Chromosome Evolution in Connection with Repetitive Sequences and Epigenetics in Plants
Abstract
:1. Introduction
2. Chromosome Evolution in Plants
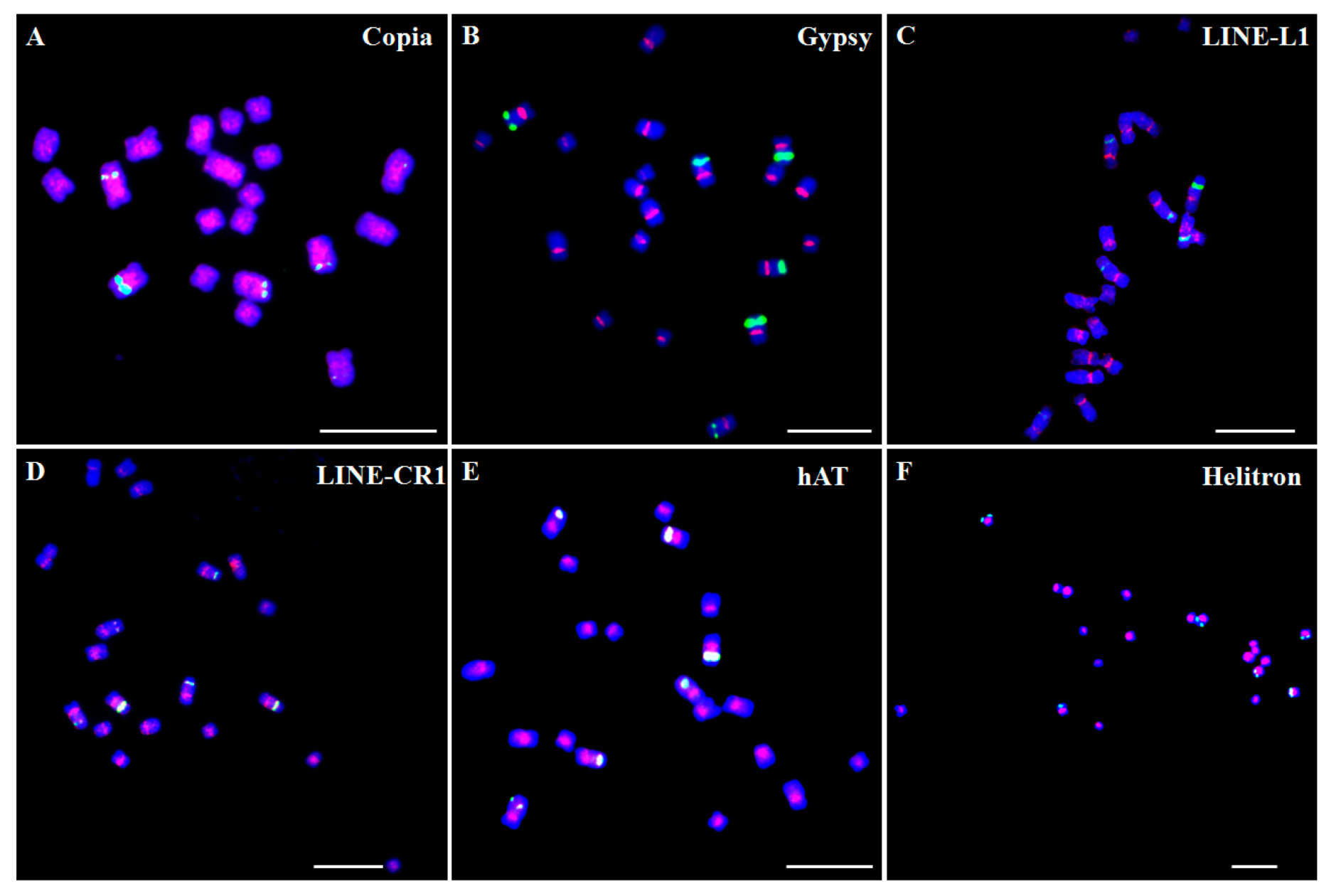
3. Characteristics and Distribution Patterns of Repetitive Sequences in Plant Chromosomes
4. Functional Aspects of Repetitive Sequences in Plant Chromosome Evolution
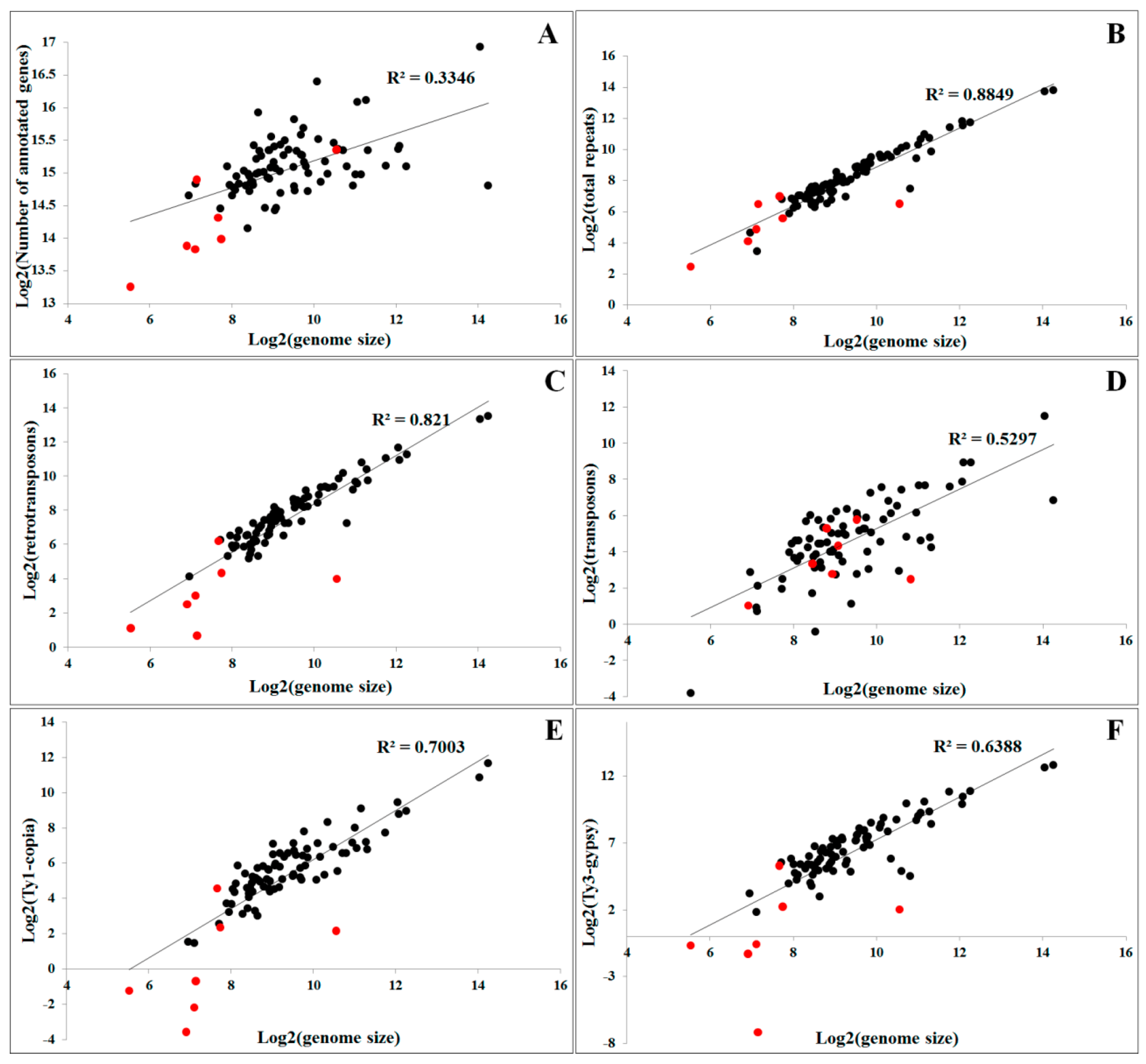
4.1. Repetitive Sequences and Evolution of Chromosome Size and DNA Composition
4.2. Repetitive Sequences and Evolution of Chromosome Structure and Shape
4.3. Repetitive Sequences and Evolution of Chromosome Number
5. Epigenetic Modification and Plant Chromosome Evolution
5.1. Epigenetic Modification and Heterochromatin
5.2. Epigenetic Modification and Centromere Function
5.3. Epigenetic Regulated Transposable Elements Reactivation and Chromosome Evolution
6. Conclusions and Perspectives
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Heslop-Harrison, J.S.; Schwarzacher, T. Organisation of the plant genome in chromosome. Plant J. 2011, 66, 18–33. [Google Scholar] [CrossRef] [PubMed]
- Richards, E.J.; Dawe, R.K. Plant centromeres: Structure and control. Curr. Opin. Plant Biol. 1998, 1, 130–135. [Google Scholar] [CrossRef]
- Schubert, I. Chromosome evolution. Curr. Opin. Plant Biol. 2007, 10, 109–115. [Google Scholar] [CrossRef]
- Weiss-Schneeweiss, H.; Schneeweiss, G. Karyotype diversity and evolutionary trends in Angiosperms. In Plant Genome Diversity; Greilhuber, J., Dolezel, J., Wendel, J.F., Eds.; Springer: Vienna, Austria, 2013; Volume 2, pp. 209–230. [Google Scholar]
- Wichman, H.A.; Payne, C.T.; Ryder, O.A.; Hamilton, M.J.; Maltbie, M.; Baker, R.J. Genomic distribution of heterochromatic sequences in Equids: Implications to rapid chromosomal evolution. J. Hered. 1991, 82, 369–377. [Google Scholar] [CrossRef] [PubMed]
