. [Google Scholar] [CrossRef] [PubMed]
- Ciemerych, M.A.; Maro, B.; Kubiak, J.Z. Control of duration of the first two mitoses in a mouse embryo. Zygote 1999, 7, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Recolin, B.; van der Laan, S.; Tsanov, N.; Maiorano, D. Molecular mechanisms of DNA replication checkpoint activation. Genes (Basel) 2014, 5, 147–175. [Google Scholar] [CrossRef] [PubMed]
- Adachi, Y.; Laemmli, U.K. Study of the cell cycle-dependent assembly of the DNA pre-replication centres in Xenopus egg extracts. EMBO J. 1994, 13, 4153–4164. [Google Scholar] [PubMed]
- Hashimoto, Y.; Takisawa, H. Xenopus Cut5 is essential for a CDK-dependent process in the initiation of DNA replication. EMBO J. 2003, 22, 2526–2535. [Google Scholar] [CrossRef] [PubMed]
- Ge, X.Q.; Han, J.; Cheng, E.C.; Yamaguchi, S.; Shima, N.; Thomas, J.L.; Lin, H. Embryonic stem cells license a high level of dormant origins to protect the genome against replication stress. Stem Cell Rep. 2015, 5, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Sonneville, R.; Craig, G.; Labib, K.; Gartner, A.; Blow, J.J. Both chromosome decondensation and condensation are dependent on DNA replication in C. elegans embryos. Cell Rep. 2015, 12, 405–417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, T.S.; Yiu, P.; Chou, M.F.; Gygi, S.; Walter, J.C. Recruitment of Xenopus Scc2 and cohesin to chromatin requires the pre-replication complex. Nat. Cell Biol. 2004, 6, 991–996. [Google Scholar] [CrossRef] [PubMed]
- Shinya, M.; Machiki, D.; Henrich, T.; Kubota, Y.; Takisawa, H.; Mimura, S. Evolutionary diversification of MCM3 genes in Xenopus laevis and danio rerio. Cell Cycle 2014, 13, 3271–3281. [Google Scholar] [CrossRef] [PubMed]
- Sible, J.C.; Erikson, E.; Hendrickson, M.; Maller, J.L.; Gautier, J. Developmental regulation of MCM replication factors in Xenopus laevis. Curr. Biol. 1998, 8, 347–350. [Google Scholar] [CrossRef]
- Tikhmyanova, N.; Coleman, T.R. Isoform switching of Cdc6 contributes to developmental cell cycle remodeling. Dev. Biol. 2003, 260, 362–375. [Google Scholar] [CrossRef]
- Silva, T.; Bradley, R.H.; Gao, Y.; Coue, M. Xenopus Cdc7/Drf1 complex is required for the initiation of DNA replication. J. Biol. Chem. 2006, 281, 11569–11576. [Google Scholar] [CrossRef] [PubMed]
- Blow, J.J.; Laskey, R.A. Initiation of DNA replication in nuclei and purified DNA by a cell-free extract of Xenopus eggs. Cell 1986, 47, 577–587. [Google Scholar] [CrossRef]
- Maiorano, D.; Krasinska, L.; Lutzmann, M.; Mechali, M. Recombinant Cdt1 induces rereplication of G2 nuclei in Xenopus egg extracts. Curr. Biol. 2005, 15, 146–153. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Takisawa, H.; Kubota, Y. Intrinsic nuclear import activity of geminin is essential to prevent re-initiation of DNA replication in Xenopus eggs. Genes Cells 2005, 10, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Arias, E.E.; Walter, J.C. Replication-dependent destruction of Cdt1 limits DNA replication to a single round per cell cycle in Xenopus egg extracts. Genes Dev. 2005, 19, 114–126. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Blow, J.J. Cdt1 downregulation by proteolysis and geminin inhibition prevents DNA re-replication in Xenopus. EMBO J. 2005, 24, 395–404. [Google Scholar] [CrossRef] [PubMed]
