BARHL1 Is Downregulated in Alzheimer’s Disease and May Regulate Cognitive Functions through ESR1 and Multiple Pathways
Abstract
:1. Introduction
2. Materials and Methods
2.1. Tissue Samples
2.2. Tissue Microarray and Immunohistochemistry
2.3. Statistical Analysis
2.4. Integrative Bioinformatics Approach
2.4.1. BARHL1 Association with Alzheimer’s Disease
2.4.2. Construction of the BARHL1 Regulatory Circuit
2.4.3. Collection of Alzheimer’s Disease, Parkinson’s Disease and Amyotrophic Lateral Sclerosis Pathway-Specific Genes
2.5. Promoter Analysis
2.6. Protein-Protein Interaction Analysis
2.7. micro RNA Analysis
2.8. Mining the BARHL1 Knockout Mouse Phenotype for Alzheimer’s Disease Symptoms
3. Results and Discussion
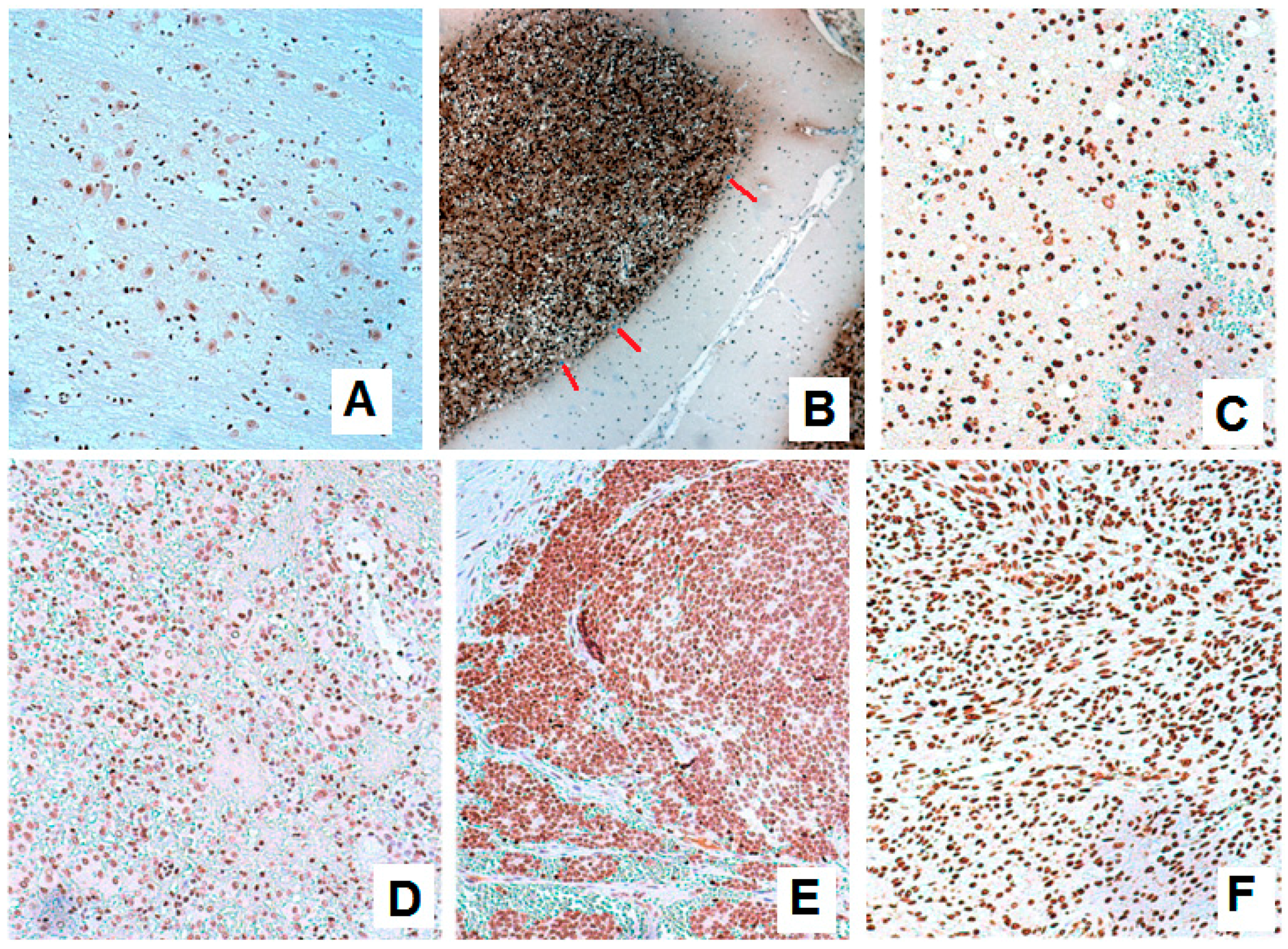
3.1. Expression of BARHL1 in the Nervous System and Breast Tumors
3.2. BARHL1 Expression in Alzheimer’s Disease
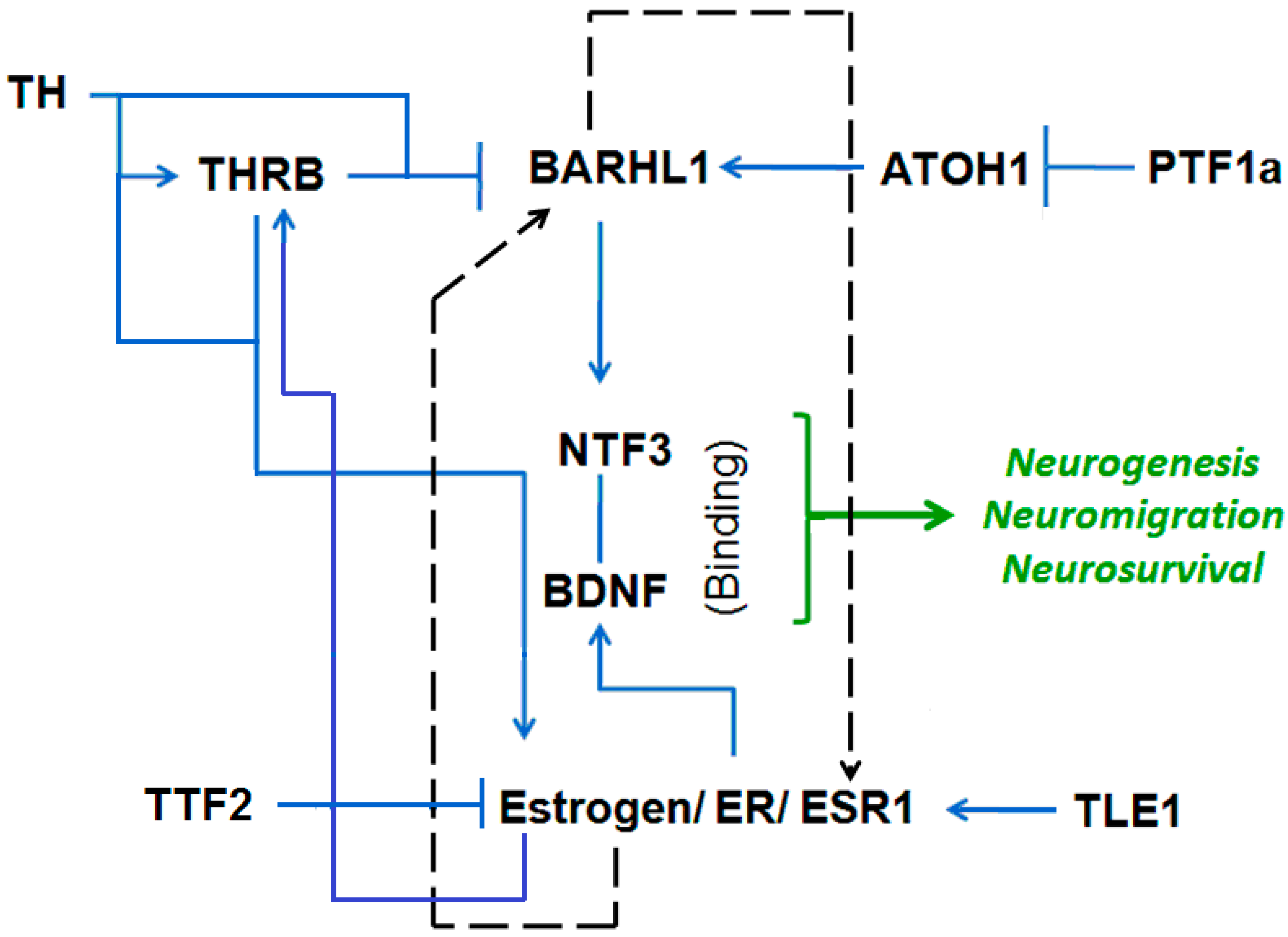
3.3. Putative Estrogen-BARHL1 Axis in Alzheimer’s Disease
3.4. Estrogen-BARHL1 Network
3.5. Estrogen and BARHL1 Regulate Each Other
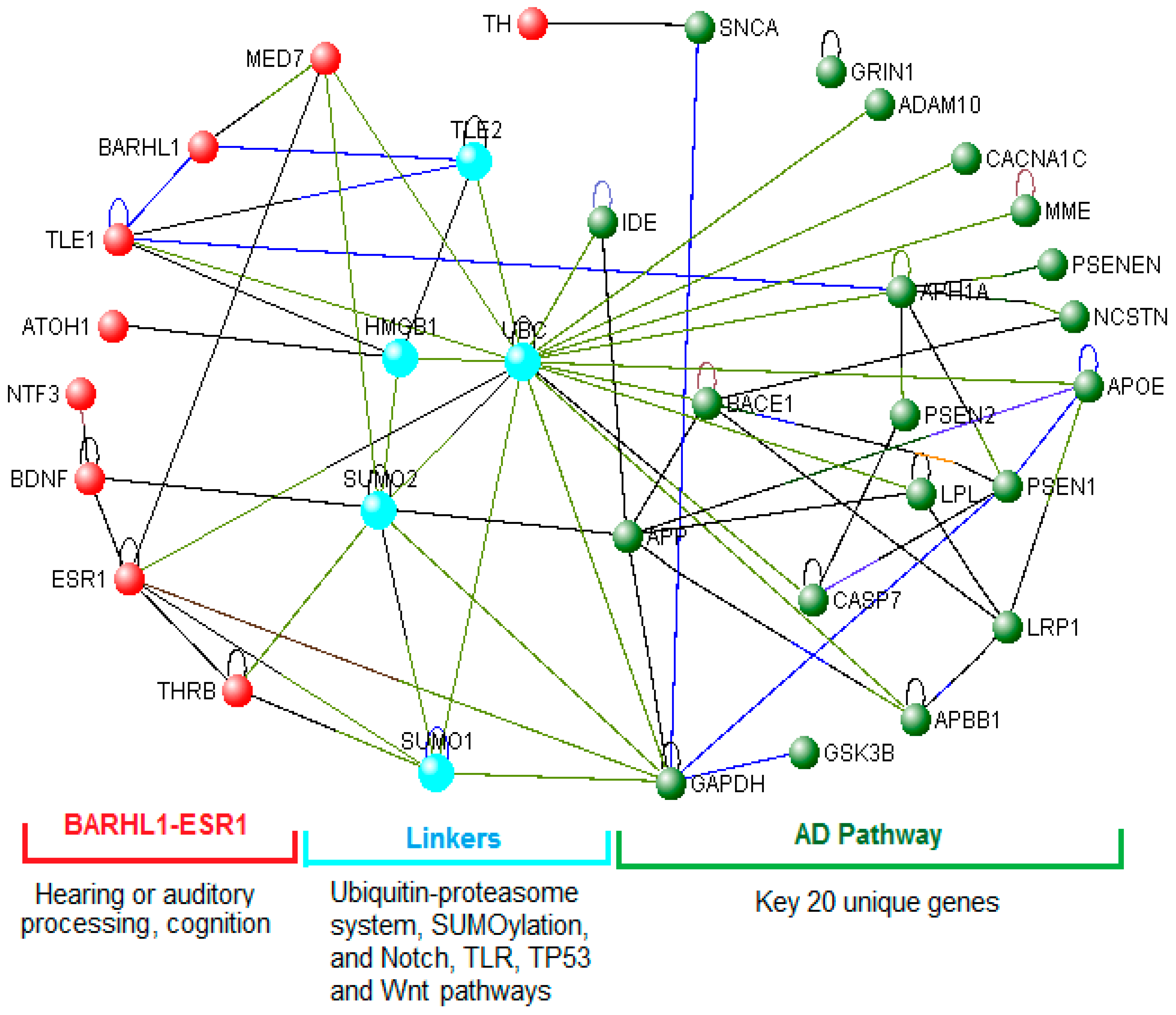
3.6. The BARHL1-ESR1 Axis May Regulate a Subset of the Alzheimer’s Disease Pathway
3.7. Role of the BARHL1-ESR1 Network in Alzheimer’s Disease
3.8. Links between the BARHL1-ESR Axis and the Alzheimer’s Disease Pathway
3.9. miRNAs Regulate the BARHL1-ESR1 Axis and the Alzheimer’s Disease Network
3.10. Functions of the BARHL1 and Alzheimer’s Disease Networks Regulating miRNAs
3.11. hsa-mir-18a May Regulate the BARHL1-AD Network and Alzheimer’s Disease Patho-Physiology
3.12. BARHL1 Knockout Mimics Alzheimer’s Disease Symptoms
3.13. Estrogen-ESR1 and Alzheimer’s Disease
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Funding
References
- Oishi, K.; Lyketsos, C.G. Alzheimer’s disease and the fornix. Front. Aging Neurosci. 2014, 6, 241. [Google Scholar] [CrossRef] [PubMed]
- Mayeux, R.; Stern, Y. Epidemiology of Alzheimer disease. Cold Spring Harb. Perspect. Med. 2012, 2. [Google Scholar] [CrossRef] [PubMed]
- Alzheimer’s Association. 2012 Alzheimer’s disease facts and figures. Alzheimers Dement. 2012, 8, 131–168. [Google Scholar]
- Kim, D.H.; Yeo, S.H.; Park, J.M.; Choi, J.Y.; Lee, T.H.; Park, S.Y.; Ock, M.S.; Eo, J.; Kim, H.S.; Cha, H.J. Genetic markers for diagnosis and pathogenesis of Alzheimer’s disease. Gene 2014, 545, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Wegiel, J.; Wisniewski, H.M.; Dziewiatkowski, J.; Badmajew, E.; Tarnawski, M.; Reisberg, B.; Mlodzik, B.; De Leon, M.J.; Miller, D.C. Cerebellar atrophy in Alzheimer’s disease-clinicopathological correlations. Brain Res. 1999, 818, 41–50. [Google Scholar] [CrossRef]
- Baloyannis, S.J.; Manolidis, S.L.; Manolidis, L.S. Synaptic alterations in the vestibulocerebellar system in Alzheimer’s disease—A Golgi and electron microscope study. Acta Oto Laryngol. 2000, 120, 247–250. [Google Scholar]
- Andersen, K.; Andersen, B.B.; Pakkenberg, B. Stereological quantification of the cerebellum in patients with Alzheimer’s disease. Neurobiol. Aging 2012, 33, 197.e11–197.e20. [Google Scholar] [CrossRef] [PubMed]
- Rekart, J.L.; Sandoval, C.J.; Bermudez-Rattoni, F.; Routtenberg, A. Remodeling of hippocampal mossy fibers is selectively induced seven days after the acquisition of a spatial but not a cued reference memory task. Learn. Mem. 2007, 14, 416–421. [Google Scholar] [CrossRef] [PubMed]
- Wilke, S.A.; Raam, T.; Antonios, J.K.; Bushong, E.A.; Koo, E.H.; Ellisman, M.H.; Ghosh, A. Specific disruption of hippocampal mossy fiber synapses in a mouse model of familial Alzheimer’s disease. PLoS ONE 2014, 9, e84349. [Google Scholar] [CrossRef] [PubMed]
- Kiktenko, A.I.; Uranova, N.A.; Orlovskaia, D.D. [Mossy fibers of the hippocampus in Alzheimer’s disease]. Zh. Nevrol. Psikhiatr. Im. S S Korsakova. 1995, 95, 43–46. (In Russian) [Google Scholar] [PubMed]