- Eichler, E.E.; Sankoff, D. Structural dynamics of eukaryotic chromosome evolution. Science 2003, 301, 793–797. [Google Scholar] [CrossRef] [PubMed]
- Dobigny, G.; Britton-Davidian, J.; Robinson, T.J. Chromosomal polymorphism in mammals: An evolutionary perspective. Biol. Rev. 2017, 92, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Mehrotra, S.; Goyal, V. Repetitive sequences in plant nuclear DNA: Types, distribution, evolution and function. Genom. Proteom. Bioinform. 2014, 12, 164–171. [Google Scholar] [CrossRef] [PubMed]
- Schnable, P.S.; Ware, D.; Fulton, R.S.; Stein, J.C.; Wei, F.; Pasternak, S.; Liang, C.; Zhang, J.; Fulton, L.; Graves, T.A.; et al. The B73 maize genome: Complexity, diversity, and dynamics. Science 2009, 326, 1112–1115. [Google Scholar] [CrossRef] [PubMed]
- Choulet, F.; Wicker, T.; Rustenholz, C.; Paux, E.; Salse, J.; Leroy, P.; Schlub, S.; Paslier, M.L.; Magdelenat, G.; Gonthier, C.; et al. Megabase level sequencing reveals contrasted organization and evolution patterns of the wheat gene and transposable element spaces. Plant Cell 2010, 22, 1686–1701. [Google Scholar] [CrossRef] [PubMed]
- Jamilloux, V.; Daron, J.; Choulet, F.; Quesneville, H. De novo annotation of transposable elements: Tacking the fat genome issue. Proc. IEEE 2017, 105, 474–481. [Google Scholar] [CrossRef]
- Zhu, Q.; Cai, Z.; Tang, Q.; Jin, W. Repetitive sequence analysis and karyotyping reveal different genome evolution and speciation of diploid and tetraploid Tripsacum dactyloides. Crop J. 2016, 4, 247–255. [Google Scholar] [CrossRef]
- Li, S.F.; Zhang, G.J.; Yuan, J.H.; Deng, C.L.; Gao, W.J. Repetitive sequences and epigenetic modification: Inseparable partners play important roles in the evolution of plant sex chromosomes. Planta 2016, 243, 1083–1095. [Google Scholar] [CrossRef] [PubMed]
