Chromosomal Evolution in Chiroptera
Abstract
:1. Introduction
2. Genomic Features of Bats and Perspectives—An Overview of Repetitive DNAs and Implications for Chromosome Evolution
3. Evolution of Genome Architecture
3.1. Conserved Karyotype Evolution
3.2. Moderate Chromosomal Evolution
3.3. Extreme Karyotype Reshuffling
4. Overview of Chromosomal Evolution within Chiropteran Families
4.1. Yinpterochiroptera (Pteropodiformes)
4.1.1. Pteropodidae
Rhinolophoidea
4.1.2. Megadermatidae
4.1.3. Hipposideridae
4.1.4. Rhinolophidae
4.2. Vespertilioniformes or Yangochiroptera (sensu Teeling et al., 2002)
4.2.1. Mormoopidae
4.2.2. Noctilionidae
4.2.3. Phyllostomidae
4.2.4. Nycteridae
4.2.5. Emballonuridae
4.2.6. Molossidae
4.2.7. Miniopteridae
4.2.8. Vespertilionidae
5. Chromosomes as Tools to Resolve Phylogenetic Problems
6. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Simmons, N.B. Order Chiroptera. In Mammal Species of the World: A Taxonomic and Geographic Reference, 3rd ed.; Wilson, D., Reeder, D., Eds.; Johns Hopkins University Press: Baltimore, MD, USA, 2005; pp. 312–529. [Google Scholar]
- Solari, S.; Martínez-Arias, V. Cambios recientes en la sistemática y taxonomía de murciélagos neotropicales (Mammalia: Chiroptera). Therya 2014, 5, 167–196. [Google Scholar] [CrossRef]
- Amador, L.I.; Arévalo, R.L.M.; Almeida, F.C.; Catalano, S.A.; Giannini, N.P. Bat systematics in the light of unconstrained analyses of a comprehensive molecular supermatrix. J. Mammal. Evol. 2016, 23, 1–34. [Google Scholar] [CrossRef]
- Fenton, M.B.; Simmons, N.B. Bats: A World of Science and Mystery; University of Chicago Press: Chicago, IL, USA, 2015; p. 303. [Google Scholar]
- Kunz, T.H.; Fenton, M.B. Bat Ecology; University of Chicago Press: Chicago, IL, USA, 2005. [Google Scholar]
- Seim, I.; Fang, X.; Xiong, Z.; Lobanov, A.V.; Huang, Z.; Ma, S.; Feng, Y.; Turanov, A.A.; Zhu, Y.; Lenz, T.L. Genome analysis reveals insights into physiology and longevity of the Brandt’s bat Myotis brandtii. Nat. Commun. 2013, 4, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, G.; Cowled, C.; Shi, Z.; Huang, Z.; Bishop-Lilly, K.A.; Fang, X.; Wynne, J.W.; Xiong, Z.; Baker, M.L.; Zhao, W. Comparative analysis of bat genomes provides insight into the evolution of flight and immunity. Science 2013, 339, 456–460. [Google Scholar] [CrossRef] [PubMed]
- Hood, C.S.; Jones, J.K., Jr. Noctilio leporinus. Mamm. Species 1984, 216, 1–7. [Google Scholar] [CrossRef]
- Rodrigues, F.H.; Reis, M.L.; Braz, V.S. Food habits of the frog-eating bat, Trachops cirrhosus, in atlantic forest of northeastern Brazil. Chiroptera Neotrop. 2014, 10, 180–182. [Google Scholar]
- Ibáñez, C.; Popa-Lisseanu, A.G.; Pastor-Beviá, D.; García-Mudarra, J.L.; Juste, J. Concealed by darkness: Interactions between predatory bats and nocturnally migrating songbirds illuminated by DNA sequencing. Mol. Ecol. 2016, 25, 5254–5263. [Google Scholar] [CrossRef] [PubMed]
- Rojas, D.; Vale, A.; Ferrero, V.; Navarro, L. When did plants become important to leaf-nosed bats? Diversification of feeding habits in the family Phyllostomidae. Mol. Ecol. 2011, 20, 2217–2228. [Google Scholar] [CrossRef] [PubMed]
- Freeman, P.W. Macroevolution in Microchiroptera: Recoupling morphology and ecology with phylogeny. Evol. Ecol. Res. 2000, 2, 317–335. [Google Scholar]
- Lekagul, B.; McNeely, J. Mammals of Thailand, 2nd ed.; The Association for the Conservation of Thailand: Bankok, Thailand, 1988; p. 758. [Google Scholar]
- Teeling, E.C.; Springer, M.S.; Madsen, O.; Bates, P.; O’brien, S.J.; Murphy, W.J. A molecular phylogeny for bats illuminates biogeography and the fossil record. Science 2005, 307, 580–584. [Google Scholar] [CrossRef] [PubMed]
- Jones, G.; Teeling, E.C. The evolution of echolocation in bats. Trends Ecol. Evol. 2006, 21, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Lack, J.B.; Roehrs, Z.P.; Stanley, C.E.; Ruedi, M.; Van Den Bussche, R.A. Molecular phylogenetics of Myotis indicate familial-level divergence for the genus Cistugo (Chiroptera). J. Mammal. 2010, 91, 976–992. [Google Scholar] [CrossRef]
- Hoofer, S.R.; Van Den Bussche, R.A. Molecular phylogenetics of the chiropteran family Vespertilionidae. Acta Chiropterol. 2003, 5, 1–63. [Google Scholar] [CrossRef]
- Miller-Butterworth, C.M.; Murphy, W.J.; O’brien, S.J.; Jacobs, D.S.; Springer, M.S.; Teeling, E.C. A family matter: Conclusive resolution of the taxonomic position of the long-fingered bats. Miniopterus. Mol. Biol. Evol. 2007, 24, 1553–1561. [Google Scholar] [CrossRef] [PubMed]
- Foley, N.M.; Thong, V.D.; Soisook, P.; Goodman, S.M.; Armstrong, K.N.; Jacobs, D.S.; Puechmaille, S.J.; Teeling, E.C. How and why overcome the impediments to resolution: Lessons from rhinolophid and hipposiderid bats. Mol. Biol. Evol. 2015, 32, 313–333. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.J.; Rabosky, D.L. Speciation dynamics during the global radiation of extant bats. Evolution 2015, 69, 1528–1545. [Google Scholar] [CrossRef] [PubMed]