- Wang, N.Y.; Su, J.F.; Dong, H.Q.; Jia, J.P.; Han, D.M. Hearing impairment in patients with mild cognitive impairment and Alzheimer’s disease. Chin. J. Otorhinolaryngol. Head Neck Surg. 2005, 40, 279–282. (In Chinese) [Google Scholar]
- Jiang, W.; Zhang, Y.; Meng, F.; Lian, B.; Chen, X.; Yu, X.; Dai, E.; Wang, S.; Liu, X.; Li, X.; et al. Identification of active transcription factor and miRNA regulatory pathways in Alzheimer’s disease. Bioinformatics 2013, 29, 2596–2602. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Qiu, F.; Xu, A.; Price, S.M.; Xiang, M. BARHL1 regulates migration and survival of cerebellar granule cells by controlling expression of the Neurotrophin-3 gene. J. Neurosci. 2004, 24, 3104–3114. [Google Scholar] [CrossRef] [PubMed]
- Poschl, J.; Lorenz, A.; Hartmann, W.; von Bueren, A.O.; Kool, M.; Li, S.; Peraud, A.; Tonn, J.C.; Herms, J.; Xiang, M.; et al. Expression of BARHL1 in medulloblastoma is associated with prolonged survival in mice and humans. Oncogene 2011, 30, 4721–4730. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Yauk, C.L.; Wade, M.G. BARHL1 is directly regulated by thyroid hormone in the developing cerebellum of mice. Biochem. Biophys. Res. Commun. 2011, 415, 157–162. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, H.; Toji, H.; Harrington, C.R.; Sasaki, K.; Izumi, Y.; Ohnuma, T.; Arai, H.; Yasuda, M.; Tanaka, C.; Emson, P.C.; et al. Lack of an association of estrogen receptor alpha gene polymorphisms and transcriptional activity with Alzheimer disease. Arch. Neurol. 2000, 57, 236–240. [Google Scholar] [CrossRef] [PubMed]
- Corbo, R.M.; Gambina, G.; Ruggeri, M.; Scacchi, R. Association of estrogen receptor α (ESR1), PvuII and XbaI polymorphisms with sporadic Alzheimer’s disease and their effect on apolipoprotein E concentrations. Dement. Geriatr. Cogn. Disord. 2006, 22, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Mattila, K.M.; Axelman, K.; Rinne, J.O.; Blomberg, M.; Lehtimaki, T.; Laippala, P.; Roytta, M.; Viitanen, M.; Wahlund, L.; Winblad, B.; et al. Interaction between estrogen receptor 1 and the epsilon4 allele of apolipoprotein E increases the risk of familial Alzheimer’s disease in women. Neurosci. Lett. 2000, 282, 45–48. [Google Scholar] [CrossRef]
- Kawas, C.; Resnick, S.; Morrison, A.; Brookmeyer, R.; Corrada, M.; Zonderman, A.; Bacal, C.; Lingle, D.D.; Metter, E. A prospective study of estrogen replacement therapy and the risk of developing Alzheimer’s disease: The Baltimore Longitudinal Study of Aging. Neurology 1997, 48, 1517–1521. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.X.; Jacobs, D.; Stern, Y.; Marder, K.; Schofield, P.; Gurland, B.; Andrews, H.; Mayeux, R. Effect of oestrogen during menopause on risk and age at onset of Alzheimer’s disease. Lancet 1996, 348, 429–432. [Google Scholar] [CrossRef]
- Paganini-Hill, A.; Henderson, V.W. Estrogen deficiency and risk of Alzheimer’s disease in women. Am. J. Epidemiol. 1994, 140, 256–261. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Solano, J.; Conesa-Zamora, P.; Trujillo-Santos, J.; Torres-Moreno, D.; Makinen, M.J.; Perez-Guillermo, M. Immunohistochemical expression profile of βcatenin, E-cadherin, P-cadherin, laminin-5γ2 chain, and SMAD4 in colorectal serrated adenocarcinoma. Hum. Pathol. 2012, 43, 1094–1102. [Google Scholar] [CrossRef] [PubMed]
- Conesa-Zamora, P.; Garcia-Solano, J.; Garcia-Garcia, F.; Turpin Mdel, C.; Trujillo-Santos, J.; Torres-Moreno, D.; Oviedo-Ramirez, I.; Carbonell-Munoz, R.; Munoz-Delgado, E.; Rodriguez-Braun, E.; et al. Expression profiling shows differential molecular pathways and provides potential new diagnostic biomarkers for colorectal serrated adenocarcinoma. Int. J. Cancer. 2013, 132, 297–307. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Chang, Y.C.; Wang, Y.; Huang, C.L.; Liu, Y.; Tian, F.; Granger, B.; Delisi, C. Visant 4.0: Integrative network platform to connect genes, drugs, diseases and therapies. Nucleic Acids Res. 2013, 41, W225–W231. [Google Scholar] [CrossRef] [PubMed]