- Ayres-Alves, T.; Cardoso, A.L.; Nagamachi, C.Y.; de Sousa, L.M.; Pieczarka, J.C.; Noronha, R.C.R. Karyotypic evolution and chromosomal organization of repetitive DNA sequences in species of Panaque, Panaqolus, and Scobinancistrus (Siluriformes and Loricariidae) from the Amazon Basin. Zebrafish 2017, 14, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.J. New evidence for the theory of chromosome organization by repetitive elements (CORE). Genes 2017, 8, 81. [Google Scholar] [CrossRef] [PubMed]
- Fedoroff, N.V. Transposable elements, epigenetics, and genome evolution. Science 2012, 338, 758–767. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, R. A comparative karyotype analysis in Haplopappus gracilis (2n = 4) and H. ravenii (2n = 8). Cytologia 1967, 32, 542–552. [Google Scholar] [CrossRef]
- Khandelwal, S. Chromosome evolution in the genus Ophioglossum L. Bot. J. Linn. Soc. 1990, 102, 205–217. [Google Scholar] [CrossRef]
- Wu, F.; Tanksley, S.D. Chromosomal evolution in the plant family Solanaceae. BMC Genom. 2010, 11, 182. [Google Scholar] [CrossRef] [PubMed]
- Schubert, I.; Lysak, M.A. Interpretation of karyotype evolution should consider chromosome structural constraints. Trends Genet. 2011, 27, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Dodsworth, S.; Chase, M.W.; Leitch, A.R. Is post-polyploidization diploidization the key to the evolutionary success of angiosperms? Bot. J. Linn. Soc. 2016, 180, 1–5. [Google Scholar] [CrossRef]
- Moraes, A.P.; Koehler, S.; Cabral, J.S.; Gomes, S.S.L.; Viccini, L.F.; Barros, F.; Felix, L.P.; Guerra, M.; Forni-Martins, E.R. Karyotype diversity and genome size variation in Neotropical Maxillariinae orchids. Plant Biol. 2017, 19, 298–308. [Google Scholar] [CrossRef] [PubMed]
- Husband, B.C.; Baldwin, S.J.; Suda, J. The incidence of polyploidy in natural plant populations: Major patterns and evolutionary processes. In Plant Genome Diversity; Greilhuber, J., Dolezel, J., Wendel, J.F., Eds.; Springer: Vienna, Austria, 2013; Volume 2, pp. 255–276. [Google Scholar]
