Targeted Next-Generation Sequencing Identification of Mutations in Disease Resistance Gene Analogs (RGAs) in Wild and Cultivated Beets
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. DNA Isolation
2.3. Library Preparation and Sequencing
2.4. Data Analysis
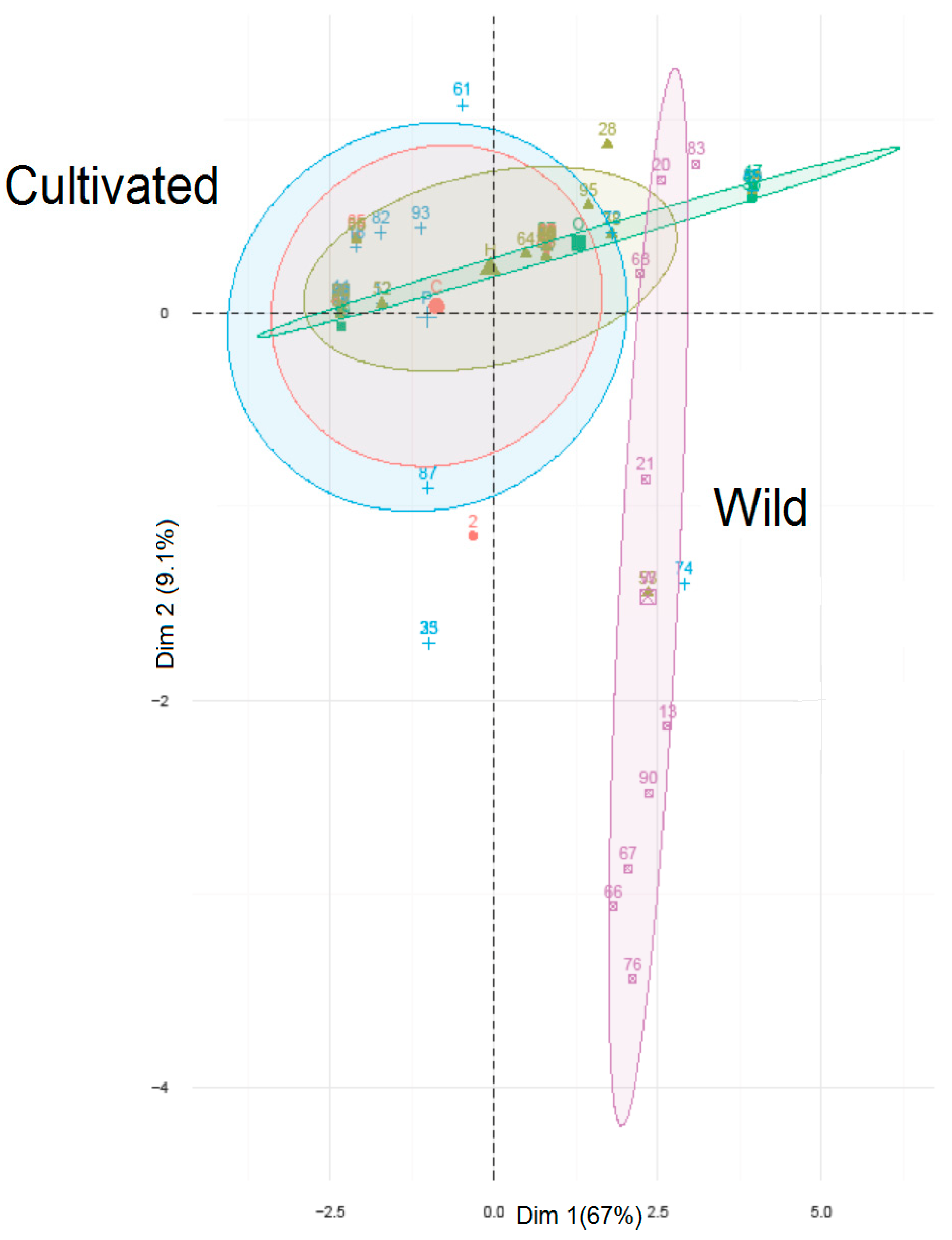
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Conflicts of Interest
References
- Food and Agriculture Organization of the United Nations (FAOSTAT). FAOSTAT Database. Available online: http://www.faostat.fao.org (accessed on 12 May 2016).