- Davis, A.P.; Murphy, C.G.; Johnson, R.; Lay, J.M.; Lennon-Hopkins, K.; Saraceni-Richards, C.; Sciaky, D.; King, B.L.; Rosenstein, M.C.; Wiegers, T.C.; et al. The comparative toxicogenomics database: Update 2013. Nucleic Acids Res. 2013, 41, D1104–D1114. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. Kegg: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Cartharius, K.; Frech, K.; Grote, K.; Klocke, B.; Haltmeier, M.; Klingenhoff, A.; Frisch, M.; Bayerlein, M.; Werner, T. MatInspector and beyond: Promoter analysis based on transcription factor binding sites. Bioinformatics 2005, 21, 2933–2942. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Bardes, E.E.; Aronow, B.J.; Jegga, A.G. ToppGene suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009, 37, W305–W311. [Google Scholar] [CrossRef] [PubMed]
- Dweep, H.; Sticht, C.; Pandey, P.; Gretz, N. MiRWalk—Database: Prediction of possible miRNAa binding sites by “walking” the genes of three genomes. J. Biomed. Inf. 2011, 44, 839–847. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Shi, B.; Wang, J.; Cao, Q.; Cui, Q. Tam: A method for enrichment and depletion analysis of a microRNA category in a list of microRNAs. BMC Bioinf. 2010, 11, 419. [Google Scholar] [CrossRef] [PubMed]
- Barh, D.; Jain, N.; Tiwari, S.; Field, J.K.; Padin-Iruegas, E.; Ruibal, A.; Lopez, R.; Herranz, M.; Bhattacharya, A.; Juneja, L.; et al. A novel in silico reverse-transcriptomics-based identification and blood-based validation of a panel of sub-type specific biomarkers in lung cancer. BMC Genom. 2013, 14 (Suppl. 6), S5. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.Y.; Qin, S.; Lu, Y.P.; Ravid, R.; Swaab, D.F.; Zhou, J.N. Decreased estrogen receptor-α expression in hippocampal neurons in relation to hyperphosphorylated tau in Alzheimer patients. Acta Neuropathol. 2003, 106, 213–220. [Google Scholar] [CrossRef] [PubMed]
- Wharton, W.; Baker, L.D.; Gleason, C.E.; Dowling, M.; Barnet, J.H.; Johnson, S.; Carlsson, C.; Craft, S.; Asthana, S. Short-term hormone therapy with transdermal estradiol improves cognition for postmenopausal women with Alzheimer’s disease: Results of a randomized controlled trial. J. Alzheimers Dis. 2011, 26, 495–505. [Google Scholar] [PubMed]
- Chellappa, R.; Li, S.; Pauley, S.; Jahan, I.; Jin, K.; Xiang, M. BARHL1 regulatory sequences required for cell-specific gene expression and autoregulation in the inner ear and central nervous system. Mol. Cell. Biol. 2008, 28, 1905–1914. [Google Scholar] [CrossRef] [PubMed]
- Pascual, M.; Abasolo, I.; Mingorance-Le Meur, A.; Martinez, A.; Del Rio, J.A.; Wright, C.V.; Real, F.X.; Soriano, E. Cerebellar gabaergic progenitors adopt an external granule cell-like phenotype in the absence of PTF1A transcription factor expression. Proc. Natl. Acad. Sci. USA 2007, 104, 5193–5198. [Google Scholar] [CrossRef] [PubMed]
- Holmes, K.A.; Hurtado, A.; Brown, G.D.; Launchbury, R.; Ross-Innes, C.S.; Hadfield, J.; Odom, D.T.; Carroll, J.S. Transducin-like enhancer protein 1 mediates estrogen receptor binding and transcriptional activity in breast cancer cells. Proc. Natl. Acad. Sci. USA 2012, 109, 2748–2753. [Google Scholar] [CrossRef] [PubMed]
- Meng, R.; Tang, H.Y.; Westfall, J.; London, D.; Cao, J.H.; Mousa, S.A.; Luidens, M.; Hercbergs, A.; Davis, F.B.; Davis, P.J.; et al. Crosstalk between integrin μavβ3 and estrogen receptor-α is involved in thyroid hormone-induced proliferation in human lung carcinoma cells. PLoS ONE 2011, 6, e27547. [Google Scholar] [CrossRef] [PubMed]
- Filby, A.L.; Thorpe, K.L.; Tyler, C.R. Multiple molecular effect pathways of an environmental oestrogen in fish. J. Mol. Endocrinol. 2006, 37, 121–134. [Google Scholar] [CrossRef] [PubMed]
- Park, E.; Gong, E.Y.; Romanelli, M.G.; Lee, K. Suppression of estrogen receptor-α transactivation by thyroid transcription factor-2 in breast cancer cells. Biochem. Biophys. Res. Commun. 2012, 421, 532–537. [Google Scholar] [CrossRef] [PubMed]