- Meredith, R.W.; Janečka, J.E.; Gatesy, J.; Ryder, O.A.; Fisher, C.A.; Teeling, E.C.; Goodbla, A.; Eizirik, E.; Simão, T.L.; Stadler, T. Impacts of the cretaceous terrestrial revolution and kpg extinction on mammal diversification. Science 2011, 334, 521–524. [Google Scholar] [CrossRef] [PubMed]
- Gunnell, G.F.; Simmons, N.B. Fossil evidence and the origin of bats. J. Mammal. Evol. 2005, 12, 209–246. [Google Scholar] [CrossRef]
- Nery, M.F.; González, D.J.; Hoffmann, F.G.; Opazo, J.C. Resolution of the laurasiatherian phylogeny: Evidence from genomic data. Mol. Phylogen. Evol. 2012, 64, 685–689. [Google Scholar] [CrossRef] [PubMed]
- Simmons, N.B. The case for chiropteran monophyly. Am. Mus. Novit. 1994, 3103, 1–54. [Google Scholar]
- Van Den Bussche, R.A.; Hoofer, S.R. Phylogenetic relationships among recent chiropteran families and the importance of choosing appropriate out-group taxa. J. Mammal. 2004, 85, 321–330. [Google Scholar] [CrossRef]
- Smith, J.; Bickham, J.; Gregory, T. Patterns of genome size diversity in bats (order Chiroptera). Genome 2013, 56, 457–472. [Google Scholar] [CrossRef] [PubMed]
- Van Den Bussche, R.; Longmire, J.; Baker, R. How bats achieve a small c-value: Frequency of repetitive DNA in Macrotus. Mamm. Genome 1995, 6, 521–525. [Google Scholar] [CrossRef] [PubMed]
- Kasai, F.; O’Brien, P.C.; Ferguson-Smith, M.A. The bat genome: GC-biased small chromosomes associated with reduction in genome size. Chromosoma 2013, 122, 535–540. [Google Scholar] [CrossRef] [PubMed]
- Ray, D.A.; Feschotte, C.; Pagan, H.J.; Smith, J.D.; Pritham, E.J.; Arensburger, P.; Atkinson, P.W.; Craig, N.L. Multiple waves of recent DNA transposon activity in the bat, Myotis lucifugus. Genome Res. 2008, 18, 717–728. [Google Scholar] [CrossRef] [PubMed]
- Ray, D.A.; Pagan, H.J.; Thompson, M.L.; Stevens, R.D. Bats with hats: Evidence for recent DNA transposon activity in genus Myotis. Mol. Biol. Evol. 2007, 24, 632–639. [Google Scholar] [CrossRef] [PubMed]
- Thomas, J.; Sorourian, M.; Ray, D.; Baker, R.J.; Pritham, E.J. The limited distribution of helitrons to vesper bats supports horizontal transfer. Gene 2011, 474, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Pace, J.K.; Feschotte, C. The evolutionary history of human DNA transposons: Evidence for intense activity in the primate lineage. Genome Res. 2007, 17, 422–432. [Google Scholar] [CrossRef] [PubMed]
- Platt, R.N.; Mangum, S.F.; Ray, D.A. Pinpointing the vesper bat transposon revolution using the Miniopterus natalensis genome. Mobile DNA 2016, 7, 12. [Google Scholar] [CrossRef] [PubMed]
- Gray, Y.H. It takes two transposons to tango: Transposable-element-mediated chromosomal rearrangements. Trends Genet. 2000, 16, 461–468. [Google Scholar] [CrossRef]
- Volleth, M.; Heller, K.-G. Varations on a theme: Karyotype comparison in eurasian Myotis species and implications for phylogeny. Vespertilio 2012, 16, 329–350. [Google Scholar]
- Cantrell, M.A.; Scott, L.; Brown, C.J.; Martinez, A.R.; Wichman, H.A. Loss of LINE-1 activity in the megabats. Genetics 2008, 178, 393–404. [Google Scholar] [CrossRef] [PubMed]
- Casavant, N.C.; Scott, L.; Cantrell, M.A.; Wiggins, L.E.; Baker, R.J.; Wichman, H.A. The end of the LINE?: Lack of recent L1 activity in a group of South American rodents. Genetics 2000, 154, 1809–1817. [Google Scholar] [PubMed]
- Erickson, I.K.; Cantrell, M.A.; Scott, L.; Wichman, H.A. Retrofitting the genome: L1 extinction follows endogenous retroviral expansion in a group of muroid rodents. J. Virol. 2011, 85, 12315–12323. [Google Scholar] [CrossRef] [PubMed]
- Grahn, R.; Rinehart, T.; Cantrell, M.; Wichman, H. Extinction of LINE-1 activity coincident with a major mammalian radiation in rodents. Cytogenet. Genome Res. 2005, 110, 407–415. [Google Scholar] [CrossRef] [PubMed]
- Parish, D.; Vise, P.; Wichman, H.; Bull, J.; Baker, R. Distribution of LINEs and other repetitive elements in the karyotype of the bat Carollia: Implications for X-chromosome inactivation. Cytogenet. Genome Res. 2002, 96, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Sotero-Caio, C.G.; Volleth, M.; Hoffmann, F.G.; Scott, L.; Wichman, H.A.; Yang, F.; Baker, R.J. Integration of molecular cytogenetics, dated molecular phylogeny, and model-based predictions to understand the extreme chromosome reorganization in the neotropical genus Tonatia (Chiroptera: Phyllostomidae). BMC Evol. Biol. 2015, 15, 220. [Google Scholar] [CrossRef] [PubMed]
- Souza, E.M.S.; Gross, M.C.; Silva, C.E.F.; Sotero-Caio, C.G.; Feldberg, E. Heterochromatin variation and LINE-1 distribution in Artibeus (Chiroptera: Phyllostomidae) from central amazon, Brazil. Comp. Cytogenet. 2017, 11, 613. [Google Scholar] [CrossRef]
- Sotero-Caio, C.G.; Cabral-de-Mello, D.; Calixto, M.S.; Valente, G.T.; Martins, C.; Loreto, V.; Souza, M.J.; Santos, N. Centromeric enrichment of LINE-1 retrotransposons and its significance for the chromosome evolution of phyllostomid bats. Chromosom. Res. 2017, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Calixto, M.S.; Andrade, I.S.; Cabral-de-Mello, D.C.; Santos, N.; Martins, C.; Loreto, V.; de Souza, M.J. Patterns of rDNA and telomeric sequences diversification: Contribution to repetitive DNA organization in Phyllostomidae bats. Genetica 2013, 142, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Baker, R.J.; Maltbie, M.; Owen, J.G.; Hamilton, M.J.; Bradley, R.D. Reduced number of ribosomal sites in bats: Evidence for a mechanism to contain genome size. J. Mammal. 1992, 73, 847–858. [Google Scholar] [CrossRef]