- Arias, E.E.; Walter, J.C. Pcna functions as a molecular platform to trigger Cdt1 destruction and prevent re-replication. Nat. Cell. Biol. 2006, 8, 84–90. [Google Scholar] [CrossRef] [PubMed]
- Wohlschlegel, J.A.; Dwyer, B.T.; Dhar, S.K.; Cvetic, C.; Walter, J.C.; Dutta, A. Inhibition of eukaryotic DNA replication by geminin binding to Cdt1. Science 2000, 290, 2309–2312. [Google Scholar] [CrossRef] [PubMed]
- Tada, S.; Li, A.; Maiorano, D.; Mechali, M.; Blow, J.J. Repression of origin assembly in metaphase depends on inhibition of RLF-B/Cdt1 by geminin. Nat. Cell. Biol. 2001, 3, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.Y.; Kaneko, K.J.; Pan, H.; DePamphilis, M.L. Geminin is essential to prevent DNA re-replication-dependent apoptosis in pluripotent cells, but not in differentiated cells. Stem Cells 2015, 33, 3239–3253. [Google Scholar] [CrossRef] [PubMed]
- McGarry, T.J.; Kirschner, M.W. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell 1998, 93, 1043–1053. [Google Scholar] [CrossRef]
- Hodgson, B.; Li, A.; Tada, S.; Blow, J.J. Geminin becomes activated as an inhibitor of Cdt1/rlf-b following nuclear import. Curr. Biol. 2002, 12, 678–683. [Google Scholar] [CrossRef]
- Maiorano, D.; Rul, W.; Mechali, M. Cell cycle regulation of the licensing activity of Cdt1 in Xenopus laevis. Exp. Cell Res. 2004, 295, 138–149. [Google Scholar] [CrossRef] [PubMed]
- Kisielewska, J.; Blow, J.J. Dynamic interactions of high Cdt1 and geminin levels regulate S phase in early Xenopus embryos. Development 2012, 139, 63–74. [Google Scholar] [CrossRef] [PubMed]
- Lutzmann, M.; Maiorano, D.; Mechali, M. A Cdt1-geminin complex licenses chromatin for DNA replication and prevents rereplication during s phase in Xenopus. EMBO J. 2006, 25, 5764–5774. [Google Scholar] [CrossRef] [PubMed]
- Chevalier, S.; Tassan, J.P.; Cox, R.; Philippe, M.; Ford, C. Both Cdc2 and Cdk2 promote S phase initiation in Xenopus egg extracts. J. Cell Sci. 1995, 108 Pt 5, 1831–1841. [Google Scholar] [PubMed]
- Blow, J.J.; Nurse, P. A Cdc2-like protein is involved in the initiation of DNA replication in Xenopus egg extracts. Cell 1990, 62, 855–862. [Google Scholar] [CrossRef]
- Murray, A.W.; Kirschner, M.W. Cyclin synthesis drives the early embryonic cell cycle. Nature 1989, 339, 275–280. [Google Scholar] [CrossRef] [PubMed]
- Hartley, R.S.; Rempel, R.E.; Maller, J.L. In vivo regulation of the early embryonic cell cycle in Xenopus. Dev. Biol. 1996, 173, 408–419. [Google Scholar] [CrossRef] [PubMed]
- Howe, J.A.; Howell, M.; Hunt, T.; Newport, J.W. Identification of a developmental timer regulating the stability of embryonic cyclin A and a new somatic A-type cyclin at gastrulation. Genes Dev. 1995, 9, 1164–1176. [Google Scholar] [CrossRef] [PubMed]
- Gotoh, T.; Shigemoto, N.; Kishimoto, T. Cyclin E2 is required for embryogenesis in Xenopus laevis. Dev. Biol. 2007, 310, 341–347. [Google Scholar] [CrossRef] [PubMed]