- Bimonte-Nelson, H.A.; Nelson, M.E.; Granholm, A.C. Progesterone counteracts estrogen-induced increases in neurotrophins in the aged female rat brain. Neuroreport 2004, 15, 2659–2663. [Google Scholar] [CrossRef] [PubMed]
- Luine, V.; Frankfurt, M. Interactions between estradiol, BDNF and dendritic spines in promoting memory. Neuroscience 2013, 239, 34–45. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Liu, X.; Senthil Kumar, S.P.; Zhang, J.; Shi, H. Central expression and anorectic effect of brain-derived neurotrophic factor are regulated by circulating estradiol levels. Horm. Behav. 2013, 63, 533–542. [Google Scholar] [CrossRef] [PubMed]
- Boverhof, D.R.; Burgoon, L.D.; Williams, K.J.; Zacharewski, T.R. Inhibition of estrogen-mediated uterine gene expression responses by dioxin. Mol. Pharmacol. 2008, 73, 82–93. [Google Scholar] [CrossRef] [PubMed]
- Uehara, T.; Ono, A.; Maruyama, T.; Kato, I.; Yamada, H.; Ohno, Y.; Urushidani, T. The Japanese toxicogenomics project: Application of toxicogenomics. Mol. Nutr. Food Res. 2010, 54, 218–227. [Google Scholar] [CrossRef] [PubMed]
- Vitvitsky, V.M.; Garg, S.K.; Keep, R.F.; Albin, R.L.; Banerjee, R. Na+ and K+ ion imbalances in Alzheimer’s disease. Biochim. Biophys. Acta 2012, 1822, 1671–1681. [Google Scholar] [CrossRef] [PubMed]
- Mata, A.M.; Berrocal, M.; Sepulveda, M.R. Impairment of the activity of the plasma membrane Ca2+-ATPase in Alzheimer’s disease. Biochem. Soc. Trans. 2011, 39, 819–822. [Google Scholar] [CrossRef] [PubMed]
- Lin, F.R.; Metter, E.J.; O’Brien, R.J.; Resnick, S.M.; Zonderman, A.B.; Ferrucci, L. Hearing loss and incident dementia. Arch. Neurol. 2011, 68, 214–220. [Google Scholar] [CrossRef] [PubMed]
- Idrizbegovic, E.; Hederstierna, C.; Dahlquist, M.; Kampfe Nordstrom, C.; Jelic, V.; Rosenhall, U. Central auditory function in early Alzheimer’s disease and in mild cognitive impairment. Age Ageing 2011, 40, 249–254. [Google Scholar] [CrossRef] [PubMed]
- Lan, Y.L.; Zhao, J.; Li, S. Estrogen receptors’ neuroprotective effect against glutamate-induced neurotoxicity. Neurol. Sci. 2014, 35, 1657–1662. [Google Scholar] [CrossRef] [PubMed]
- Jamshed, N.; Ozair, F.F.; Aggarwal, P.; Ekka, M. Alzheimer disease in post-menopausal women: Intervene in the critical window period. J. Mid Life Health 2014, 5, 38–40. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Saunders, A.J. The role of ubiquitin-proteasome in the metabolism of amyloid precursor protein (APP): Implications for novel therapeutic strategies for Alzheimer’s disease. Discov. Med. 2014, 18, 41–50. [Google Scholar] [PubMed]
- Riederer, B.M.; Leuba, G.; Vernay, A.; Riederer, I.M. The role of the ubiquitin proteasome system in Alzheimer’s disease. Exp. Biol. Med. 2011, 236, 268–276. [Google Scholar] [CrossRef] [PubMed]
- Lee, L.; Sakurai, M.; Matsuzaki, S.; Arancio, O.; Fraser, P. Sumo and Alzheimer’s disease. Neuromol. Med. 2013, 15, 720–736. [Google Scholar] [CrossRef] [PubMed]
- Woo, H.N.; Park, J.S.; Gwon, A.R.; Arumugam, T.V.; Jo, D.G. Alzheimer’s disease and Notch signaling. Biochem. Biophys. Res. Commun. 2009, 390, 1093–1097. [Google Scholar] [CrossRef] [PubMed]
- Landreth, G.E.; Reed-Geaghan, E.G. Toll-like receptors in Alzheimer’s disease. Curr. Top. Microbiol. Immunol. 2009, 336, 137–153. [Google Scholar] [PubMed]
- Inestrosa, N.C.; Montecinos-Oliva, C.; Fuenzalida, M. Wnt signaling: Role in Alzheimer disease and schizophrenia. J. Neuroimmune Pharmacol. 2012, 7, 788–807. [Google Scholar] [CrossRef] [PubMed]
- Hooper, C.; Meimaridou, E.; Tavassoli, M.; Melino, G.; Lovestone, S.; Killick, R. p53 is upregulated in Alzheimer’s disease and induces Tau phosphorylation in HEK293A cells. Neurosci. Lett. 2007, 418, 34–37. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.X.; Huang, Q.; Hu, Y.; Stromberg, A.J.; Nelson, P.T. Patterns of microRNA expression in normal and early Alzheimer’s disease human temporal cortex: White matter versus gray matter. Acta Neuropathol. 2011, 121, 193–205. [Google Scholar] [CrossRef] [PubMed]