- Santos, N.; Fagundes, V.; Yonenaga-Yassuda, Y.; Souza, M.J. Localization of rRNA genes in phyllostomidae bats reveals silent NORs in Artibeus cinereus. Hereditas 2002, 136, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Santos, N.; Fagundes, V.; Yonenaga-Yassuda, Y.; Souza, M.J. Comparative karyology of brazilian vampire bats Desmodus rotundus and Diphylla ecaudata (Phyllostomidae, Chiroptera): Banding patterns, base-specific fluorochromes and FISH of ribosomal genes. Hereditas 2001, 134, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Faria, K.; Marchesin, S.; Moreira, P.; Beguelini, M.; Morielle-Versute, E. New insights into telomeric DNA sequence (TTAGGG)n location in bat chromosomes. Genet. Mol. Res. 2009, 8, 1079–1084. [Google Scholar] [CrossRef] [PubMed]
- Ono, T.; Yoshida, M.C. Differences in the chromosomal distribution of telomeric (TTAGGG)n sequences in two species of the vespertilionid bats. Chromosom. Res. 1997, 5, 203–205. [Google Scholar] [CrossRef]
- Finato, A.O.; Varella-Garcia, M.; Tajara, E.H.; Taddei, V.A.; Morielle-Versute, E. Intrachromosomal distribution of telomeric repeats in Eumops glaucinus and Eumops perotis (Molossidae, Chiroptera). Chromosom. Res. 2000, 8, 563–569. [Google Scholar] [CrossRef]
- Meyne, J.; Baker, R.J.; Hobart, H.H.; Hsu, T.C.; Ryder, O.A.; Ward, O.G.; Wiley, J.E.; Wurster-Hill, D.H.; Yates, T.L.; Moyzis, R.K. Distribution of non-telomeric sites of the (TTAGGG)n telomeric sequence in vertebrate chromosomes. Chromosoma 1990, 99, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Multani, A.; Ozen, M.; Furlong, C.; Zhao, Y.-J.; Hsu, T.; Pathak, S. Heterochromatin and interstitial telomeric DNA homology. Chromosoma 2001, 110, 214–220. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.K.; Simmons, M.J. Gross chromosome rearrangements mediated by transposable elements in Drosophila melanogaster. Bioessays 1994, 16, 269–275. [Google Scholar] [CrossRef] [PubMed]
- Hedges, D.; Deininger, P. Inviting instability: Transposable elements, double-strand breaks, and the maintenance of genome integrity. Mutat. Res. 2007, 616, 46–59. [Google Scholar] [CrossRef] [PubMed]
- Carbone, L.; Harris, R.A.; Gnerre, S.; Veeramah, K.R.; Lorente-Galdos, B.; Huddleston, J.; Meyer, T.J.; Herrero, J.; Roos, C.; Aken, B. Gibbon genome and the fast karyotype evolution of small apes. Nature 2014, 513, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Volleth, M.; Heller, K. Chromosome number reduction accompanied by extensive heterochromatin addition in the bat Glauconycteris beatrix (Mammalia; Chiroptera, Vespertilionidae). Cytogenet. Genome Res. 2007, 119, 245–247. [Google Scholar] [CrossRef] [PubMed]
- Naidu, K.N.; Gururaj, M.E. Karyotypic architecture of the false vampire bat Megaderma lyra. Cytologia 1985, 50, 913–919. [Google Scholar] [CrossRef]
- Baker, R.J.; Bickham, J.W. Speciation by monobrachial centric fusions. Proc. Natl. Acad. Sci. USA 1986, 83, 8245–8248. [Google Scholar] [CrossRef] [PubMed]
- Pieczarka, J.C.; Nagamachi, C.Y.; O’Brien, P.C.; Yang, F.; Rens, W.; Barros, R.M.S.; Noronha, R.C.; Rissino, J.; de Oliveira, E.H.; Ferguson-Smith, M.A. Reciprocal chromosome painting between two South American bats: Carollia brevicauda and Phyllostomus hastatus (Phyllostomidae, Chiroptera). Chromosom. Res. 2005, 13, 339–347. [Google Scholar] [CrossRef] [PubMed]
- Ribas, T.F.; Rodrigues, L.R.; Nagamachi, C.Y.; Gomes, A.J.; Benathar, T.C.; Yang, F.; Ferguson-Smith, M.A.; Pieczarka, J.C. Two new cytotypes reinforce that Micronycteris hirsuta Peters, 1869 does not represent a monotypic taxon. BMC Genet. 2013, 14, 119. [Google Scholar] [CrossRef] [PubMed]
- Ribas, T.; Rodrigues, L.; Nagamachi, C.; Gomes, A.; Rissino, J.; O’Brien, P.; Yang, F.; Ferguson-Smith, M.; Pieczarka, J. Phylogenetic reconstruction by cross-species chromosome painting and G-banding in four species of Phyllostomini tribe (Chiroptera, Phyllostomidae) in the Brazilian Amazon: An independent evidence for monophyly. PLoS ONE 2015, 10, e0122845. [Google Scholar] [CrossRef] [PubMed]
- Volleth, M.; Heller, K.-G.; Pfeiffer, R.A.; Hameister, H.A. Comparative Zoo-FISH analysis in bats elucidates the phylogenetic relationships between Megachiroptera and five Microchiropteran families. Chromosom. Res. 2002, 10, 477–497. [Google Scholar] [CrossRef]
- Baker, R.J.; Bickham, J.W. Karyotypic evolution in bats: Evidence of extensive and conservative chromosomal evolution in closely related taxa. Syst. Zool. 1980, 29, 239–253. [Google Scholar] [CrossRef]
- Hoffmann, F.G.; McGuire, L.P.; Counterman, B.A.; Ray, D.A. Transposable elements and small RNAs: Genomic fuel for species diversity. Mob. Genet. Elem. 2015, 5, 63–66. [Google Scholar] [CrossRef] [PubMed]