- Biancardi, E.; McGrath, J.M.; Panella, L.W.; Lewellen, R.T.; Stevanato, P. Sugar beet. In Tuber and Root Crops; Springer: New York, NY, USA, 2010; pp. 173–219. [Google Scholar]
- Pavli, O.I.; Stevanato, P.; Biancardi, E.; Skaracis, G.N. Achievements and prospects in breeding for rhizomania resistance in sugar beet. Field Crops Res. 2011, 122, 165–172. [Google Scholar] [CrossRef]
- Stevanato, P.; Trebbi, D.; Panella, L.; Richardson, K.; Broccanello, C.; Pakish, L.; Fenwick, A.L.; Saccomani, M. Identification and validation of a SNP marker linked to the gene HsBvm-1 for nematode resistance in sugar beet. Plant Mol. Biol. Rep. 2015, 33, 474–479. [Google Scholar] [CrossRef]
- Stevanato, P.; De Biaggi, M.; Skaracis, G.; Colombo, M.; Mandolino, G.; Biancardi, E. The sea beet (Beta vulgaris L. ssp. maritima) of the Adriatic coast as source of resistance for sugar beet. Sugar Tech. 2001, 3, 77–82. [Google Scholar] [CrossRef]
- Biancardi, E.; Lewellen, R.T.; De Biaggi, M.; Erichsen, A.W.; Stevanato, P. The origin of rhizomania resistance in sugar beet. Euphytica 2002, 127, 383–397. [Google Scholar] [CrossRef]
- Sekhwal, M.K.; Li, P.; Lam, I.; Wang, X.; Cloutier, S.; Yo, F.M. Disease resistance gene analogs (RGAs) in plants. Int. J. Mol. Sci. 2015, 16, 19248–19290. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Liu, X.; Dai, L.; Wang, G. Recent progress in elucidating the structure, function and evolution of disease resistance genes in plants. J. Genet. Genom. 2007, 34, 765–776. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, 1178–1186. [Google Scholar] [CrossRef] [PubMed]
- Monaco, M.K.; Stein, J.; Naithani, S.; Wei, S.; Dharmawardhana, P.; Kumari, S.; Amarasinghe, V.; Youens-Clark, K.; Thomason, J.; Preece, J.; et al. Gramene 2013: Comparative plant genomics resources. Nucleic Acids Res. 2014, 42, 1193–1199. [Google Scholar] [CrossRef] [PubMed]
- Nordberg, H.; Cantor, M.; Dusheyko, S.; Hua, S.; Poliakov, A.; Shabalov, I.; Smirnova, T.; Grigoriev, I.V.; Dubchak, I. The genome portal of the department of energy joint genome institute: 2014 Updates. Nucleic Acids Res. 2014, 42, 26–31. [Google Scholar] [CrossRef] [PubMed]
- Cai, D.; Kleine, M.; Kifle, S.; Harloff, H.J.; Sandal, N.N.; Marcker, K.A.; Klein-Lankhorst, R.M.; Salentijn, M.J.; Lange, W.; Stiekema, W.J.; et al. Positional cloning of a gene for nematode resistance in sugar beet. Science 1997, 275, 832–834. [Google Scholar] [CrossRef] [PubMed]
- Dohm, J.C.; Minoche, A.E.; Holtgräwe, D.; Capella-Gutiérrez, S.; Zakrzewski, F.; Tafer, H.; Rupp, O.; Sörensen, T.R.; Stracke, R.; Reinhardt, R.; et al. The genome of the recently domesticated crop plant sugar beet (Beta vulgaris). Nature 2014, 505–546. [Google Scholar] [CrossRef] [PubMed]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Nat. Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Li, Y.; Li, S.; Hu, N.; He, Y.; Pong, R.; Lin, D.; Lu, L.; Law, M. Comparison of Next-Generation sequencing systems. J. Biomed. Biotechnol. 2012, 11. [Google Scholar] [CrossRef] [PubMed]