- Dickson, D.W. Apoptotic mechanisms in Alzheimer neurofibrillary degeneration: Cause or effect? J. Clin. Investig. 2004, 114, 23–27. [Google Scholar] [CrossRef] [PubMed]
- Dean, D.C., 3rd.; Jerskey, B.A.; Chen, K.; Protas, H.; Thiyyagura, P.; Roontiva, A.; O’Muircheartaigh, J.; Dirks, H.; Waskiewicz, N.; Lehman, K.; et al. Brain differences in infants at differential genetic risk for late-onset Alzheimer disease: A cross-sectional imaging study. JAMA Neurol. 2014, 71, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Langbaum, J.B.; Chen, K.; Caselli, R.J.; Lee, W.; Reschke, C.; Bandy, D.; Alexander, G.E.; Burns, C.M.; Kaszniak, A.W.; Reeder, S.A.; et al. Hypometabolism in Alzheimer-affected brain regions in cognitively healthy latino individuals carrying the apolipoprotein A ε4 allele. Arch. Neurol. 2010, 67, 462–468. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Qiu, C.; Tu, J.; Geng, B.; Yang, J.; Jiang, T.; Cui, Q. HMDD v2.0: A database for experimentally supported human microRNA and disease associations. Nucleic Acids Res. 2014, 42, D1070–D1074. [Google Scholar] [CrossRef] [PubMed]
- Beyer, K.; Domingo-Sabat, M.; Lao, J.I.; Carrato, C.; Ferrer, I.; Ariza, A. Identification and characterization of a new α-synuclein isoform and its role in Lewy body diseases. Neurogenetics 2008, 9, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Faghihi, M.A.; Modarresi, F.; Khalil, A.M.; Wood, D.E.; Sahagan, B.G.; Morgan, T.E.; Finch, C.E.; St Laurent, G., 3rd.; Kenny, P.J.; Wahlestedt, C. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of β-secretase. Nat. Med. 2008, 14, 723–730. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.S.; Dunckley, T.; Beach, T.G.; Grover, A.; Mastroeni, D.; Ramsey, K.; Caselli, R.J.; Kukull, W.A.; McKeel, D.; Morris, J.C.; et al. Altered neuronal gene expression in brain regions differentially affected by Alzheimer’s disease: A reference data set. Physiol. Genom. 2008, 33, 240–256. [Google Scholar] [CrossRef] [PubMed]
- Hebert, S.S.; Horre, K.; Nicolai, L.; Bergmans, B.; Papadopoulou, A.S.; Delacourte, A.; De Strooper, B. MicroRNA regulation of Alzheimer’s amyloid precursor protein expression. Neurobiol. Dis. 2009, 33, 422–428. [Google Scholar] [CrossRef] [PubMed]
- Jaeger, S.; Pietrzik, C.U. Functional role of lipoprotein receptors in Alzheimer’s disease. Curr. Alzheimer Res. 2008, 5, 15–25. [Google Scholar] [PubMed]
- Dong, S.; Duan, Y.; Hu, Y.; Zhao, Z. Advances in the pathogenesis of Alzheimer’s disease: A re-evaluation of amyloid cascade hypothesis. Trans. Neurodegener. 2012, 1, 18. [Google Scholar]
- Mazzola, J.L.; Sirover, M.A. Reduction of glyceraldehyde-3-phosphate dehydrogenase activity in Alzheimer’s disease and in Huntington’s disease fibroblasts. J. Neurochem. 2001, 76, 442–449. [Google Scholar] [CrossRef] [PubMed]
- Gong, H.; Dong, W.; Rostad, S.W.; Marcovina, S.M.; Albers, J.J.; Brunzell, J.D.; Vuletic, S. Lipoprotein lipase (LPL) is associated with neurite pathology and its levels are markedly reduced in the dentate gyrus of Alzheimer’s disease brains. J. Histochem. Cytochem. 2013, 61, 857–868. [Google Scholar] [CrossRef] [PubMed]
- Dermaut, B.; Theuns, J.; Sleegers, K.; Hasegawa, H.; Van den Broeck, M.; Vennekens, K.; Corsmit, E.; St George-Hyslop, P.; Cruts, M.; van Duijn, C.M.; et al. The gene encoding Nicastrin, a major γ-secretase component, modifies risk for familial early-onset Alzheimer disease in a Dutch population-based sample. Am. J. Hum. Genet. 2002, 70, 1568–1574. [Google Scholar] [CrossRef] [PubMed]
- Sesele, K.; Thanopoulou, K.; Paouri, E.; Tsefou, E.; Klinakis, A.; Georgopoulos, S. Conditional inactivation of Nicastrin restricts amyloid deposition in an Alzheimer’s disease mouse model. Aging Cell 2013, 12, 1032–1040. [Google Scholar] [CrossRef] [PubMed]
- Schipper, H.M.; Maes, O.C.; Chertkow, H.M.; Wang, E. MicroRNA expression in Alzheimer blood mononuclear cells. Gene Regul. Syst. Biol. 2007, 1, 263–274. [Google Scholar]