- Bickham, J.W. Chromosomal variation and evolutionary relationships of vespertilionid bats. J. Mammal. 1979, 60, 350–363. [Google Scholar] [CrossRef]
- Volleth, M.; Bronner, G.; Göpfert, M.; Heller, K.-G.; Von Helversen, O.; Yong, H.-S. Karyotype comparison and phylogenetic relationships of Pipistrellus-like bats (Vespertilionidae; Chiroptera; Mammalia). Chromosom. Res. 2001, 9, 25–46. [Google Scholar] [CrossRef]
- Rautenbach, I.; Bronner, G.; Schlitter, D. Karyotypic data and attendant systematic implications for the bats of southern Africa. Koedoe 1993, 36, 87–104. [Google Scholar] [CrossRef]
- Mao, X.; Nie, W.; Wang, J.; Su, W.; Feng, Q.; Wang, Y.; Dobigny, G.; Yang, F. Comparative cytogenetics of bats (Chiroptera): The prevalence of Robertsonian translocations limits the power of chromosomal characters in resolving interfamily phylogenetic relationships. Chromosom. Res. 2008, 16, 155–170. [Google Scholar] [CrossRef] [PubMed]
- Hood, C.S.; Baker, R.J. G- and C-banding chromosomal studies of bats of the family Emballonuridae. J. Mammal. 1986, 67, 705–711. [Google Scholar] [CrossRef]
- Araújo, R.E.F.; Nagamachi, C.Y.; Costa, M.J.R.; Noronha, R.C.R.; Rodrigues, L.R.R.; Pieczarka, J.C. First description of multivalent ring structures in eutherian mammalian meiosis: New chromosomal characterization of Cormura brevirostris (Emballonuridae, Chiroptera). Genetica 2016, 144, 407–415. [Google Scholar] [CrossRef] [PubMed]
- Sreepada, K.; Naidu, K.N.; Gururaj, M. Chromosomal variations in four indian species of Taphozous (Chiroptera: Mammalia). Biol. Zentralbl. 1995, 114, 307–314. [Google Scholar]
- Naidu, K.N.; Gururaj, M.E. Karyotypic characteristics of the tomb bat Taphozous saccolaimus. Curr. Sci. 1986, 55, 469–471. [Google Scholar]
- Baker, R.J.; Genoways, H.H.; Seyfarth, P.A. Results of the Alcoa Foundation-Suriname Expeditions. VI. Additional Chromosomal Data for Bats (Mammalia: Chiroptera) from Suriname. Ann. Carnegie Mus. 1981, 50, 333–344. [Google Scholar]
- Ao, L.; Mao, X.; Nie, W.; Gu, X.; Feng, Q.; Wang, J.; Su, W.; Wang, Y.; Volleth, M.; Yang, F. Karyotypic evolution and phylogenetic relationships in the order Chiroptera as revealed by G-banding comparison and chromosome painting. Chromosom. Res. 2007, 15, 257–268. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Nie, W.; Wang, J.; Su, W.; Ao, L.; Feng, Q.; Wang, Y.; Volleth, M.; Yang, F. Karyotype evolution in Rhinolophus bats (Rhinolophidae, Chiroptera) illuminated by cross-species chromosome painting and G-banding comparison. Chromosom. Res. 2007, 15, 835–848. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.-G.; Wang, J.-H.; Su, W.-T.; Wang, Y.; Yang, F.; Nie, W. Karyotypic evolution in family Hipposideridae (Chiroptera, Mammalia) revealed by comparative chromosome painting, G- and C-banding. Zool. Res. 2010, 31, 453–460. [Google Scholar] [PubMed]
- Sreepada, K.; Naidu, K.N.; Gururaj, M.E. Trends of karyotypic evolution in the genus Hipposideros (Chiroptera: Mammalia). Cytobios 1993, 75, 49–57. [Google Scholar] [PubMed]
- Hood, C.S.; Schlitter, D.A.; Georgudaki, J.I.; Yenbutra, S.; Baker, R.J. Chromosomal studies of bats (Mammalia: Chiroptera) from Thailand. Ann. Carnegie Mus. 1988, 57, 99–109. [Google Scholar]
- Ao, L.; Gu, X.; Feng, Q.; Wang, J.; O’Brien, P.C.M.; Fu, B.; Mao, X.; Su, W.; Wang, Y.; Volleth, M. Karyotype relationships of six bat species (Chiroptera, Vespertilionidae) from China revealed by chromosome painting and G-banding comparison. Cytogenet. Genome Res. 2006, 115, 145–153. [Google Scholar] [CrossRef] [PubMed]
- Richards, L.R.; Rambau, R.V.; Lamb, J.M.; Taylor, P.J.; Yang, F.; Schoeman, M.C.; Goodman, S.M. Cross-species chromosome painting in bats from Madagascar: The contribution of Myzopodidae to revealing ancestral syntenies in Chiroptera. Chromosom. Res. 2010, 18, 635–653. [Google Scholar] [CrossRef] [PubMed]
- Bickham, J.W.; Hafner, J.C. A chromosomal banding study of three species of vespertilionid bats from Yugoslavia. Genetica 1978, 48, 1–3. [Google Scholar] [CrossRef]
- Sites, J.W., Jr.; Bickham, J.W.; Haiduk, M.W. Conservative chromosomal change in the bat family Mormoopidae. Can. J. Genet. Cytol. 1981, 23, 459–467. [Google Scholar] [CrossRef]
- Patton, J.C.; Baker, R.J. Chromosomal homology and evolution of phyllostomatoid bats. Syst. Zool. 1978, 27, 449–462. [Google Scholar] [CrossRef]
- Bickham, J.; Daniel, M.; Haiduk, M. Karyotype of Mystacina tuberculata (Chiroptera: Mystacinidae). J. Mammal. 1980, 61, 322–324. [Google Scholar] [CrossRef]
- Linares, O.; Löbig-A, I. El cariotipo del murciélago cavernícola Natalus tumidirostris, del norte de Venezuela, y observaciones sobre las afinidades de esta especie con N. stramineus (Chiroptera: Natalidae). Bol. Soc. Venez. Espeleol. 1973, 4, 89–95. [Google Scholar]
- Baker, R.; Jordan, R. Chromosomal studies of some Neotropical bats of the families Emballonuridae, Noctilionidae, Natalidae and Vespertilionidae. Caryologia 1970, 23, 595–604. [Google Scholar] [CrossRef]