- Nystedt, B.; Street, N.R.; Wetterbom, A.; Zuccolo, A.; Lin, Y.C.; Scofield, D.G.; Vezzi, F.; Delhomme, N.; Giacomello, S.; Alexeyenko, A.; et al. The Norway spruce genome sequence and conifer genome evolution. Nature 2013, 497, 579–584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Casacuberta, J.M.; Santiago, N. Plant LTR-retrotransposons and MITEs: Control of transposition and impact on the evolution of plant genes and genomes. Gene 2003, 311, 1–11. [Google Scholar] [CrossRef]
- Sharma, S.; Raina, S.N. Organization and evolution of highly repeated satellite DNA sequences in plant chromosomes. Cytogenet. Genome Res. 2005, 109, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Neumann, P.; Nouzová, M.; Macas, J. Molecular and cytogenetic analysis of repetitive DNA in pea (Pisum sativum L.). Genome 2001, 44, 716–728. [Google Scholar] [CrossRef] [PubMed]
- Macas, J.; Neumann, P.; Navrátilová, A. Repetitive DNA in the pea (Pisum sativum L.) genome: Comprehensive characterization using 454 sequencing and comparison to soybean and Medicago truncatula. BMC Genom. 2007, 8, 427. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Cai, Z.; Hu, T.; Liu, H.; Bao, C.; Mao, W.; Jin, W. Repetitive sequence analysis and karyotyping reveals centromere-associated DNA sequences in radish (Raphanus sativus L.). BMC Plant Biol. 2015, 15, 105. [Google Scholar] [CrossRef] [PubMed]
- Novák, P.; Ávila Robledillo, L.; Koblížková, A.; Vrbová, I.; Neumann, P.; Macas, J. TAREAN: A computational tool for identification and characterization of satellite DNA from unassembled short reads. Nucleic Acids Res. 2017, 45, e111. [Google Scholar] [CrossRef] [PubMed]
- Li, S.F.; Deng, C.L.; Gao, W.J. Repetitive sequence analysis in the genome of Humulus scandens. Manuscript in preparation.