- Geng, Y.; Yu, Q.; Sicinska, E.; Das, M.; Schneider, J.E.; Bhattacharya, S.; Rideout, W.M.; Bronson, R.T.; Gardner, H.; Sicinski, P. Cyclin E ablation in the mouse. Cell 2003, 114, 431–443. [Google Scholar] [CrossRef]
- Ortega, S.; Prieto, I.; Odajima, J.; Martin, A.; Dubus, P.; Sotillo, R.; Barbero, J.L.; Malumbres, M.; Barbacid, M. Cyclin-dependent kinase 2 is essential for meiosis but not for mitotic cell division in mice. Nat. Genet. 2003, 35, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Fang, F.; Newport, J.W. Evidence that the G1-S and G2-M transitions are controlled by different Cdc2 proteins in higher eukaryotes. Cell 1991, 66, 731–742. [Google Scholar] [CrossRef]
- Jackson, P.K.; Chevalier, S.; Philippe, M.; Kirschner, M.W. Early events in DNA replication require cyclin E and are blocked by p21CIP1. J. Cell Biol. 1995, 130, 755–769. [Google Scholar] [CrossRef] [PubMed]
- Strausfeld, U.P.; Howell, M.; Descombes, P.; Chevalier, S.; Rempel, R.E.; Adamczewski, J.; Maller, J.L.; Hunt, T.; Blow, J.J. Both cyclin A and cyclin E have S-phase promoting (SPF) activity in Xenopus egg extracts. J. Cell Sci. 1996, 109 Pt 6, 1555–1563. [Google Scholar]
- Fisher, D.; Nurse, P. A single fission yeast mitotic cyclin B-p34cdc2 kinase promotes both S-phase and mitosis in the absence of G1-cyclins. EMBO J. 1996, 15, 850–860. [Google Scholar]
- Moore, J.D.; Kirk, J.A.; Hunt, T. Unmasking the S-phase-promoting potential of cyclin B1. Science 2003, 300, 987–990. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Kumagai, A.; Schlacher, K.; Shevchenko, A.; Shevchenko, A.; Dunphy, W.G. Interaction of chk1 with treslin negatively regulates the initiation of chromosomal DNA replication. Mol. Cell 2015, 57, 492–505. [Google Scholar] [CrossRef] [PubMed]
- Oswald, J.; Engemann, S.; Lane, N.; Mayer, W.; Olek, A.; Fundele, R.; Dean, W.; Reik, W.; Walter, J. Active demethylation of the paternal genome in the mouse zygote. Curr. Biol. 2000, 10, 475–478. [Google Scholar] [CrossRef]
- Santos, F.; Hendrich, B.; Reik, W.; Dean, W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev. Biol. 2002, 241, 172–182. [Google Scholar] [CrossRef] [PubMed]
- Foe, V.E.; Alberts, B.M. Studies of nuclear and cytoplasmic behaviour during the five mitotic cycles that precede gastrulation in Drosophila embryogenesis. J. Cell Sci. 1983, 61, 31–70. [Google Scholar] [PubMed]
- McKnight, S.L.; Miller, O.L., Jr. Electron microscopic analysis of chromatin replication in the cellular blastoderm Drosophila melanogaster embryo. Cell 1977, 12, 795–804. [Google Scholar] [CrossRef]
- Shermoen, A.W.; McCleland, M.L.; O’Farrell, P.H. Developmental control of late replication and S phase length. Curr. Biol. 2010, 20, 2067–2077. [Google Scholar] [CrossRef] [PubMed]
- Yuan, K.; Shermoen, A.W.; O’Farrell, P.H. Illuminating DNA replication during Drosophila development using tale-lights. Curr. Biol. 2014, 24, R144–R145. [Google Scholar] [CrossRef] [PubMed]
- McCleland, M.L.; Shermoen, A.W.; O’Farrell, P.H. DNA replication times the cell cycle and contributes to the mid-blastula transition in Drosophila embryos. J. Cell Biol. 2009, 187, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Kimelman, D.; Kirschner, M.; Scherson, T. The events of the midblastula transition in Xenopus are regulated by changes in the cell cycle. Cell 1987, 48, 399–407. [Google Scholar] [CrossRef]