- Stevanato, P.; Biscarini, F. Digital PCR as new approach to SNP genotyping in sugar beet. Sugar Tech. 2016, 18, 429–432. [Google Scholar] [CrossRef]
- Metzker, M.L. Sequencing technologies—The next generation. Nat. Rev. Genet. 2010, 11, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Capistrano-Gossmann, G.G.; Ries, D.; Holtgrawe, D.; Minoche, A.; Kraft, T.; Frerichmann, S.L.M.; Soerensen, T.R.; Dohm, J.C.; Gonzalez, I.; Schilhable, M.; et al. Crop wild relative populations of Beta vulgaris allow direct mapping of agronomically important genes. Nat. Commun. 2017, 8, 15708. [Google Scholar] [CrossRef] [PubMed]
- Grimmer, M.K.; Bean, K.M.R.; Luterbacher, M.C.; Stevens, M.; Asher, M.J.C. Beet mild yellowing virus resistance derived from wild and cultivated Beta germplasm. Plant Breeding 2008, 127, 315–318. [Google Scholar] [CrossRef]
- Chang, F.; Li, M.M. Clinical application of amplicon-based next-generation sequencing in cancer. Cancer Genet. 2013, 206, 413–419. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Zhang, E.; Ye, H.; Nandakumar, V.; Wang, Z.; Chen, L.; Tang, C.; Li, J.; Li, H.; Zhang, W.; et al. PIK3CA and TP53 gene mutations in human breast cancer tumors frequently detected by Ion Torrent DNA sequencing. PLoS ONE 2014, 9, 99306. [Google Scholar] [CrossRef] [PubMed]
- Hunger, S.; Di Gaspero, G.; Möhring, S.; Bellin, D.; Schäfer-Pregl, R.; Borchardt, D.C.; Durel, C.E.; Werber, M.; Weisshaar, B.; Salamini, F.; et al. Isolation and linkage analysis of expressed disease-resistance gene analogues of sugar beet (Beta vulgaris L.). Genome 2003, 46, 70–82. [Google Scholar] [CrossRef] [PubMed]
- Aulchenko, Y.S.; Ripke, S.; Isaacs, A.; van Duijn, C.M. GenABEL: An R library for genome-wide association analysis. Bioinformatics 2007, 23, 1294–1296. [Google Scholar] [CrossRef] [PubMed]
- Rousseeuw, P.J.; Ruts, I.; Tukey, J.W. The Bagplot: A Bivariate Boxplot. Am. Stat. 1999, 53, 382–387. [Google Scholar] [CrossRef]
- Bortz, J.; Lienert, G.; Boehnke, K. Verteilungsfreie Methoden in der Biostatistik; Springer: Berlin, Germany, 2000; ISBN 3-540-67590-6. [Google Scholar]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Statsoft STATISTICA. Available online: http://www.statsoft.com/Products/STATISTICA-Features/Version-12 (accessed on 18 March 2016).
- Sahu, K.K.; Chattopadhyay, D. Genome-wide sequence variations between wild and cultivated tomato species revisited by whole genome sequence mapping. BMC Genomics 2017, 18, 430. [Google Scholar] [CrossRef] [PubMed]
- Raamsdonk, L.W.D. Wild and cultivated plants: The parallelism between evolution and domestication. Evol. Trends Pl. 1993, 7, 73–84. [Google Scholar]
- Letscher, J.P.W.; Frese, L. Analysis of morphological variation in wild beet (Beta vulgaris L.) from Sicily. Genet. Resour. Crop Evol. 1993, 40, 15–24. [Google Scholar] [CrossRef]