- Brandes, A.; Heslop-Harrison, J.S.; Kamm, A.; Kubis, S.; Doudrick, R.L.; Schmidt, T. Comparative analysis of the chromosomal and genomic organization of Ty1-copia-like retrotranposons in pteridophytes, gymnosperms and angiosperms. Plant Mol. Biol. 1997, 33, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Heslop-Harrison, J.S.; Brandes, A.; Taketa, S.; Schmidt, T.; Vershinin, A.V.; Alknimova, E.G.; Kamm, A.; Doudrick, R.L.; Schwarzacher, T.; Katsiotis, A.; et al. The chromosomal distributions of Ty1-copia group retrotransposable elements in higher plants and their implications for genome evolution. Genetica 1997, 100, 197–204. [Google Scholar] [CrossRef] [PubMed]
- The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 2000, 408, 796–815. [Google Scholar]
- Nagaki, K.; Neumann, P.; Zhang, D.; Ouyang, S.; Buell, C.R.; Cheng, Z.; Jiang, J. Structure, divergence, and distribution of the CRR centromeric retrotransposon family in rice. Mol. Biol. Evol. 2005, 22, 845–855. [Google Scholar] [CrossRef] [PubMed]
- Kejnovsky, E.; Kubat, Z.; Macas, J.; Hobza, R.; Mracek, J.; Vyskot, B. Retand: A novel family of gypsy-like retrotransposon harboring an amplified tandem repeat. Mol. Genet. Genom. 2006, 276, 254–263. [Google Scholar] [CrossRef] [PubMed]
- Hřibová, E.; Neumann, P.; Matsumoto, T.; Roux, N.; Macas, J.; Doležel, J. Repetitive part of the banana (Musa acuminate) genome investigated by low-depth 454 sequencing. BMC Plant Biol. 2010, 10, 204. [Google Scholar] [CrossRef] [PubMed]
- Nagaki, K.; Tanaka, K.; Yamaji, N.; Kobayashi, H.; Murata, M. Sunflower centromeres consist of a centromere-specific LINE and a chromosome-specific tandem repeat. Front. Plant Sci. 2015, 6, 912. [Google Scholar] [CrossRef] [PubMed]
- Heitkam, T.; Petrasch, S.; Zakrzewski, F.; Kögler, A.; Wenke, T.; Wanke, S.; Schmidt, T. Next-generation sequencing reveals differentially amplified tandem repeats as a major genome component of Northern Europe’s oldest Camellia japonica. Chromosome Res. 2015, 23, 791–806. [Google Scholar] [CrossRef] [PubMed]
- Steflova, P.; Tokan, V.; Vogel, I.; Lexa, M.; Macas, J.; Novak, P.; Hobza, R.; Vyskot, B.; Kejnovsky, E. Contrasting patterns transposable element and satellite distribution on sex chromosome (XY1Y2) in the dioecious plant Rumex acetosa. Genome Biol. Evol. 2013, 5, 769–782. [Google Scholar] [CrossRef] [PubMed]
- Shibata, F.; Hizume, M.; Kuroki, Y. Chromosome painting of Y chromosomes and isolation of a Y chromosome-specific repetitive sequence in the dioecious plant Rumex acetosa. Chromosoma 1999, 108, 266–270. [Google Scholar] [CrossRef] [PubMed]