- Lau, P.; Bossers, K.; Janky, R.; Salta, E.; Frigerio, C.S.; Barbash, S.; Rothman, R.; Sierksma, A.S.; Thathiah, A.; Greenberg, D.; et al. Alteration of the microRNA network during the progression of Alzheimer’s disease. EMBO Mol. Med. 2013, 5, 1613–1634. [Google Scholar] [CrossRef] [PubMed]
- Phillips, H.S.; Hains, J.M.; Armanini, M.; Laramee, G.R.; Johnson, S.A.; Winslow, J.W. BDNF mRNA is decreased in the hippocampus of individuals with Alzheimer’s disease. Neuron 1991, 7, 695–702. [Google Scholar] [CrossRef]
- Davis, J.D.; Podolanczuk, A.; Donahue, J.E.; Stopa, E.; Hennessey, J.V.; Luo, L.G.; Lim, Y.P.; Stern, R.A. Thyroid hormone levels in the prefrontal cortex of post-mortem brains of Alzheimer’s disease patients. Curr. Aging Sci. 2008, 1, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Knusel, B.; Beck, K.D.; Winslow, J.W.; Rosenthal, A.; Burton, L.E.; Widmer, H.R.; Nikolics, K.; Hefti, F. Brain-derived neurotrophic factor administration protects basal forebrain cholinergic but not nigral dopaminergic neurons from degenerative changes after axotomy in the adult rat brain. J. Neurosci. 1992, 12, 4391–4402. [Google Scholar] [PubMed]
- Lindvall, O.; Kokaia, Z.; Bengzon, J.; Elmer, E.; Kokaia, M. Neurotrophins and brain insults. Trends Neurosci. 1994, 17, 490–496. [Google Scholar] [CrossRef]
- Arancibia, S.; Silhol, M.; Mouliere, F.; Meffre, J.; Hollinger, I.; Maurice, T.; Tapia-Arancibia, L. Protective effect of BDNF against β-amyloid induced neurotoxicity in vitro and in vivo in rats. Neurobiol. Dis. 2008, 31, 316–326. [Google Scholar] [CrossRef] [PubMed]
- Tan, Z.S.; Vasan, R.S. Thyroid function and Alzheimer’s disease. J. Alzheimers Dis. 2009, 16, 503–507. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Fici, G.J.; Mao, C.A.; Myers, R.L.; Shuang, R.; Donoho, G.P.; Pauley, A.M.; Himes, C.S.; Qin, W.; Kola, I.; et al. Positive and negative regulation of the γ-secretase activity by Nicastrin in a murine model. J. Biol. Chem. 2003, 278, 33445–33449. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Ma, G.; Cai, H.; Price, D.L.; Wong, P.C. Nicastrin is required for assembly of presenilin/γ-secretase complexes to mediate Notch signaling and for processing and trafficking of β-amyloid precursor protein in mammals. J. Neurosci. 2003, 23, 3272–3277. [Google Scholar] [PubMed]
- Ma, S.L.; Tang, N.L.; Leung, G.T.; Fung, A.W.; Lam, L.C. Estrogen receptor α polymorphisms and the risk of cognitive decline: A 2-year follow-up study. Am. J. Geriatr. Psychiatry 2014, 22, 489–498. [Google Scholar] [CrossRef] [PubMed]
- Cheng, D.; Liang, B.; Hao, Y.; Zhou, W. Estrogen receptor α gene polymorphisms and risk of Alzheimer’s disease: Evidence from a meta-analysis. Clin. Interv. Aging 2014, 9, 1031–1038. [Google Scholar] [CrossRef] [PubMed]
- Leuba, G.; Saini, K. Pathology of subcortical visual centres in relation to cortical degeneration in Alzheimer’s disease. Neuropathol. Appl. Neurobiol. 1995, 21, 410–422. [Google Scholar] [CrossRef] [PubMed]
- Cardenas-Aguayo, M.d.C.; Gomez-Virgilio, L.; DeRosa, S.; Meraz-Rios, M.A. The role of Tau oligomers in the onset of Alzheimer’s disease neuropathology. ACS Chem. Neurosci. 2014, 5, 1178–1191. [Google Scholar] [CrossRef] [PubMed]
- Dugger, B.N.; Tu, M.; Murray, M.E.; Dickson, D.W. Disease specificity and pathologic progression of Tau pathology in brainstem nuclei of Alzheimer’s disease and progressive supranuclear palsy. Neurosci. Lett. 2011, 491, 122–126. [Google Scholar] [CrossRef] [PubMed]
- Kaltschmidt, B.; Heinrich, M.; Kaltschmidt, C. Stimulus-dependent activation of NF-κB specifies apoptosis or neuroprotection in cerebellar granule cells. Neuromol. Med. 2002, 2, 299–309. [Google Scholar] [CrossRef]