- Quail, M.A.; Smith, M.; Coupland, P.; Otto, T.D.; Harris, S.R.; Connor, T.R.; Bertoni, A.; Swerdlow, H.P.; Gu, Y. A tale of three next generation sequencing platforms: Comparison of Ion Torrent, pacific biosciences and Illumnia MIseq sequencers. BMC Genomics 2012, 13, 341. [Google Scholar] [CrossRef] [PubMed]
- Rothberg, J.M.; Hinz, W.; Rearick, T.M.; Schultz, J.; Mileski, W.; Davey, M.; Leamon, J.H.; Johnson, K.; Milgrew, M.J.; Edwards, M.; et al. An integrated semiconductor device enabling non-optical genome sequencing. Nature 2011, 475, 348–352. [Google Scholar] [CrossRef] [PubMed]
- Beseli, A.; Noar, R.; Daub, M.E. Characterization of Cercospora nicotianae hypothetical proteins in Cercoporin resistance. PLoS ONE 2015. [Google Scholar] [CrossRef] [PubMed]
- Hammond-Kosack, K.E.; Jones, J.D. Plant disease resistance genes. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1997, 48, 575–607. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.W.; Han, S.W.; Sririyanum, M.; Park, C.J.; Seo, Y.S.; Ronald, P.C. A type I-secreted, sulfated peptide triggers Xa21-mediated innate immunity. Science 2009, 326, 850–853. [Google Scholar] [CrossRef] [PubMed]
| Sample ID | Number of SNP | Genotype | Sample ID | Number of SNP | Genotype | Sample ID | Number of SNP | Genotype |
|---|---|---|---|---|---|---|---|---|
| 33 | 278 | P | 83 | 151 | W | 5 | 120 | O |
| 82 | 248 | P | 9 | 150 | O | 53 | 118 | H |
| 67 | 225 | W | 25 | 150 | P | 17 | 116 | O |
| 90 | 222 | W | 46 | 149 | H | 16 | 114 | P |
| 96 | 218 | H | 76 | 149 | W | 58 | 113 | O |
| 88 | 206 | P | 55 | 148 | H | 50 | 112 | O |
| 73 | 190 | C | 62 | 148 | H | 56 | 112 | H |
| 80 | 189 | P | 92 | 148 | H | 70 | 110 | H |
| 2 | 188 | C | 75 | 147 | H | 79 | 109 | P |
| 42 | 187 | C | 22 | 145 | H | 86 | 103 | H |
| 63 | 184 | H | 14 | 144 | H | 91 | 103 | P |
| 8 | 180 | H | 77 | 144 | P | 45 | 98 | O |
| 23 | 180 | H | 49 | 143 | C | 65 | 96 | C |
| 74 | 176 | P | 6 | 142 | P | 39 | 92 | H |
| 59 | 173 | H | 3 | 141 | P | 78 | 90 | H |
| 7 | 171 | H | 41 | 139 | O | 26 | 86 | P |
| 1 | 168 | O | 48 | 139 | P | 37 | 86 | H |
| 15 | 168 | H | 18 | 138 | O | 12 | 85 | P |
| 66 | 168 | W | 27 | 137 | O | 93 | 84 | P |
| 71 | 166 | P | 31 | 136 | H | 10 | 82 | P |
| 32 | 165 | H | 34 | 135 | P | 36 | 81 | H |
| 40 | 163 | P | 94 | 134 | P | 19 | 75 | O |
| 24 | 160 | H | 30 | 132 | H | 21 | 71 | W |
| 52 | 160 | H | 47 | 132 | H | 38 | 71 | H |
| 72 | 157 | P | 89 | 132 | P | 87 | 71 | P |
| 13 | 155 | W | 11 | 129 | P | 35 | 57 | O |
| 43 | 155 | H | 64 | 129 | H | 60 | 57 | P |
| 20 | 154 | W | 95 | 129 | H | 28 | 31 | H |
| 4 | 152 | P | 51 | 127 | C | 61 | 31 | P |
| 54 | 152 | H | 81 | 126 | C | - | - | - |
| 68 | 152 | W | 57 | 121 | C | - | - | - |
| Gene ID | NCBI Accession Number | Scaffold | Length (bp) | SNP (n) | MNP (n) | CDS | Non CDS | Freq. of SNP (%) | Freq. of MNP (%) |
|---|---|---|---|---|---|---|---|---|---|
| Bv7_171470_ojty | BU089572 | Bvchr7.sca021 | 7827 | 57 | 2 | 31 | 18 | 0.73 | 0.03 |