- Mariotti, B.; Manzano, S.; Kejnovský, E.; Vyskot, B.; Jamilena, M. Accumulation of Y-specific satellite DNAs during the evolution of Rumex acetosa sex chromosomes. Mol. Genet. Genom. 2009, 281, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Na, J.K.; Wang, J.; Ming, R. Accumulation of interspersed and sex-specific repeats in the non-recombining region of papaya sex chromosomes. BMC Genom. 2014, 15, 335. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yazaki, J.; Sundaresan, A.; Cokus, S.; Chan, S.W.L.; Chen, H.; Henderson, I.R.; Shinn, P.; Pellegrini, M.; Jacobsen, S.E. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 2006, 126, 1189–1201. [Google Scholar] [CrossRef] [PubMed]
- Vergara, Z.; Sequeira-Mendes, J.; Morata, J.; Peiró, R.; Hénaff, E.; Costas, C.; Casacuberta, J.M.; Gutierrez, C. Retrotransposons are specified as DNA replication origins in the gene-poor regions of Arabidopsis heterochromatin. Nucleic Acids Res. 2017. [Google Scholar] [CrossRef] [PubMed]
- Martin, A.; Troadec, C.; Boualem, A.; Rajab, M.; Fernandez, R.; Morin, H.; Pitrat, M.; Dogimont, C.; Bendahmane, A. A transposon-induced epigenetic change leads to sex determination in melon. Nature 2009, 461, 1135–1138. [Google Scholar] [CrossRef] [PubMed]
- Mao, H.; Wang, H.; Liu, S.; Li, Z.; Yang, X.; Yan, J.; Li, J.; Tran, L.S.P.; Qin, F. A transposable element in a NAC gene is associated with drought tolerance in maize seedlings. Nat. Commun. 2015, 6, 8326. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Liu, J.; Xie, K.; Xing, F.; Xiong, F.; Xiao, J.; Li, X.; Xiong, L. Translational repression by a miniature inverted-repeat transposable element in the 3′ untranslated region. Nat. Commun. 2017, 8, 14651. [Google Scholar] [CrossRef] [PubMed]
- Levin, D.A.; Funderburg, S.W. Genome size in angiosperms: Temperate versus tropical species. Am. Nat. 1979, 114, 784–795. [Google Scholar] [CrossRef]
- Uozu, S.; Ikehashi, H.; Ohmido, N.; Ohtsubo, H.; Ohtsubo, E.; Fukui, K. Repetitive sequences: Cause for variation in genome size and chromosome morphology in the genus Oryza. Plant Mol. Biol. 1997, 35, 791–799. [Google Scholar] [CrossRef] [PubMed]
- Schubert, I.; Oud, J.L. There is an upper limit of chromosome size for normal development of an organism. Cell 1997, 88, 515–520. [Google Scholar] [CrossRef]
- Bennetzen, J.L. Mechanisms and rates of genome expansion and contraction in flowering plants. Genetica 2002, 115, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Sessegolo, C.; Burlet, N.; Haudry, A. Strong phylogenetic inertia on genome size and transposable element content among 26 species of flies. Biol. Lett. 2016, 12, 20160407. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, J.S.; Kim, H.; Nason, J.D.; Wing, R.A.; Wendel, J.F. Differential lineage-specific amplification of transposable elements is responsible for genome size variation in Gossypium. Genome Res. 2006, 16, 1252–1261. [Google Scholar] [CrossRef] [PubMed]
- Piegu, B.; Guyot, R.; Picault, N.; Roulin, A.; Saniyal, A.; Kim, H.; Collura, K.; Brar, D.S.; Jackson, S.; Wing, R.A.; et al. Doubling genome size without polyploidization: Dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wile relative of rice. Genome Res. 2006, 16, 1262–1269. [Google Scholar] [CrossRef] [PubMed]