- Edgar, B.A.; Schubiger, G. Parameters controlling transcriptional activation during early Drosophila development. Cell 1986, 44, 871–877. [Google Scholar] [CrossRef]
- Kane, D.A.; Kimmel, C.B. The zebrafish midblastula transition. Development 1993, 119, 447–456. [Google Scholar] [PubMed]
- Kane, D.A.; Hammerschmidt, M.; Mullins, M.C.; Maischein, H.M.; Brand, M.; van Eeden, F.J.; Furutani-Seiki, M.; Granato, M.; Haffter, P.; Heisenberg, C.P.; et al. The zebrafish epiboly mutants. Development 1996, 123, 47–55. [Google Scholar] [PubMed]
- Dalle Nogare, D.E.; Pauerstein, P.T.; Lane, M.E. G2 acquisition by transcription-independent mechanism at the zebrafish midblastula transition. Dev. Biol. 2009, 326, 131–142. [Google Scholar] [CrossRef] [PubMed]
- Hamatani, T.; Ko, M.; Yamada, M.; Kuji, N.; Mizusawa, Y.; Shoji, M.; Hada, T.; Asada, H.; Maruyama, T.; Yoshimura, Y. Global gene expression profiling of preimplantation embryos. Hum. Cell. 2006, 19, 98–117. [Google Scholar] [CrossRef] [PubMed]
- Takashima, Y.; Guo, G.; Loos, R.; Nichols, J.; Ficz, G.; Krueger, F.; Oxley, D.; Santos, F.; Clarke, J.; Mansfield, W.; et al. Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell 2014, 158, 1254–1269. [Google Scholar] [CrossRef] [PubMed]
- Desmarais, J.A.; Hoffmann, M.J.; Bingham, G.; Gagou, M.E.; Meuth, M.; Andrews, P.W. Human embryonic stem cells fail to activate Chk1 and commit to apoptosis in response to DNA replication stress. Stem Cells 2012, 30, 1385–1393. [Google Scholar] [CrossRef] [PubMed]
- Van der Laan, S.; Tsanov, N.; Crozet, C.; Maiorano, D. High dub3 expression in mouse escs couples the G1/S checkpoint to pluripotency. Mol. Cell 2013, 52, 366–379. [Google Scholar] [CrossRef] [PubMed]
- Rempel, R.E.; Sleight, S.B.; Maller, J.L. Maternal Xenopus Cdk2-Cyclin E complexes function during meiotic and early embryonic cell cycles that lack a G1 phase. J. Biol. Chem. 1995, 270, 6843–6855. [Google Scholar] [PubMed]
- Guadagno, T.M.; Newport, J.W. Cdk2 kinase is required for entry into mitosis as a positive regulator of Cdc2-Cyclin B kinase activity. Cell 1996, 84, 73–82. [Google Scholar] [CrossRef]
- Strausfeld, U.P.; Howell, M.; Rempel, R.; Maller, J.L.; Hunt, T.; Blow, J.J. Cip1 blocks the initiation of DNA replication in Xenopus extracts by inhibition of cyclin-dependent kinases. Curr. Biol. 1994, 4, 876–883. [Google Scholar] [CrossRef]
- Howe, J.A.; Newport, J.W. A developmental timer regulates degradation of cyclin E1 at the midblastula transition during Xenopus embryogenesis. Proc. Natl. Acad. Sci. USA 1996, 93, 2060–2064. [Google Scholar] [CrossRef] [PubMed]
- Su, J.Y.; Rempel, R.E.; Erikson, E.; Maller, J.L. Cloning and characterization of the Xenopus cyclin-dependent kinase inhibitor p27XIC1. Proc. Natl. Acad. Sci. USA 1995, 92, 10187–10191. [Google Scholar] [CrossRef] [PubMed]
- Hartley, R.S.; Sible, J.C.; Lewellyn, A.L.; Maller, J.L. A role for cyclin E/Cdk2 in the timing of the midblastula transition in Xenopus embryos. Dev. Biol. 1997, 188, 312–321. [Google Scholar] [CrossRef] [PubMed]