| Bv2_043450_zhxk | BU089578 | Bvchr2.sca016 | 6687 | 0 | 0 | 0 | 0 | 0.00 | 0.00 |
| Bv9_225140_hpxq | BH897910 | Bvchr9.sca026 | 4738 | 0 | 0 | 0 | 0 | 0.00 | 0.00 |
| Bv6_147620_tzmc | BU089568 | Bvchr6.sca027 | 5232 | 0 | 0 | 0 | 0 | 0.00 | 0.00 |
| Bv6_152400_uaqg | BU089560 | Bvchr6.sca028 | 4164 | 9 | 1 | 0 | 10 | 0.22 | 0.02 |
| Bv7_172060_pnhe | BU089566 | Bvchr7.sca021 | 6568 | 29 | 1 | 12 | 18 | 0.44 | 0.02 |
| Bv8_184910_pkon | BU089558 | Bvchr8.sca001 | 4504 | 51 | 1 | 23 | 29 | 1.13 | 0.02 |
| Bv1_004790_mxoz | BU089550 | Bvchr1.sca001 | 5299 | 44 | 5 | 16 | 33 | 0.83 | 0.09 |
| Bv3_055670_fxjn | BU089554 | Bvchr3.sca003 | 6902 | 35 | 1 | 15 | 21 | 0.51 | 0.01 |
| Bv3_061550_ctrc | BU089552 | Bvchr3.sca008 | 4365 | 42 | 3 | 22 | 23 | 0.96 | 0.07 |
| Bv3_063740_usyf | BH897908 | Bvchr3.sca010 | 3986 | 116 | 11 | 61 | 66 | 2.91 | 0.28 |
| Bv5_094270_ijae | BU089565 | Bvchr5.sca002 | 5117 | 51 | 2 | 28 | 25 | 1.00 | 0.04 |
| Bv5_103960_ophq | BU089548 | Bvchr5.sca008 | 1783 | 0 | 0 | 0 | 0 | 0.00 | 0.00 |
| Bv6_148520_dzyz | BU089547 | Bvchr6.sca027 | 3628 | 0 | 0 | 0 | 0 | 0.00 | 0.00 |
| Bv7_157960_wiwi | BU089579 | Bvchr7.sca001 | 3891 | 14 | 1 | 15 | 0 | 0.36 | 0.03 |
| Bv7_160190_sdkg | BU089562 | Bvchr7.sca002 | 2767 | 0 | 0 | 0 | 0 | 0,00 | 0.00 |
| Bv7_171340_mgxu | BU089575 | Bvchr7.sca021 | 4685 | 74 | 9 | 44 | 39 | 1.58 | 0.19 |
| Bv7_175730_zcms | BU089561 | Bvchr7.sca021 | 3479 | 0 | 0 | 0 | 0 | 0.00 | 0.00 |
| Bv7u_181380_zcug | BH897904 | Bvchr7_un.sca003 | 3667 | 0 | 0 | 0 | 0 | 0.00 | 0.00 |
| Bv9_205550_geqf | BH897907 | Bvchr9.sca001 | 4424 | 0 | 0 | 0 | 0 | 0.00 | 0.00 |
| Bv9_218680_gqnp | BU089549 | Bvchr9.sca025 | 7235 | 0 | 0 | 0 | 0 | 0.00 | 0.00 |
| Gene ID | Sample ID | MNP/Indel | RGA Class | Annotation |
|---|---|---|---|---|
| Bv7_171470_ojty | 16 | AG/GA | NLcc | Disease resistance protein At4g27190 |
| 16 | AGT/TTC | |||
| 52 | AGA/CGC | |||
| 95 | AA/CT | |||
| 95 | AG/GC | |||
| Bv8_184910_pkon | 13 | CCCTCC/- | RLK | hypothetical protein |
| Bv1_004790_mxoz | 8 | GCCCC/CCCCT | Pto | U-box domain-containing protein 33 AltName: Full = Plant U-box protein 33; Includes: RecName: Full = E3 ubiquitin ligase; EC = 6.3.2.-; Includes: RecName: Full = Serine/threonine-protein kinase; EC = 2.7.11.-; |
| Bv1_004790_mxoz | 71 | -/AT | Pto | U-box domain-containing protein 33 AltName: Full = Plant U-box protein 33; Includes: RecName: Full = E3 ubiquitin ligase; EC = 6.3.2.-; Includes: RecName: Full = Serine/threonine-protein kinase; EC = 2.7.11.-; |
| Bv3_055670_fxjn | 77 | CA/- | Pto | U-box domain-containing protein 32 EC = 6.3.2.-; AltName: Full = Plant U-box protein 32; |
| Bv3_061550_ctrc | 11 | AT/GG | Pto | hypothetical protein |
| 31 | GAC/GAC | |||
| 35 | CT/AC | |||
| 54 | TTG/TTG | |||
| Bv3_063740_usyf | 9 | CG/GA | NLcc | Putative disease resistance protein RGA3 AltName: Full = RGA1-blb; AltName: Full = Blight resistance protein B149; |
| 9 | GG/AC | |||
| 38 | CT/GC | |||