- Baker, R.J.; Haiduk, M.W.; Robbins, L.W.; Cadena, A.; Koop, B. Chromosomal studies of South American bats and their systematic implications. In Mammalian biology in South America; Mares, M.A., Genoways, H.H., Eds.; Special Publication Series; Pymatuning Laboratory of Evology VI: Pittsburgh, PA, USA, 1982; Volume 4, pp. 303–327. [Google Scholar]
- Varella-Garcia, M.; Versute, E.M.; Taddei, V.A. A survey of cytogenetic data on Brazilian bats. Rev. Bras. Genét. 1989, 12, 761–793. [Google Scholar]
- Denys, C.; Kadjo, B.; Missoup, A.D.; Monadjem, A.; Aniskine, V. New records of bats (Mammalia: Chiroptera) and karyotypes from Guinean Mount Nimba (West Africa). Ital. J. Zool. 2013, 80, 279–290. [Google Scholar] [CrossRef]
- Moratelli, R.; Morielle-Versute, E. Métodos e aplicações da citogenética na taxonomia de morcegos brasileiros. In Morcegos do Brasil; Reis, N.R., Peracchi, A.L., Pedro, W.A., Lima, I.P., Eds.; Univesidade Estadual de Londrina: Londrina, Brazil, 2007; pp. 197–218. [Google Scholar]
- Haiduk, M.W.; Baker, R.J. Cladistical analysis of G-banded chromosomes of nectar feeding bats (Glossophaginae: Phyllostomidae). Syst. Zool. 1982, 31, 252–265. [Google Scholar] [CrossRef]
- Volleth, M.; Klett, C.; Kollak, A.; Dixkens, C.; Winter, Y.; Just, W.; Vogel, W.; Hameister, H. Zoo-FISH analysis in a species of the order Chiroptera: Glossophaga soricina (Phyllostomidae). Chromosom. Res. 1999, 7, 57–64. [Google Scholar] [CrossRef]
- Gomes, A.J.B.; Nagamachi, C.Y.; Rodrigues, L.R.R.; Benathar, T.C.M.; Ribas, T.F.A.; O’Brien, P.C.M.; Yang, F.; Ferguson-Smith, M.A.; Pieczarka, J.C. Chromosomal phylogeny of vampyressine bats (Chiroptera, Phyllostomidae) with description of two new sex chromosome systems. BMC Evol. Biol. 2016, 16, 119. [Google Scholar] [CrossRef] [PubMed]
- Pieczarka, J.C.; Gomes, A.J.; Nagamachi, C.Y.; Rocha, D.C.; Rissino, J.D.; O’Brien, P.C.; Yang, F.; Ferguson-Smith, M.A. A phylogenetic analysis using multidirectional chromosome painting of three species (Uroderma magnirostrum, U. bilobatum and Artibeus obscurus) of subfamily Stenodermatinae (Chiroptera-Phyllostomidae). Chromosom. Res. 2013, 21, 383–392. [Google Scholar] [CrossRef] [PubMed]
- Sotero-Caio, C.G.; Pieczarka, J.C.; Nagamachi, C.Y.; Gomes, A.J.B.; Lira, T.C.; O’Brien, P.C.M.; Ferguson-Smith, M.A.; Souza, M.J.; Santos, N. Chromosomal homologies among vampire bats revealed by chromosome painting (Phyllostomidae, Chiroptera). Cytogenet. Genome Res. 2011, 132, 156–164. [Google Scholar] [CrossRef] [PubMed]
- Sotero-Caio, C.G.; Volleth, M.; Gollahon, L.S.; Fu, B.; Cheng, W.; Ng, B.L.; Yang, F.; Baker, R.J. Chromosomal evolution among leaf-nosed nectarivorous bats—Evidence from cross-species chromosome painting (Phyllostomidae, Chiroptera). BMC Evol. Biol. 2013, 13, 276. [Google Scholar] [CrossRef] [PubMed]
- Richards, L.R.; Rambau, R.V.; Goodman, S.M.; Taylor, P.J.; Schoeman, M.C.; Yang, F.; Lamb, J.M. Karyotypic evolution in Malagasy Flying Foxes (Pteropodidae, Chiroptera) and their hipposiderid relatives as determined by comparative chromosome painting. Cytogenet. Genome Res. 2016, 148, 185–198. [Google Scholar] [CrossRef] [PubMed]
- Volleth, M.; Biedermann, M.; Schorcht, W.; Heller, K.-G. Evidence for two karyotypic variants of the lesser horseshoe bat (Rhinolophus hipposideros, Chiroptera, Mammalia) in central Europe. Cytogenet. Genome Res. 2013, 140, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Kacprzyk, J.; Teeling, E.C.; Kelleher, C.; Volleth, M. Wing membrane biopsies for bat cytogenetics: Finding of 2n = 54 in Irish Rhinolophus hipposideros (Rhinolophidae, Chiroptera, Mammalia) supports two geographically separated chromosomal variants in Europe. Cytogenet. Genome Res. 2016, 148, 279–283. [Google Scholar] [CrossRef] [PubMed]
- Volleth, M.; Heller, K.-G.; Yong, H.-S.; Müller, S. Karyotype evolution in the horseshoe bat Rhinolophus sedulus by whole-arm reciprocal translocation (WART). Cytogenet. Genome Res. 2014, 143, 241–250. [Google Scholar] [CrossRef] [PubMed]
- Volleth, M.; Loidl, J.; Mayer, F.; Yong, H.-S.; Müller, S.; Heller, K.-G. Surprising genetic diversity in Rhinolophus luctus (Chiroptera: Rhinolophidae) from peninsular Malaysia: Description of a new species based on genetic and morphological characters. Acta Chiropt. 2015, 17, 1–20. [Google Scholar] [CrossRef]
- Volleth, M.; Son, N.T.; Wu, Y.; Li, Y.; Yu, W.; Lin, L.-K.; Arai, S.; Trifonov, V.; Liehr, T.; Harada, M. Comparative chromosomal studies in Rhinolophus formosae and R. luctus from China and Vietnam: Elevation of R. l. lanosus to species rank. Acta Chiropt. 2017, 19, 41–50. [Google Scholar] [CrossRef]
- Dulić, B.; Mutere, F. Chromosomes of some East African bats. Säugetierkd. Mitt. 1977, 25, 231–233. [Google Scholar]
- Handa, S.; Kaur, S. Chromosomes of Rhinopoma microphyllum kinneari (Rhinopomatidae). Microb. Lett. 1979, 12, 47–48. [Google Scholar]
- Qumsiyeh, M.B.; Baker, R.J. G- and C-banded karyotypes of the Rhinopomatidae (Microchiroptera). J. Mammal. 1985, 66, 541–544. [Google Scholar] [CrossRef]
- Kulemzina, A.; Nie, W.; Trifonov, V.; Staroselec, Y.; Vasenkov, D.; Volleth, M.; Yang, F.; Graphodatsky, A. Comparative chromosome painting of four siberian Vespertilionidae species with Aselliscus stoliczkanus and human probes. Cytogenet. Genome Res. 2011, 134, 200–205. [Google Scholar] [CrossRef] [PubMed]