- Hirsch, E.C.; Graybiel, A.M.; Duyckaerts, C.; Javoy-Agid, F. Neuronal loss in the pedunculopontine tegmental nucleus in Parkinson disease and in progressive supranuclear palsy. Proc. Natl. Acad. Sci. USA 1987, 84, 5976–5980. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, M.S. The role of Tau in neurodegenerative diseases and its potential as a therapeutic target. Scientifica 2012, 2012, 796024. [Google Scholar] [CrossRef] [PubMed]
- Morishima, Y.; Gotoh, Y.; Zieg, J.; Barrett, T.; Takano, H.; Flavell, R.; Davis, R.J.; Shirasaki, Y.; Greenberg, M.E. Beta-amyloid induces neuronal apoptosis via a mechanism that involves the c-jun n-terminal kinase pathway and the induction of fas ligand. J. Neurosci. 2001, 21, 7551–7560. [Google Scholar] [PubMed]
- Awasthi, A.; Matsunaga, Y.; Yamada, T. Amyloid-β causes apoptosis of neuronal cells via caspase cascade, which can be prevented by amyloid-β-derived short peptides. Exp. Neurol. 2005, 196, 282–289. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.S.; Suen, K.C.; Kwok, N.S.; So, K.F.; Hugon, J.; Chang, R.C. β-amyloid peptides induces neuronal apoptosis via a mechanism independent of unfolded protein responses. Apoptosis 2006, 11, 687–700. [Google Scholar] [CrossRef] [PubMed]
- Einstein, G.; Buranosky, R.; Crain, B.J. Dendritic pathology of granule cells in Alzheimer’s disease is unrelated to neuritic plaques. J. Neurosci. 1994, 14, 5077–5088. [Google Scholar] [PubMed]
- Toledano, A.; Alvarez, M.I.; Rivas, L.; Lacruz, C.; Martinez-Rodriguez, R. Amyloid precursor proteins in the cerebellar cortex of Alzheimer’s disease patients devoid of cerebellar β-amyloid deposits: Immunocytochemical study of five cases. J. Neural Transm. 1999, 106, 1151–1169. [Google Scholar] [CrossRef] [PubMed]
- Galli, C.; Piccini, A.; Ciotti, M.T.; Castellani, L.; Calissano, P.; Zaccheo, D.; Tabaton, M. Increased amyloidogenic secretion in cerebellar granule cells undergoing apoptosis. Proc. Natl. Acad. Sci. USA 1998, 95, 1247–1252. [Google Scholar] [CrossRef] [PubMed]
- Blazquez-Llorca, L.; Garcia-Marin, V.; Merino-Serrais, P.; Avila, J.; DeFelipe, J. Abnormal Tau phosphorylation in the thorny excrescences of CA3 hippocampal neurons in patients with Alzheimer’s disease. J. Alzheimers Dis. 2011, 26, 683–698. [Google Scholar] [PubMed]
- Li, S.; Xiang, M. Barhl1 is required for maintenance of a large population of neurons in the zonal layer of the superior colliculus. Dev. Dyn. 2006, 235, 2260–2265. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Price, S.M.; Cahill, H.; Ryugo, D.K.; Shen, M.M.; Xiang, M. Hearing loss caused by progressive degeneration of cochlear hair cells in mice deficient for the BARHL1 homeobox gene. Development 2002, 129, 3523–3532. [Google Scholar] [PubMed]
- Albers, K. Hearing loss and dementia: New insights. Minn. Med. 2012, 95, 52–54. [Google Scholar] [PubMed]
- Sinha, U.K.; Hollen, K.M.; Rodriguez, R.; Miller, C.A. Auditory system degeneration in Alzheimer’s disease. Neurology 1993, 43, 779–785. [Google Scholar] [CrossRef] [PubMed]
- Janicki, S.C.; Park, N.; Cheng, R.; Clark, L.N.; Lee, J.H.; Schupf, N. Estrogen receptor α variants affect age at onset of Alzheimer’s disease in a multiethnic female cohort. Dement. Geriatr. Cogn. Disord. 2014, 38, 200–213. [Google Scholar] [CrossRef] [PubMed]
- Boada, M.; Antunez, C.; Lopez-Arrieta, J.; Caruz, A.; Moreno-Rey, C.; Ramirez-Lorca, R.; Moron, F.J.; Hernandez, I.; Mauleon, A.; Rosende-Roca, M.; et al. Estrogen receptor α gene variants are associated with Alzheimer’s disease. Neurobiol. Aging 2012, 33, 198e115–198e124. [Google Scholar] [CrossRef] [PubMed]
- Scacchi, R.; Gambina, G.; Broggio, E.; Corbo, R.M. Sex and ESR1 genotype may influence the response to treatment with Donepezil and Rivastigmine in patients with Alzheimer’s disease. Int. J. Geriatr. Psychiatry 2014, 29, 610–615. [Google Scholar] [CrossRef] [PubMed]
- Harte-Hargrove, L.C.; Maclusky, N.J.; Scharfman, H.E. Brain-derived neurotrophic factor-estrogen interactions in the hippocampal mossy fiber pathway: Implications for normal brain function and disease. Neuroscience 2013, 239, 46–66. [Google Scholar] [CrossRef] [PubMed]