| Bv5_094270_ijae | 71 | TA/AC | RLK | hypothetical protein |
| Bv7_157960_wiwi Bv7_157960_wiwi Bv7_157960_wiwi | 41 | TT/CC | CNL | Putative disease resistance protein RGA3 AltName: Full = RGA1-blb; AltName: Full = Blight resistance protein B149; |
| 57 | CCA/CCA | |||
| 67 | GG/GG | |||
| Bv7_171340_mgxu | 12 | AATAACCC/AATAACCC | Ncc | Probable disease resistance protein At4g27220 |
| 40 | AGT/CAC | |||
| 40 | AT/TC | |||
| 40 | ATG/CCA | |||
| 68 | -/TCCTTCA |
| Beet Groups | ||||||||
|---|---|---|---|---|---|---|---|---|
| Gene ID | Gene Name | C | H | O | P | W | Total | CV (%) |
| 1 | Bv7_171470_ojty | 14 | 12 | 7 | 8 | 15 | 56 | 31.0 |
| 2 | Bv2_043450_zhxk | 0 | 0 | 0 | 0 | 0 | 0 | 0.0 |
| 3 | Bv9_225140_hpxq | 0 | 0 | 0 | 0 | 0 | 0 | 0.0 |
| 4 | Bv6_147620_tzmc | 0 | 0 | 0 | 0 | 0 | 0 | 0.0 |
| 5 | Bv6_152400_uaqg | 3 | 3 | 2 | 3 | 3 | 14 | 15.9 |
| 6 | Bv7_172060_pnhe | 3 | 1 | 3 | 1 | 1 | 9 | 60.0 |
| 7 | Bv8_184910_pkon | 12 | 11 | 6 | 11 | 9 | 49 | 24.3 |
| 8 | Bv1_004790_mxoz | 22 | 11 | 17 | 13 | 11 | 74 | 31.8 |
| 9 | Bv3_055670_fxjn | 4 | 3 | 4 | 2 | 3 | 16 | 26.1 |
| 10 | Bv3_061550_ctrc | 8 | 4 | 7 | 3 | 4 | 26 | 41.6 |
| 11 | Bv3_063740_usyf | 22 | 26 | 18 | 34 | 29 | 129 | 23.9 |
| 12 | Bv5_094270_ijae | 17 | 12 | 13 | 10 | 15 | 67 | 20.1 |
| 13 | Bv5_103960_ophq | 0 | 0 | 0 | 0 | 0 | 0 | 0.0 |
| 14 | Bv6_148520_dzyz | 0 | 0 | 0 | 0 | 0 | 0 | 0.0 |
| 15 | Bv7_157960_wiwi | 3 | 1 | 3 | 2 | 1 | 10 | 50.0 |
| 16 | Bv7_160190_sdkg | 0 | 0 | 0 | 0 | 0 | 0 | 0.0 |
| 17 | Bv7_171340_mgxu | 24 | 20 | 15 | 12 | 26 | 97 | 30.4 |
| 18 | Bv7_175730_zcms | 0 | 0 | 0 | 0 | 0 | 0 | 0.0 |
| 19 | Bv7u_181380_zcug | 0 | 0 | 0 | 0 | 0 | 0 | 0.0 |
| 20 | Bv9_205550_geqf | 0 | 0 | 0 | 0 | 0 | 0 | 0.0 |
| 21 | Bv9_218680_gqnp | 0 | 0 | 0 | 0 | 0 | 0 | 0.0 |
| Total | 132 | 104 | 95 | 99 | 117 | 621 | ||
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stevanato, P.; Broccanello, C.; Pajola, L.; Biscarini, F.; Richards, C.; Panella, L.; Hassani, M.; Formentin, E.; Chiodi, C.; Concheri, G.; et al. Targeted Next-Generation Sequencing Identification of Mutations in Disease Resistance Gene Analogs (RGAs) in Wild and Cultivated Beets. Genes 2017, 8, 264. https://doi.org/10.3390/genes8100264
Stevanato P, Broccanello C, Pajola L, Biscarini F, Richards C, Panella L, Hassani M, Formentin E, Chiodi C, Concheri G, et al. Targeted Next-Generation Sequencing Identification of Mutations in Disease Resistance Gene Analogs (RGAs) in Wild and Cultivated Beets. Genes. 2017; 8(10):264. https://doi.org/10.3390/genes8100264
Chicago/Turabian StyleStevanato, Piergiorgio, Chiara Broccanello, Luca Pajola, Filippo Biscarini, Chris Richards, Lee Panella, Mahdi Hassani, Elide Formentin, Claudia Chiodi, Giuseppe Concheri, and et al. 2017. "Targeted Next-Generation Sequencing Identification of Mutations in Disease Resistance Gene Analogs (RGAs) in Wild and Cultivated Beets" Genes 8, no. 10: 264. https://doi.org/10.3390/genes8100264