- Teeling, E.C.; Madsen, O.; Van Den Bussche, R.A.; de Jong, W.W.; Stanhope, M.J.; Springer, M.S. Microbat paraphyly and the convergent evolution of a key innovation in Old World rhinolophoid microbats. Proc. Natl. Acad. Sci. USA 2002, 99, 1431–1436. [Google Scholar] [CrossRef] [PubMed]
- Hutcheon, J.M.; Kirsch, J.A. A moveable face: Deconstructing the Microchiroptera and a new classification of extant bats. Acta Chiropt. 2006, 8, 1–10. [Google Scholar] [CrossRef]
- Eick, G.N.; Jacobs, D.S.; Matthee, C.A. A nuclear DNA phylogenetic perspective on the evolution of echolocation and historical biogeography of extant bats (Chiroptera). Mol. Biol. Evol. 2005, 22, 1869–1886. [Google Scholar] [CrossRef] [PubMed]
- Volleth, M.; Yang, F.; Müller, S. High-resolution chromosome painting reveals the first genetic signature for the chiropteran suborder Pteropodiformes (Mammalia: Chiroptera). Chromosom. Res. 2011, 19, 507–519. [Google Scholar] [CrossRef] [PubMed]
- Bergmans, W. Taxonomy and biogeography of African fruit bats (Mammalia, Megachiroptera). 5. The genera Lissonycteris Andersen, 1912, Myonycteris Matschie, 1899 and Megaloglossus Pagenstecher, 1885; general remarks and conclusions; annex: Key to all species. Beaufortia 1997, 47, 11–90. [Google Scholar]
- Volleth, M. Of bats and molecules: Chromosomal characters for judging phylogenetic relationships. In Bat Evolution, Ecology, and Conservation; Adams, R.A., Pedersen, S.C., Eds.; Springer Science Business Media: New York, NY, USA, 2013; pp. 129–146. [Google Scholar]
- Haiduk, M.; Baker, R.; Robbins, L.; Schlitter, D. Chromosomal evolution in African Megachiroptera: G- and C-band assessment of the magnitude of change in similar standard karyotypes. Cytogenet. Genome Res. 1981, 29, 221–232. [Google Scholar] [CrossRef]
- Peterson, R.L.; Nagorsen, D.W. Chromosomes of fifteen species of bats (Chiroptera) from Kenya and Rhodesia. Roy. Ont. Mus. Life Sci. Occ. Pap. 1975, 27, 1–14. [Google Scholar]
- Soisook, P.; Prajakjitr, A.; Karapan, S.; Francis, C.M.; Bates, P. A new genus and species of false vampire (Chiroptera: Megadermatidae) from peninsular Thailand. Zootaxa 2015, 3931, 528–550. [Google Scholar] [CrossRef] [PubMed]
- Rickart, E.A.; Mercier, J.A.; Heaney, L.R. Cytogeography of philippine bats (Mammalia: Chiroptera). Proc. Biol. Soc. Wash. 1999, 112, 453–469. [Google Scholar]
- Harada, M.; Kobayashi, T. Studies on the small mammal fauna of Sabah, East Malaysia II. Karyological analysis of some Sabahan Mammals (Primates, Rodentia, Chiroptera). Contr. Biol. Lab. Kyoto Univ. 1980, 26, 83–95. [Google Scholar]
- Foley, N.M.; Goodman, S.M.; Whelan, C.V.; Puechmaille, S.J.; Teeling, E. Towards Navigating the Minotaur’s Labyrinth: Cryptic Diversity and Taxonomic Revision within the Speciose Genus Hipposideros (Hipposideridae). Acta Chiropt. 2017, 19, 1–18. [Google Scholar] [CrossRef]
- Koubínová, D.; Sreepada, K.; Koubek, P.; Zima, J. Karyotypic variation in rhinolophid and hipposiderid bats (Chiroptera: Rhinolophidae, Hipposideridae). Acta Chiropt. 2010, 12, 393–400. [Google Scholar] [CrossRef]
- Stoffberg, S.; Jacobs, D.S.; Mackie, I.J.; Matthee, C.A. Molecular phylogenetics and historical biogeography of Rhinolophus bats. Mol. Phylogenet. Evol. 2010, 54, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Dool, S.E.; Puechmaille, S.J.; Foley, N.M.; Allegrini, B.; Bastian, A.; Mutumi, G.L.; Maluleke, T.G.; Odendaal, L.J.; Teeling, E.C.; Jacobs, D.S. Nuclear introns outperform mitochondrial DNA in inter-specific phylogenetic reconstruction: Lessons from horseshoe bats (Rhinolophidae: Chiroptera). Mol. Phylogen. Evol. 2016, 97, 196–212. [Google Scholar] [CrossRef] [PubMed]
- Zima, J.; Volleth, M.; Horácek, I.; Cerveny, J.; Cervena, A.; Prucha, K.; Macholan, M. Comparative karyology of rhinolophid bats (Chiroptera: Rhinolophidae). In Prague Studies in Mammalogy; Horácek, I., Vohralik, V., Eds.; Charles University Press: Prague, Czech Republic, 1992; pp. 229–236. [Google Scholar]
- Guillén-Servent, A.; Francis, C.; Ricklefs, R. Phylogeny and biogeography of the horseshoe bats. In Horseshoe Bats of the World (Chiroptera: Rhinolophidae); Csorba, G., Ujhelyi, P., Thomas, N., Eds.; Alana Books: Shropshire, UK, 2003. [Google Scholar]
- Pavan, A.C.; Marroig, G. Integrating multiple evidences in taxonomy: Species diversity and phylogeny of mustached bats (Mormoopidae: Pteronotus). Mol. Phylogenet. Evol. 2016, 103, 184–198. [Google Scholar] [CrossRef] [PubMed]
- Pavan, A.C.; Marroig, G. Timing and patterns of diversification in the neotropical bat genus Pteronotus (Mormoopidae). Mol. Phylogenet. Evol. 2017, 108, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Pavan, A.C.; Martins, F.M.; Morgante, J.S. Evolutionary history of bulldog bats (genus Noctilio): Recent diversification and the role of the caribbean in Neotropical biogeography. Biol. J. Linn. Soc. 2013, 108, 210–224. [Google Scholar] [CrossRef]