- Cizas, P.; Jekabsone, A.; Borutaite, V.; Morkuniene, R. Prevention of amyloid-beta oligomer-induced neuronal death by EGTA, estradiol, and endocytosis inhibitor. Medicina 2011, 47, 107–112. [Google Scholar] [PubMed]
- Napolitano, M.; Costa, L.; Piacentini, R.; Grassi, C.; Lanzone, A.; Gulino, A. 17β-estradiol protects cerebellar granule cells against β-amyloid-induced toxicity via the apoptotic mitochondrial pathway. Neurosci. Lett. 2014, 561, 134–139. [Google Scholar] [CrossRef] [PubMed]


| Samples | Types | Number of Samples |
|---|---|---|
| Breast tumors | HR−/HER2− | 23 |
| HR−/HER2+ | 21 | |
| HR+/HER2− | 20 | |
| HR+/HER2+ | 13 | |
| Total | 77 | |
| Nervous system tumors | Neuroblastoma | 4 |
| Meningioma | 5 | |
| Glioma | 4 | |
| Peripheral nerve sheath | 3 | |
| Total | 16 | |
| Neurodegenerative diseases | Alzheimer’s disease | 10 |
| Lateral amyotrophic sclerosis | 6 | |
| Parkinson’s disease | 1 | |
| Total | 17 | |
| Total | 110 |
| Percentage of Cells (%) | Intensity | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Type of tissue | 0 | <50 | 50–75 | >75 | 100 | A | B | C | D | |
| Control | Hippocampus | 0 | 0 | 0 | 2 | 2 | 0 | 1 | 1 | 2 |
| Olfactory bulb | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 0 | |
| Medulla | 1 | 0 | 1 | 1 | 1 | 1 | 0 | 2 | 1 | |
| Disease | Alzheimer’s disease | 5 | 3 | 2 | 0 | 0 | 5 | 2 | 2 | 0 |
| Parkinson’s disease | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | |
| Lateral amyotrophic sclerosis | 0 | 1 | 2 | 0 | 3 | 0 | 2 | 2 | 2 | |
| Regulation of BARHL1 and ESR1 | References |
|---|---|
| BARHL1 upregulates NT-3 (neuro tropin 3 in mouse cerebellum) and thereby regulates the survival of cerebellar granule cells. | Li et al., 2004 [13] |
| The BARHL1 promoter has a TRβ binding site, and T3 (thyroid hormone) inhibits the expression of BARHL1. Thus, Brahl1 plays a role in impaired neuro-development caused by hypothyroidism. | Dong et al., 2011 [15] |
| ATOH1/MATH1 upregulates BARHL1 in inner ear and central nervous system. | Chellappa et al., 2008 [34] |
| ATOH1/MATH1 is repressed by PTF1a. | Pascual et al., 2007 [35] |
| TLE1 positively regulates ER-mediated gene expression and cell division. | Holmes et al., 2012 [36] |
| Thyroid hormone (T3) phosphorylates and activates ERα. | Meng et al., 2011 [37] |
| Estrogen positively regulates THRB in fish. | Filby et al., 2006 [38] |
| TTF2 inhibits transactivation of estrogen receptor-alpha in breast cancer cells. | Park et al., 2012 [39] |
| Estrogen increased the expression of NTF3, BDNF and NGF proteins. | Bimonte et al., 2004 [40] |
| Estrogens increase BDNF levels in the medial prefrontal cortex (PFC) and the hippocampus. | Luine et al., 2013 [41] |
| Estradiol induces the BDNF expression and positively regulates dendritic growth, spinogenesis and synaptogenesis in the developing Purkinje cell. | Zhu et al., 2013 [42] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barh, D.; García-Solano, M.E.; Tiwari, S.; Bhattacharya, A.; Jain, N.; Torres-Moreno, D.; Ferri, B.; Silva, A.; Azevedo, V.; Ghosh, P.; et al. BARHL1 Is Downregulated in Alzheimer’s Disease and May Regulate Cognitive Functions through ESR1 and Multiple Pathways. Genes 2017, 8, 245. https://doi.org/10.3390/genes8100245
Barh D, García-Solano ME, Tiwari S, Bhattacharya A, Jain N, Torres-Moreno D, Ferri B, Silva A, Azevedo V, Ghosh P, et al. BARHL1 Is Downregulated in Alzheimer’s Disease and May Regulate Cognitive Functions through ESR1 and Multiple Pathways. Genes. 2017; 8(10):245. https://doi.org/10.3390/genes8100245
Chicago/Turabian StyleBarh, Debmalya, María E. García-Solano, Sandeep Tiwari, Antaripa Bhattacharya, Neha Jain, Daniel Torres-Moreno, Belén Ferri, Artur Silva, Vasco Azevedo, Preetam Ghosh, and et al. 2017. "BARHL1 Is Downregulated in Alzheimer’s Disease and May Regulate Cognitive Functions through ESR1 and Multiple Pathways" Genes 8, no. 10: 245. https://doi.org/10.3390/genes8100245