- Wroble, B.N.; Finkielstein, C.V.; Sible, J.C. Wee1 kinase alters cyclin E/Cdk2 and promotes apoptosis during the early embryonic development of Xenopus laevis. BMC Dev. Biol. 2007. [Google Scholar] [CrossRef] [PubMed]
- Shechter, D.; Costanzo, V.; Gautier, J. Atr and atm regulate the timing of DNA replication origin firing. Nat. Cell. Biol. 2004, 6, 648–655. [Google Scholar] [CrossRef] [PubMed]
- Van, C.; Yan, S.; Michael, W.M.; Waga, S.; Cimprich, K.A. Continued primer synthesis at stalled replication forks contributes to checkpoint activation. J. Cell Biol. 2010, 189, 233–246. [Google Scholar] [CrossRef] [PubMed]
- Betous, R.; Pillaire, M.J.; Pierini, L.; van der Laan, S.; Recolin, B.; Ohl-Seguy, E.; Guo, C.; Niimi, N.; Gruz, P.; Nohmi, T.; et al. DNA polymerase kappa-dependent DNA synthesis at stalled replication forks is important for chk1 activation. EMBO J. 2013, 32, 2172–2185. [Google Scholar] [CrossRef] [PubMed]
- DeStephanis, D.; McLeod, M.; Yan, S. Rev1 is important for the ATR-Chk1 DNA damage response pathway in Xenopus egg extracts. Biochem. Biophys. Res. Commun. 2015, 460, 609–615. [Google Scholar] [CrossRef] [PubMed]
- Costanzo, V.; Shechter, D.; Lupardus, P.J.; Cimprich, K.A.; Gottesman, M.; Gautier, J. An ATR- and CDC7-dependent DNA damage checkpoint that inhibits initiation of DNA replication. Mol. Cell 2003, 11, 203–213. [Google Scholar] [CrossRef]
- Dasso, M.; Newport, J.W. Completion of DNA replication is monitored by a feedback system that controls the initiation of mitosis in vitro: Studies in Xenopus. Cell 1990, 61, 811–823. [Google Scholar] [CrossRef]
- Kappas, N.C.; Savage, P.; Chen, K.C.; Walls, A.T.; Sible, J.C. Dissection of the Xchk1 signaling pathway in Xenopus laevis embryos. Mol Biol Cell 2000, 11, 3101–3108. [Google Scholar] [CrossRef] [PubMed]
- Kermi, C.; Prieto, S.; van der Laan, S.; Tsanov, N.; Recolin, B.; Uro-Coste, E.; Delisle, M-B.; Maiorano, D. RAD18 is a maternal limiting factor that suppresses the UV-dependent DNA damge checkpoint in Xenopus embryos. Dev. Cell 2015, 34, 364–372. [Google Scholar] [CrossRef] [PubMed]
- Hartman, P.S.; Herman, R.K. Radiation-sensitive mutants of Caenorhabditis elegans. Genetics 1982, 102, 159–178. [Google Scholar] [PubMed]
- Holway, A.H.; Kim, S.H.; La Volpe, A.; Michael, W.M. Checkpoint silencing during the DNA damage response in Caenorhabditis elegans embryos. J. Cell Biol. 2006, 172, 999–1008. [Google Scholar] [CrossRef] [PubMed]
- Newport, J.; Dasso, M. On the coupling between DNA replication and mitosis. J. Cell Sci. Suppl. 1989, 12, 149–160. [Google Scholar] [CrossRef] [PubMed]
- Clute, P.; Masui, Y. Microtubule dependence of chromosome cycles in Xenopus laevis blastomeres under the influence of a DNA synthesis inhibitor, aphidicolin. Dev. Biol. 1997, 185, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Fogarty, P.; Campbell, S.D.; Abu-Shumays, R.; Phalle, B.S.; Yu, K.R.; Uy, G.L.; Goldberg, M.L.; Sullivan, W. The Drosophila grapes gene is related to checkpoint gene Chk1/Rad27 and is required for late syncytial division fidelity. Curr. Biol. 1997, 7, 418–426. [Google Scholar] [CrossRef]