- Khan, F.A.A.; Phillips, C.D.; Baker, R.J. Timeframes of speciation, reticulation, and hybridization in the bulldog bat explained through phylogenetic analyses of all genetic transmission elements. Syst. Biol. 2013, 63, 96–110. [Google Scholar] [CrossRef] [PubMed]
- Noronha, R.C.; Nagamachi, C.Y.; O’Brien, P.C.; Ferguson-Smith, M.A.; Pieczarka, J.C. Meiotic analysis of XX/XY and neo-XX/XY sex chromosomes in Phyllostomidae by cross-species chromosome painting revealing a common chromosome 15-XY rearrangement in Stenodermatinae. Chromosom. Res. 2010, 18, 667–676. [Google Scholar] [CrossRef] [PubMed]
- Noronha, R.; Nagamachi, C.; O’brien, P.; Ferguson-Smith, M.; Pieczarka, J. Neo-XY body: An analysis of XY1Y2 meiotic behavior in Carollia (Chiroptera, Phyllostomidae) by chromosome painting. Cytogenet. Genome Res. 2009, 124, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Baker, R.J. Karyology. In Biology of bats of the New World family Phyllostomatidae; Part, III; Baker, R.J., Jones, J.K., Carter, D.C., Eds.; Special Publications; The Museum: Lubbock, TX, USA, 1979; pp. 107–155. [Google Scholar]
- Noronha, R.C.R.; Nagamachi, C.Y.; Pieczarka, J.C.; Marques-Aguiar, S.; Assis, M.D.F.L.D.; Barros, R.M.D.S. Meiotic analyses of the sex chromosomes in Carolliinae-Phyllostomidae (Chiroptera): NOR separates the XY1Y2 into two independent parts. Caryologia 2004, 57, 1–9. [Google Scholar] [CrossRef]
- Solari, S.; Baker, J.R. Mitochondrial DNA sequence, karyotypic, and morphological variation in the Carollia castena species complex (Chiroptera: Phyllostomidae) with description of a new species. Occas. Pap. Tex. Tech Univ. Mus. 2006, 254, 1–16. [Google Scholar]
- Hsu, T.; Baker, R.; Utakoji, T. The multiple sex chromosome system of american leaf-nosed bats (Chiroptera, Phyllostomidae). Cytogenet. Genome Res. 1968, 7, 27–38. [Google Scholar] [CrossRef]
- Sreepada, K.; Koubínová, D.; Konecny, A.; Koubek, P.; Ráb, P.; Rábová, M.; Zima, J. Karyotypes of three species of molossid bats (Molossidae, Chiroptera) from India and Western Africa. Folia Zool. 2008, 57, 347–357. [Google Scholar]
- Freitas, T.; Bogo, M.; Christoff, A. G-, C-bands and NOR studies in two species of bats from southern Brazil (Chiroptera: Vespertilionidae, Molossidae). Z. Säugetierkd. 1992, 57, 330–334. [Google Scholar]
- Morielle-Versute, E.; Varella-Garcia, M.; Taddei, V. Karyotypic patterns of seven species of molossid bats (Molossidae, Chiroptera). Cytogenet. Genome Res. 1996, 72, 26–33. [Google Scholar] [CrossRef]
- Leite-Silva, C.; Santos, N.; Fagundes, V.; Yonenaga-Yassuda, Y.; Souza, M.J. Karyotypic characterization of the bat species Molossus ater, M. molossus and Molossops planirostris (Chiroptera, Molossidae) using FISH and banding techniques. Hereditas 2003, 138, 94–100. [Google Scholar] [CrossRef] [PubMed]
- McDonough, M.M.; Ammerman, L.K.; Timm, R.M.; Genoways, H.H.; Larsen, P.A.; Baker, R.J. Speciation within bonneted bats (genus Eumops): The complexity of morphological, mitochondrial, and nuclear data sets in systematics. J. Mammal. 2008, 89, 1306–1315. [Google Scholar] [CrossRef]
- Warner, J.W.; Patton, J.L.; Gardner, A.L.; Baker, R.J. Karyotypic analyses of twenty-one species of molossid bats (Molossidae: Chiroptera). Can. J. Genet. Cytol. 1974, 16, 165–176. [Google Scholar] [CrossRef] [PubMed]
- Ruedas, L.A.; Lee, T.E.; Bickham, J.W.; Schlitter, D.A. Chromosomes of five species of vespertilionid bats from Africa. J. Mammal. 1990, 71, 94–100. [Google Scholar] [CrossRef]
- Volleth, M.; Heller, K.G. Phylogenetic relationships of vespertilionid genera (Mammalia: Chiroptera) as revealed by karyological analysis. J. Zool. Syst. Evol. Res. 1994, 32, 11–34. [Google Scholar] [CrossRef]
- Kearney, T.C.; Volleth, M.; Contrafatto, G.; Taylor, P.J. Systematic implications of chromosome GTG-band and bacula morphology for southern african Eptesicus and Pipistrellus and several other species of Vespertilioninae (Chiroptera: Vespertilionidae). Acta Chiropt. 2002, 4, 55–76. [Google Scholar] [CrossRef]
- Son, N.T.; Csorba, G.; Tu, V.T.; Thong, V.D.; Wu, Y.; Harada, M.; Oshida, T.; Endo, H.; Motokawa, M. A new species of the genus Murina (Chiroptera: Vespertilionidae) from the central highlands of Vietnam with a review of the subfamily Murininae in Vietnam. Acta Chiropt. 2015, 17, 201–232. [Google Scholar] [CrossRef]
- Bickham, J.W.; Baker, R.J. Implications of chromosomal variation in Rhogeessa (Chiroptera: Vespertilionidae). J. Mammal. 1977, 58, 448–453. [Google Scholar] [CrossRef]
- Khan, F.A.A.; Solari, S.; Swier, V.J.; Larsen, P.A.; Abdullah, M.; Baker, R.J. Systematics of Malaysian woolly bats (Vespertilionidae: Kerivoula) inferred from mitochondrial, nuclear, karyotypic, and morphological data. J. Mammal. 2010, 91, 1058–1072. [Google Scholar] [CrossRef]
- Lack, J.B.; Van Den Bussche, R.A. Identifying the confounding factors in resolving phylogenetic relationships in Vespertilionidae. J. Mammal. 2010, 91, 1435–1448. [Google Scholar] [CrossRef]
- Volleth, M.; Eick, G. Chromosome evolution in bats as revealed by FISH: The ongoing search for the ancestral chiropteran karyotype. Cytogenet. Genome Res. 2012, 137, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Ferguson-Smith, M.A.; Trifonov, V. Mammalian karyotype evolution. Nat. Rev. Genet. 2007, 8, 950–962. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.E., Jr.; Hoofer, S.R.; Van Den Bussche, R.A. Molecular phylogenetics and taxonomic revision of the genus Tonatia (Chiroptera: Phyllostomidae). J. Mammal. 2002, 83, 49–57. [Google Scholar] [CrossRef]
- Parlos, J.A.; Timm, R.M.; Swier, V.J.; Zeballos, H.; Baker, R.J. Evaluation of the paraphyletic assemblages within Lonchophyllinae, with description of a new tribe and genus. Occ. Pap. Tex. Tech Mus. 2014, 320, 1–23. [Google Scholar]
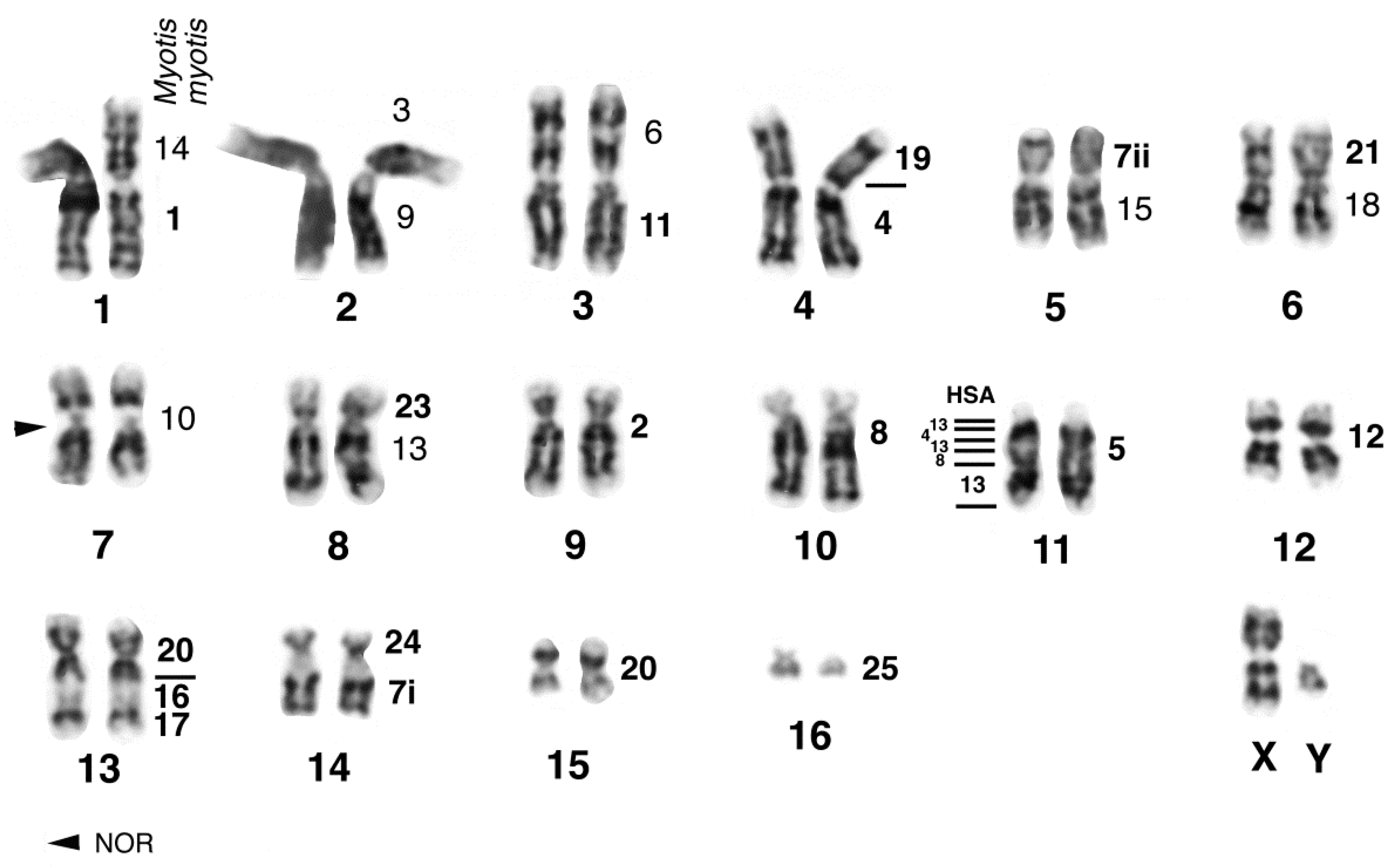
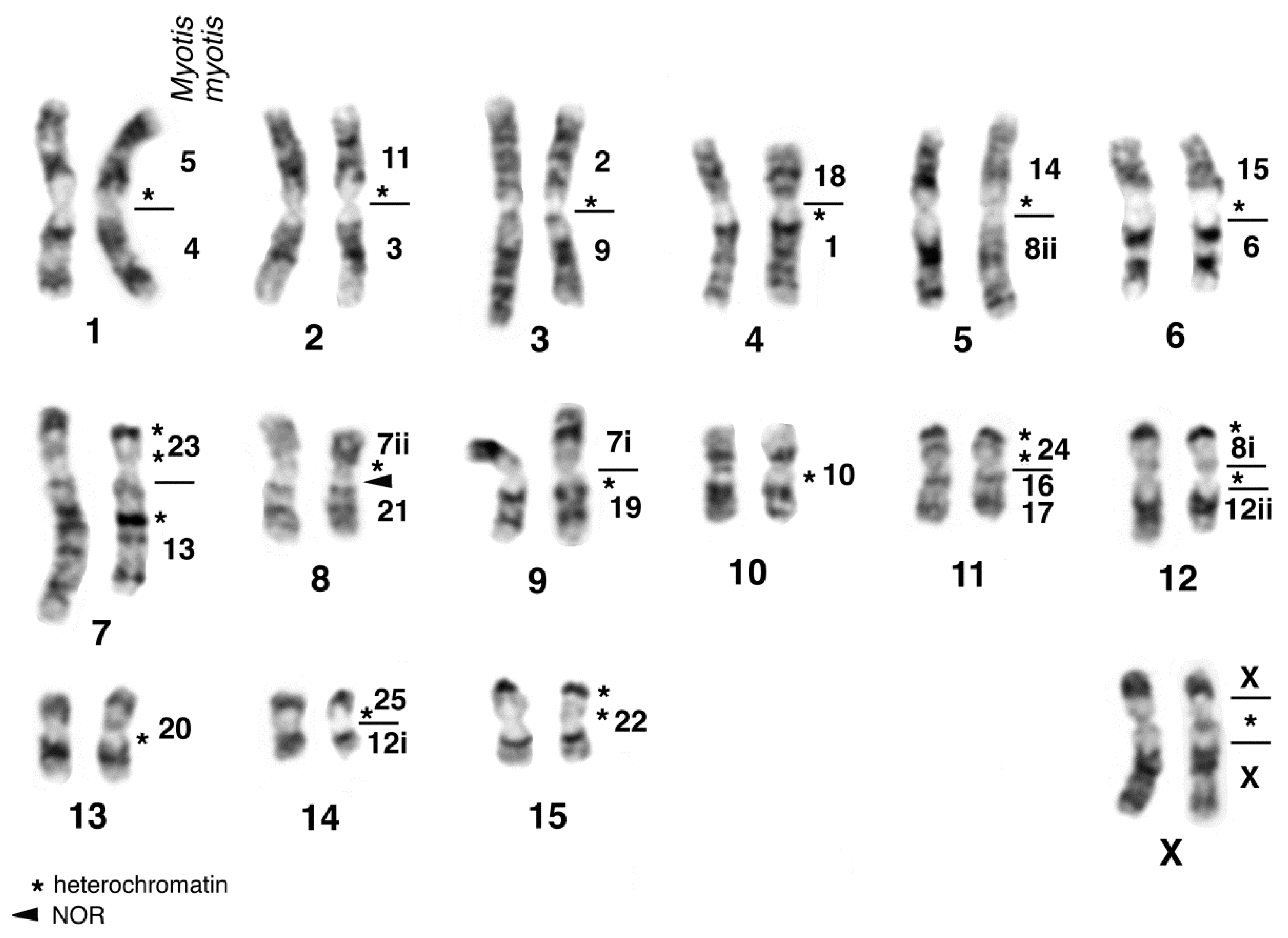

| Family | 2n | N | Homogen | G-Banded | Zoo-FISH | References |
|---|---|---|---|---|---|---|
| Cistugidae | 50 | 2 | 2 | - | - | [67] |
| Craseonycteridae | nk | 1 | - | - | - | - |
| Emballonuridae | 22–48 | 53 | 20 | 11 | 1 | [68,69,70,71,72] |
| Furipteridae | 34 | 2 | 1 | - | - | [73] |
| Hipposideridae | 24–52 | 81 | 27 | 11 | 7 | [62,74,75,76,77] |
| Megadermatidae | 38–54 | 6 | 3 | 2 | 1 | [57,68,78] |
| Miniopteridae | 46 | 25 | 10 | 3 | 2 | [79,80,81] |
| Molossidae | 34–52 | 110 | 50 | 11 | 3 | [62,68,80] |
| Mormoopidae | 38 | 15 | 5 | 8 | - | [82,83] |
| Mystacinidae | 36 | 2 | 1 | - | - | [84] |
| Myzopodidae | 26 | 2 | 1 | 1 | 1 | [80] |
| Natalidae | 36 | 8 | 2 | - | - | [85,86] |
| Noctilionidae | 34 | 2 | 2 | 2 | - | [86,87,88] |
| Nycteridae | 34–42 | 16 | 6 | - | - | [89] |
| Phyllostomidae | 14–46 | 204 | 106 | 74 | 25 | [41,59,60,61,63,87,90,91,92,93,94,95,96] |
| Pteropodidae | 24–58 | 192 | 45 | 13 | 4 | [62,68,74,75,97] |
| Rhinolophidae | 28–62 | 96 | 48 | 20 | 13 | [62,74,75,79,97,98,99,100,101,102] |
| Rhinonycteridae | 36–40 | 9 | 2 | - | - | [67,103] |
| Rhinopomatidae | 36–42 | 4 | 3 | 2 | - | [104,105] |
| Thyropteridae | 32–40 | 5 | 2 | 1 | - | [87] |
| Vespertilionidae | 18–52 | 436 | 189 | 71 | 12 | [62,75,79,80,106] |
| Total | 14–62 | 1271 | 525 | 230 | 69 |
| Species 2n | RHI | RPE | RSI | RRO | RSE | RMO | RLU | RLA | RFO | AST | HLA | ESP |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 54 | 44 | 36 | 56 | 28 | 32 | 32 | 32 | 52 | 30 | 32 | 36 | |
| MMY homology | ||||||||||||
| 1/3 | o | x | ||||||||||
| 1/11 | x | x | ||||||||||
| 1/14 | x | x | ||||||||||
| 1/16-17 | x | x | ||||||||||
| 1/18 | x | |||||||||||
| 1/20 * | x | |||||||||||
| 2/5 | x | x | ||||||||||
| 2/6 | x | x | ||||||||||
| 2/8ii | x | |||||||||||
| 2/9 | x | |||||||||||
| 2/19 | x | |||||||||||
| 2/25 | x | |||||||||||
| 3/6 | x | |||||||||||
| 3/7ii | x | |||||||||||
| 3/8i | x | o | ||||||||||
| 3/9 | x | |||||||||||
| 3/11 | x | |||||||||||
| 3/13 | x | x | ||||||||||
| 3/15 | x | x | ||||||||||
| 4/5 | x | x | x | x | ||||||||
| 4/8ii | x | x | x | x | ||||||||
| 4/14 | x | |||||||||||
| 4/18 | x | |||||||||||
| 4/19 | x | |||||||||||
| 5/6 | x | |||||||||||
| 5/9 | x | |||||||||||
| 5/10i | x | |||||||||||
| 5/1 | x | |||||||||||
| 5/18 | x | |||||||||||
| 6/11 | x | x | ||||||||||
| 6/15 | x | x | ||||||||||
| 6/25 | x | |||||||||||
| 7i/7ii | x | |||||||||||
| 7i/12ii | x | x | x | |||||||||
| 7i/18 | x | |||||||||||
| 7i/19 | x | x | ||||||||||
| 7i/22i | x | |||||||||||
| 7ii/9 | x | |||||||||||
| 7ii/15 | x | |||||||||||
| 7ii/19 | x | x | x | |||||||||
| 7ii/21 | x | |||||||||||
| 7ii/23 | x | |||||||||||
| 8i/12ii | x | |||||||||||
| 8i/20 * | o | |||||||||||
| 8i/22i | x | x | x | x | ||||||||
| 8i/24 | x | |||||||||||
| 8ii/11 | x | |||||||||||
| 8ii/14 | x | x | ||||||||||
| 8ii/22ii | x | |||||||||||
| 9/11 | x | |||||||||||
| 9/14 | x | |||||||||||
| 9/15 | x | x | x | |||||||||
| 9/19 | x | |||||||||||
| 10i/12i | x | |||||||||||
| 10i/12ii | x | |||||||||||
| 10i/16-17 | x | |||||||||||
| 10i/22ii | x | x | ||||||||||
| 10i/24 | x | x | ||||||||||
| 10ii/12ii | x | |||||||||||
| 10ii/18 | x | |||||||||||
| 10ii/21 | x | x | x | |||||||||
| 10ii/22ii | o | |||||||||||
| 11/15 | x | |||||||||||
| 11/20i + ii | x | |||||||||||
| 12i/16-17 | x | |||||||||||
| 12i/25 | x | x | x | x | x | |||||||
| 12ii/20 * | x | |||||||||||
| 12ii/22ii | x | |||||||||||
| 13/14 | x | |||||||||||
| 13/16-17 | x | x | ||||||||||
| 13/18 | x | |||||||||||
| 13/23 | x | x | x | |||||||||
| 14/18 | x | x | ||||||||||
| 14/23 | x | |||||||||||
| 16-17/21 | x | |||||||||||
| 16-17/24 | x | x | ||||||||||
| 18/21 | x | |||||||||||
| 19/24 | x | |||||||||||
| 20i/22ii | x | |||||||||||
| 20i/24 | x | x | x | |||||||||
| 20ii/23 | x | x | x | |||||||||
| 20ii/25 | x | |||||||||||
| 21/22i + ii | x | |||||||||||
| 22ii/24 | o |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sotero-Caio, C.G.; Baker, R.J.; Volleth, M. Chromosomal Evolution in Chiroptera. Genes 2017, 8, 272. https://doi.org/10.3390/genes8100272
Sotero-Caio CG, Baker RJ, Volleth M. Chromosomal Evolution in Chiroptera. Genes. 2017; 8(10):272. https://doi.org/10.3390/genes8100272
Chicago/Turabian StyleSotero-Caio, Cibele G., Robert J. Baker, and Marianne Volleth. 2017. "Chromosomal Evolution in Chiroptera" Genes 8, no. 10: 272. https://doi.org/10.3390/genes8100272