- Yuan, K.; Farrell, J.A.; O’Farrell, P.H. Different cyclin types collaborate to reverse the S-phase checkpoint and permit prompt mitosis. J. Cell Biol. 2012, 198, 973–980. [Google Scholar] [CrossRef] [PubMed]
- Sibon, O.C.; Stevenson, V.A.; Theurkauf, W.E. DNA-replication checkpoint control at the Drosophila midblastula transition. Nature 1997, 388, 93–97. [Google Scholar] [PubMed]
- Stumpff, J.; Duncan, T.; Homola, E.; Campbell, S.D.; Su, T.T. Drosophila Wee1 kinase regulates Cdk1 and mitotic entry during embryogenesis. Curr. Biol. 2004, 14, 2143–2148. [Google Scholar] [CrossRef] [PubMed]
- Farrell, J.A.; Shermoen, A.W.; Yuan, K.; O’Farrell, P.H. Embryonic onset of late replication requires cdc25 down-regulation. Genes Dev. 2012, 26, 714–725. [Google Scholar] [CrossRef] [PubMed]
- Edgar, B.A.; Sprenger, F.; Duronio, R.J.; Leopold, P.; O’Farrell, P.H. Distinct molecular mechanism regulate cell cycle timing at successive stages of Drosophila embryogenesis. Genes Dev. 1994, 8, 440–452. [Google Scholar] [CrossRef] [PubMed]
- Farrell, J.A.; O’Farrell, P.H. Mechanism and regulation of Cdc25/twine protein destruction in embryonic cell-cycle remodeling. Curr. Biol. 2013, 23, 118–126. [Google Scholar] [CrossRef] [PubMed]
- Di Talia, S.; She, R.; Blythe, S.A.; Lu, X.; Zhang, Q.F.; Wieschaus, E.F. Posttranslational control of Cdc25 degradation terminates Drosophila’s early cell-cycle program. Curr. Biol. 2013, 23, 127–132. [Google Scholar] [CrossRef] [PubMed]
- Conn, C.W.; Lewellyn, A.L.; Maller, J.L. The DNA damage checkpoint in embryonic cell cycles is dependent on the DNA-to-cytoplasmic ratio. Dev. Cell 2004, 7, 275–281. [Google Scholar] [CrossRef] [PubMed]
- Ohmori, H.; Friedberg, E.C.; Fuchs, R.P.; Goodman, M.F.; Hanaoka, F.; Hinkle, D.; Kunkel, T.A.; Lawrence, C.W.; Livneh, Z.; Nohmi, T.; et al. The y-family of DNA polymerases. Mol. Cell 2001, 8, 7–8. [Google Scholar] [CrossRef]
- Roerink, S.F.; Koole, W.; Stapel, L.C.; Romeijn, R.J.; Tijsterman, M. A broad requirement for tls polymerases eta and kappa, and interacting sumoylation and nuclear pore proteins, in lesion bypass during C. elegans embryogenesis. PLoS Genet. 2012, 8, e1002800. [Google Scholar] [CrossRef] [PubMed]
- Butuci, M.; Williams, A.B.; Wong, M.M.; Kramer, B.; Michael, W.M. Zygotic genome activation triggers chromosome damage and checkpoint signaling in C. elegans primordial germ cells. Dev. Cell 2015, 34, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Blythe, S.A.; Wieschaus, E.F. Zygotic genome activation triggers the DNA replication checkpoint at the midblastula transition. Cell 2015, 160, 1169–1181. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Li, J.M.; Elemento, O.; Tavazoie, S.; Wieschaus, E.F. Coupling of zygotic transcription to mitotic control at the Drosophila mid-blastula transition. Development 2009, 136, 2101–2110. [Google Scholar] [CrossRef] [PubMed]
- Almouzni, G.; Mechali, M. Assembly of spaced chromatin promoted by DNA synthesis in extracts from Xenopus eggs. EMBO J. 1988, 7, 665–672. [Google Scholar] [PubMed]
- Almouzni, G.; Wolffe, A.P. Constraints on transcriptional activator function contribute to transcriptional quiescence during early Xenopus embryogenesis. EMBO J. 1995, 14, 1752–1765. [Google Scholar] [PubMed]
- Prioleau, M.N.; Huet, J.; Sentenac, A.; Mechali, M. Competition between chromatin and transcription complex assembly regulates gene expression during early development. Cell 1994, 77, 439–449. [Google Scholar] [CrossRef]
- Amodeo, A.A.; Jukam, D.; Straight, A.F.; Skotheim, J.M. Histone titration against the genome sets the DNA-to-cytoplasm threshold for the Xenopus midblastula transition. Proc. Natl. Acad. Sci. USA 2015, 112, E1086–E1095. [Google Scholar] [CrossRef] [PubMed]
- Stack, J.H.; Newport, J.W. Developmentally regulated activation of apoptosis early in Xenopus gastrulation results in cyclin a degradation during interphase of the cell cycle. Development 1997, 124, 3185–3195. [Google Scholar] [PubMed]
- Harrison, M.M.; Li, X.Y.; Kaplan, T.; Botchan, M.R.; Eisen, M.B. Zelda binding in the early Drosophila melanogaster embryo marks regions subsequently activated at the maternal-to-zygotic transition. PLoS Genet. 2011, 7, e1002266. [Google Scholar] [CrossRef] [PubMed]
- Nien, C.Y.; Liang, H.L.; Butcher, S.; Sun, Y.; Fu, S.; Gocha, T.; Kirov, N.; Manak, J.R.; Rushlow, C. Temporal coordination of gene networks by zelda in the early Drosophila embryo. PLoS Genet 2011, 7, e1002339. [Google Scholar] [CrossRef] [PubMed]
- Satija, R.; Bradley, R.K. The tagteam motif facilitates binding of 21 sequence-specific transcription factors in the Drosophila embryo. Genome Res. 2012, 22, 656–665. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Chen, H.; Ling, J.; Yu, D.; Struffi, P.; Small, S. Impacts of the ubiquitous factor zelda on bicoid-dependent DNA binding and transcription in Drosophila. Genes Dev. 2014, 28, 608–621. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Zhu, Y.; Zhang, K.; Yeung, M.; Durocher, D.; Xiao, W. Rad6-rad18 mediates a eukaryotic SOS response by ubiquitinating the 9-1-1 checkpoint clamp. Cell 2008, 133, 601–611. [Google Scholar] [CrossRef] [PubMed]
- Ten Bosch, J.R.; Benavides, J.A.; Cline, T.W. The tagteam DNA motif controls the timing of Drosophila pre-blastoderm transcription. Development 2006, 133, 1967–1977. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.L.; Nien, C.Y.; Liu, H.Y.; Metzstein, M.M.; Kirov, N.; Rushlow, C. The zinc-finger protein zelda is a key activator of the early zygotic genome in Drosophila. Nature 2008, 456, 400–403. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.W.; Spangenberg, S.; Vogt, N.; Grosshans, J. Number of nuclear divisions in the Drosophila blastoderm controlled by onset of zygotic transcription. Curr. Biol. 2013, 23, 133–138. [Google Scholar] [CrossRef] [PubMed]
- Benoit, B.; He, C.H.; Zhang, F.; Votruba, S.M.; Tadros, W.; Westwood, J.T.; Smibert, C.A.; Lipshitz, H.D.; Theurkauf, W.E. An essential role for the RNA-binding protein smaug during the Drosophila maternal-to-zygotic transition. Development 2009, 136, 923–932. [Google Scholar] [CrossRef] [PubMed]
- Stead, E.; White, J.; Faast, R.; Conn, S.; Goldstone, S.; Rathjen, J.; Dhingra, U.; Rathjen, P.; Walker, D.; Dalton, S. Pluripotent cell division cycles are driven by ectopic cdk2, cyclin A/E and E2F activities. Oncogene 2002, 21, 8320–8333. [Google Scholar] [CrossRef] [PubMed]
- Turinetto, V.; Orlando, L.; Sanchez-Ripoll, Y.; Kumpfmueller, B.; Storm, M.P.; Porcedda, P.; Minieri, V.; Saviozzi, S.; Accomasso, L.; Cibrario Rocchietti, E.; et al. High basal gammah2ax levels sustain self-renewal of mouse embryonic and induced pluripotent stem cells. Stem Cells 2012, 30, 1414–1423. [Google Scholar] [CrossRef] [PubMed]
- Banath, J.P.; Banuelos, C.A.; Klokov, D.; MacPhail, S.M.; Lansdorp, P.M.; Olive, P.L. Explanation for excessive DNA single-strand breaks and endogenous repair foci in pluripotent mouse embryonic stem cells. Exp. Cell Res. 2009, 315, 1505–1520. [Google Scholar] [CrossRef] [PubMed]
- Vanneste, E.; Voet, T.; Le Caignec, C.; Ampe, M.; Konings, P.; Melotte, C.; Debrock, S.; Amyere, M.; Vikkula, M.; Schuit, F.; et al. Chromosome instability is common in human cleavage-stage embryos. Nat. Med. 2009, 15, 577–583. [Google Scholar] [CrossRef] [PubMed]
- Kort, D.H.; Chia, G.; Treff, N.R.; Tanaka, A.J.; Xing, T.; Vensand, L.B.; Micucci, S.; Prosser, R.; Lobo, R.A.; Sauer, M.V.; et al. Human embryos commonly form abnormal nuclei during development: A mechanism of DNA damage, embryonic aneuploidy, and developmental arrest. Hum. Reprod. 2016, 31, 312–323. [Google Scholar] [CrossRef] [PubMed]
- Blasco, M.A.; Serrano, M.; Fernandez-Capetillo, O. Genomic instability in iPS: Time for a break. EMBO J. 2011, 30, 991–993. [Google Scholar] [CrossRef] [PubMed]
- Hensey, C.; Gautier, J. A developmental timer that regulates apoptosis at the onset of gastrulation. Mech. Dev. 1997, 69, 183–195. [Google Scholar] [CrossRef]
- Anderson, J.A.; Lewellyn, A.L.; Maller, J.L. Ionizing radiation induces apoptosis and elevates cyclin A1-Cdk2 activity before but not after the midblastula transition in Xenopus. Mol. Biol. Cell 1997, 8, 1195–1206. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, W.; Fogarty, P.; Theurkauf, W. Mutations affecting the cytoskeletal organization of syncytial Drosophila embryos. Development 1993, 118, 1245–1254. [Google Scholar] [PubMed]
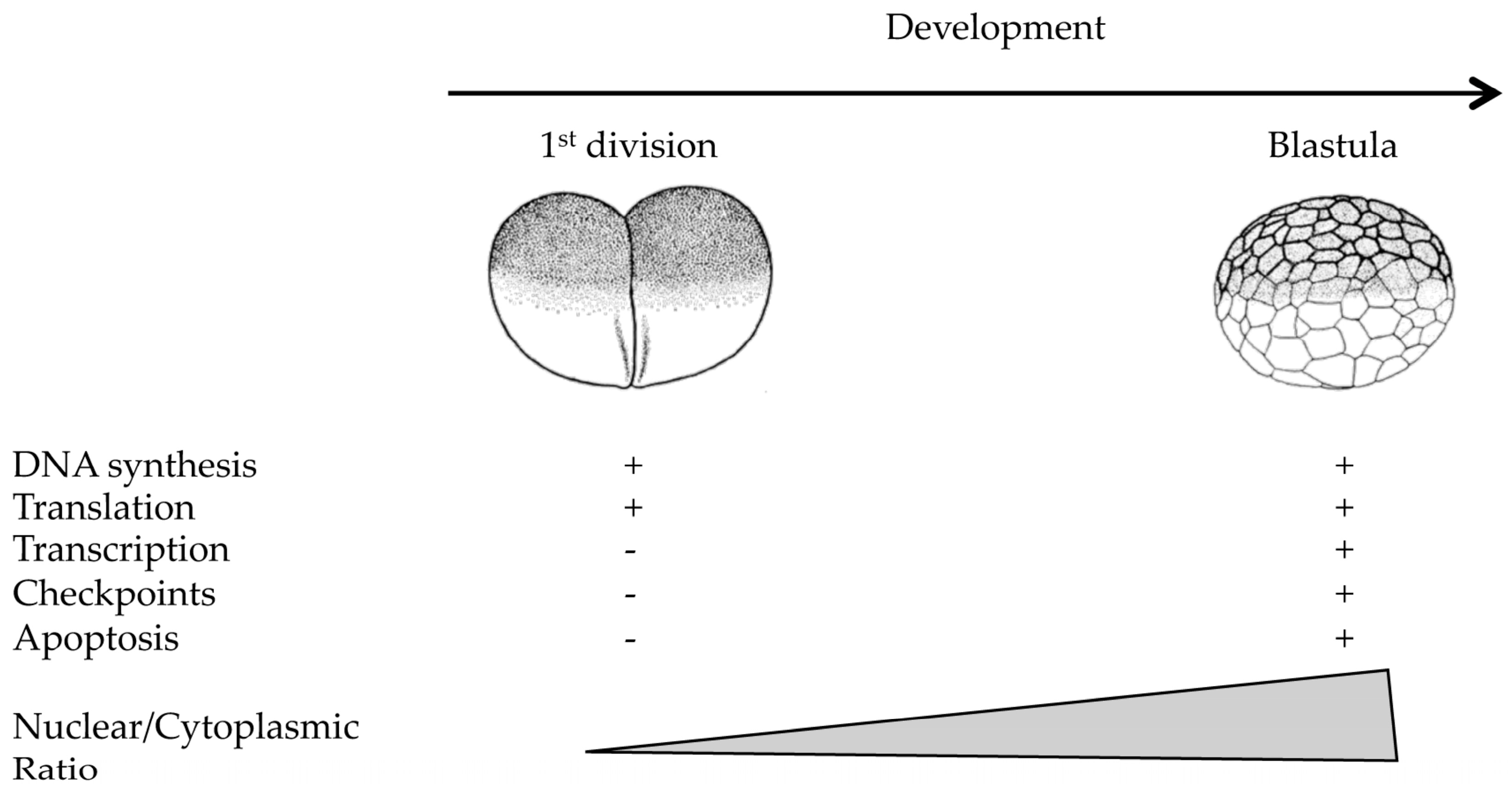
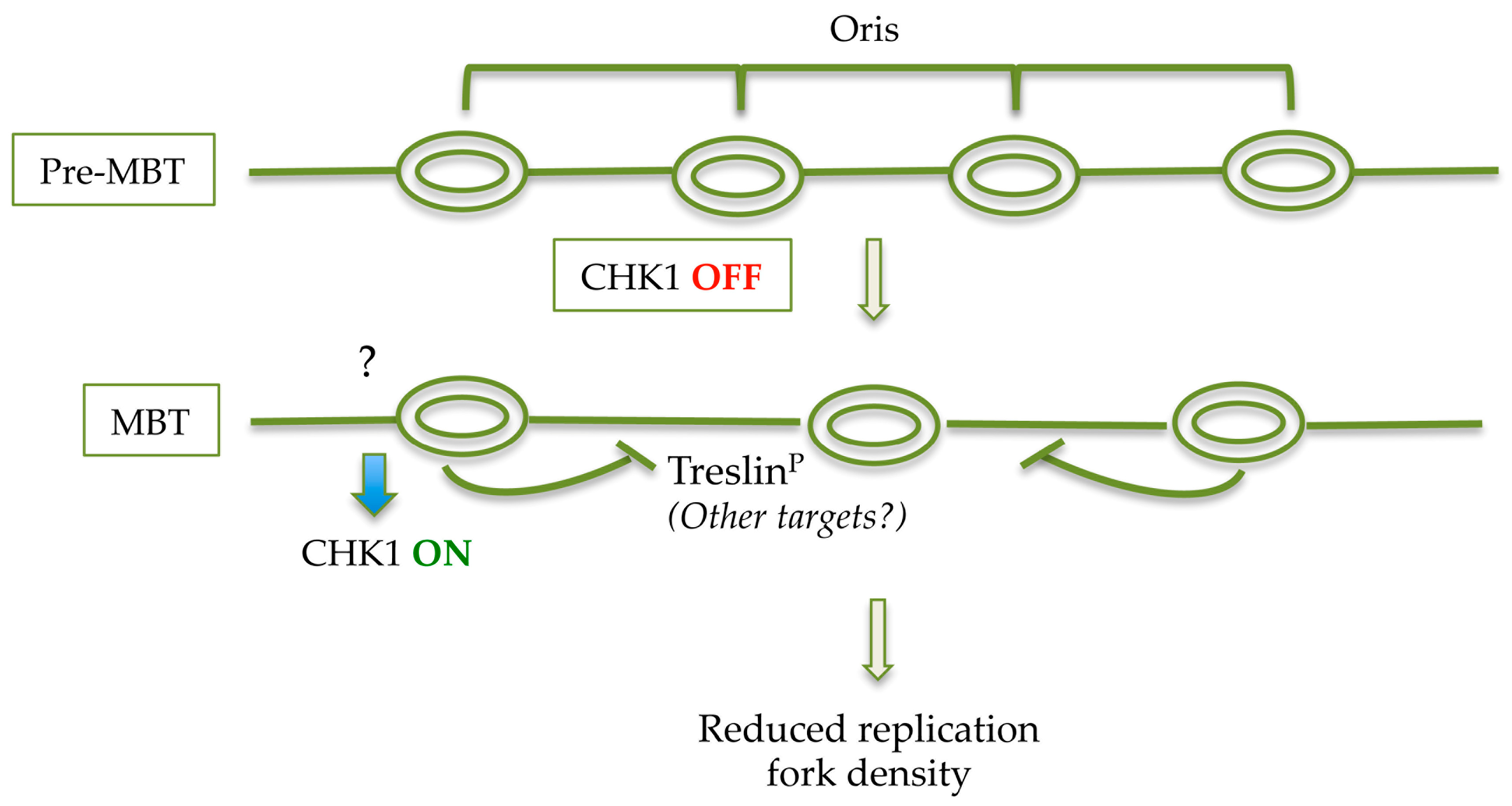

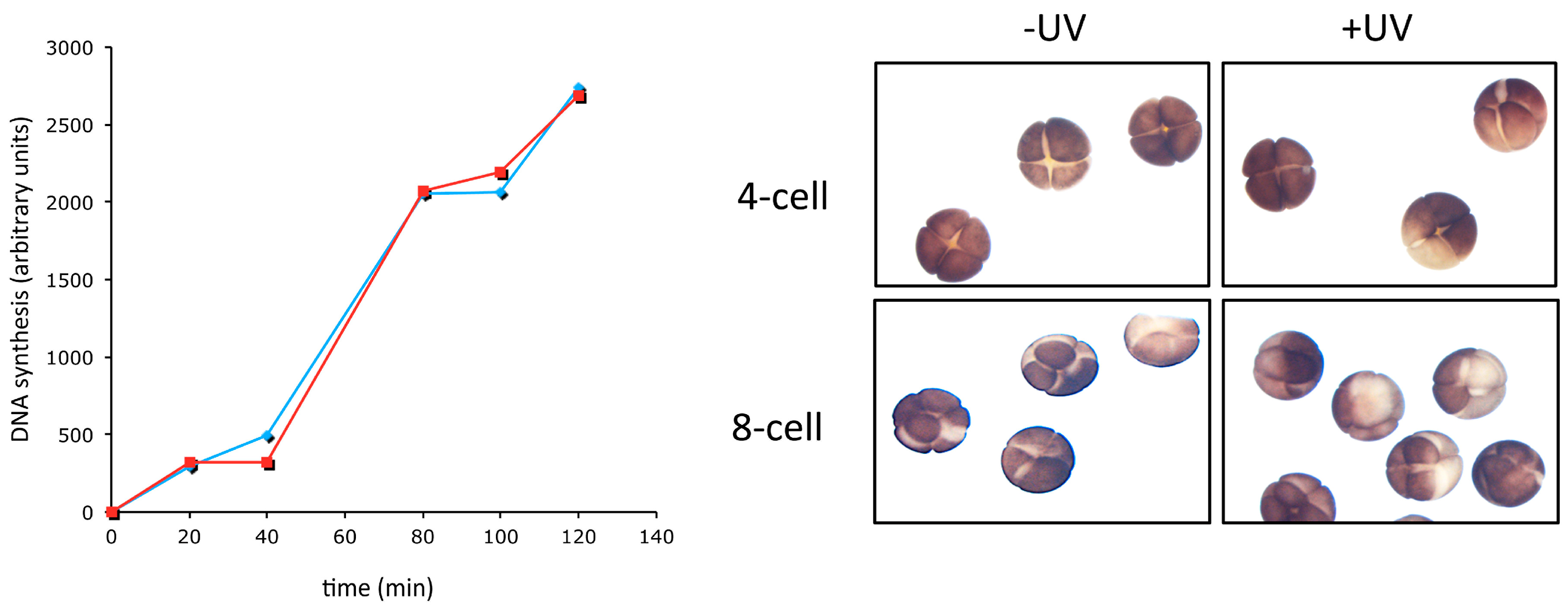
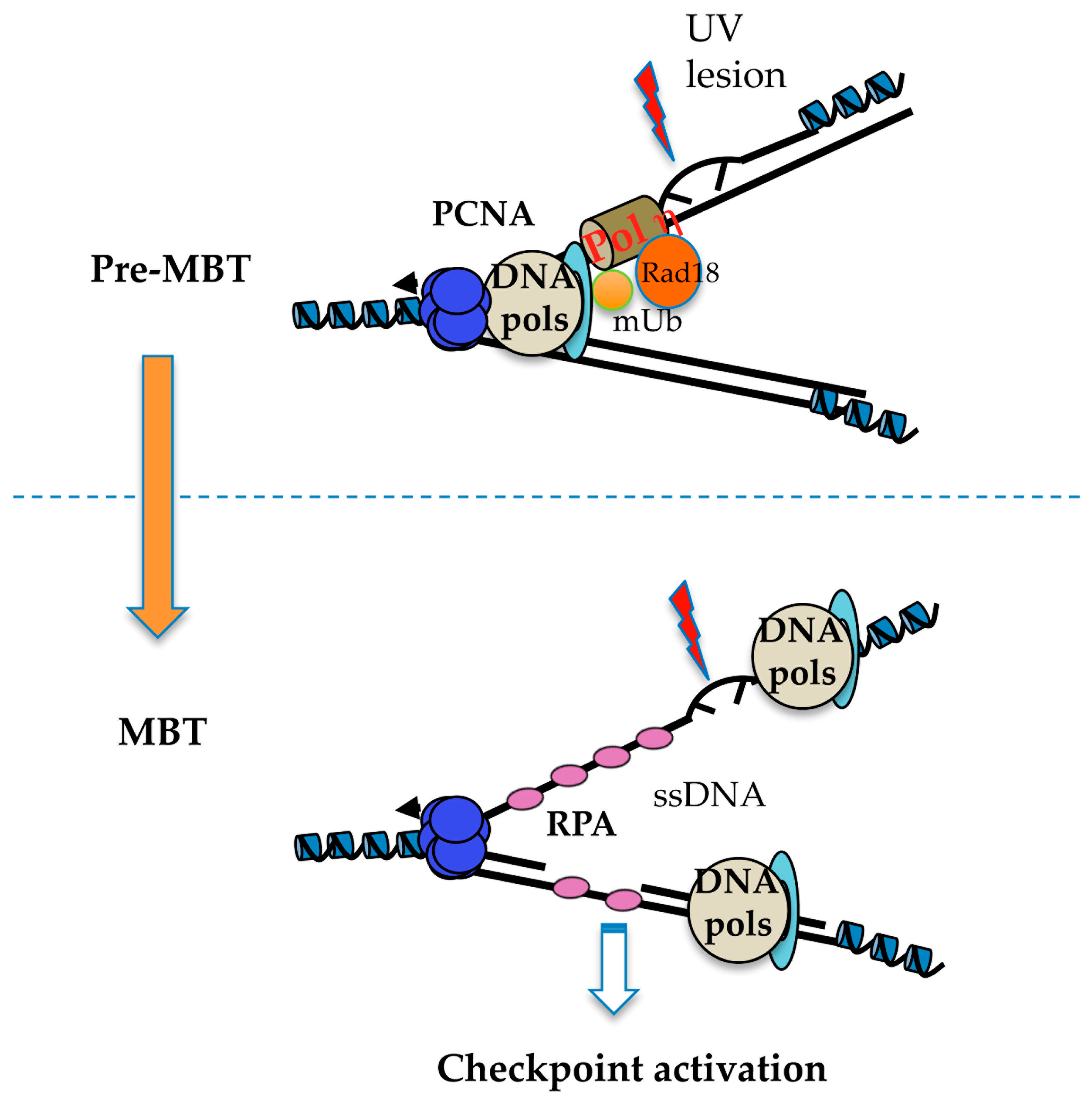
| Organism | Cycle 1 | Cycle 2 | Cycle 3–4 | Blastula/Blastocyst | Somatic Cell |
|---|---|---|---|---|---|
| Drosophila [3] | 3.4 min | 3.4 min | 3.4 min | 50 min (ZGA) | 8 h |
| Xenopus [15] | 20 min | 20 min | 20 min | 210 min (ZGA) | 8 h |
| Mouse [10] | 4–7 h | 1–5 h (ZGA) | n.d. | 8 h | 8 h |
| Human [35] | 7–8 h | n.d. | (ZGA, n.d.) | 8 h | 8 h |
© 2017 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kermi, C.; Lo Furno, E.; Maiorano, D. Regulation of DNA Replication in Early Embryonic Cleavages. Genes 2017, 8, 42. https://doi.org/10.3390/genes8010042
Kermi C, Lo Furno E, Maiorano D. Regulation of DNA Replication in Early Embryonic Cleavages. Genes. 2017; 8(1):42. https://doi.org/10.3390/genes8010042
Chicago/Turabian StyleKermi, Chames, Elena Lo Furno, and Domenico Maiorano. 2017. "Regulation of DNA Replication in Early Embryonic Cleavages" Genes 8, no. 1: 42. https://doi.org/10.3390/genes8010042