- Harkess, A.; Mercati, F.; Abbate, L.; McKain, M.; Pires, J.C.; Sala, T.; Sunseri, F.; Falavigna, A.; Leebens-Mack, J. Retrotransposon proliferation coincident with the evolution of dioecy in Asparagus. G3 Genes Genom. Genet. 2016, 6, 2679–2685. [Google Scholar] [CrossRef] [PubMed]
- Cegan, R.; Vyskot, B.; Kejnovsky, E.; Kubat, Z.; Blavet, H.; Šafář, J.; Doležel, J.; Blavet, N.; Hobza, R. Genomic diversity in two related plant species with and without sex chromosomes—Silene latifolia and S. vulgaris. PLoS ONE 2012, 7, e31898. [Google Scholar] [CrossRef] [PubMed]
- Kidwell, M.G. Transposable elements and the evolution of genome size in eukaryotes. Genetica 2002, 115, 49–63. [Google Scholar] [CrossRef] [PubMed]
- Bergero, R.; Forrest, A.; Charlesworth, D. Active miniature transposons from a plant genome and its nonrecombing Y chromosome. Genetics 2008, 178, 1085–1092. [Google Scholar] [CrossRef] [PubMed]
- Sousa, A.; Bellot, S.; Fuchs, J.; Houben, A.; Renner, S.S. Analysis of transposable elements and organellar DNA in male and female genomes of a species with a huge Y chromosome reveals distinct Y centromeres. Plant J. 2016, 88, 387–396. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, K.; Akiyama, Y.; Fukui, K.; Kamada, H.; Satoh, S. Characterization, genome sizes and morphology of sex chromosomes in hemp. Cytologia 1998, 63, 459–464. [Google Scholar] [CrossRef]
- Neumann, P.; Koblížková, A.; Navrátilová, A.; Macas, J. Significant expansion of Vicia pannonica genome size mediated by amplification of a single type of giant retroelement. Genetics 2006, 173, 1047–1056. [Google Scholar] [CrossRef] [PubMed]
- Park, M.; Park, J.; Kim, S.; Kwon, J.K.; Par, H.M.; Bae, I.H.; Yang, T.J.; Lee, Y.H.; Kang, B.C.; Choi, D. Evolution of the large genome in Capsicum annuum occurred through accumulation of single-type long terminal repeat retrotransposons and their derivatives. Plant J. 2012, 69, 1018–1029. [Google Scholar] [CrossRef] [PubMed]
- Estep, M.C.; DeBarry, J.D.; Bennetzen, J.L. The dynamics of LTR retrotransposon accumulation across 25 million years of panicoid grass evolution. Heredity 2013, 110, 194–204. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, J.S.; Proulx, S.R.; Rapp, R.A.; Wendel, J.F. Rapid DNA loss as a counterbalance to genome expansion through retrotransposon proliferation in plants. Proc. Natl. Acad. Sci. USA 2009, 106, 17811–17816. [Google Scholar] [CrossRef] [PubMed]
- Vicient, C.M.; Casacuberta, J.M. Impact of transposable elements on polyploidy plant genomes. Ann. Bot. 2017, 120, 195–207. [Google Scholar] [CrossRef] [PubMed]
- VanBuren, R.; Ming, R. Dynamic transposable element accumulation in the nascent sex chromosomes of papaya. Mob. Genet. Elem. 2013, 3, e23462. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, T.; Heslop-Harrison, J.S. Genomes, genes and junk: The large-scale organization of plant chromosomes. Trends Plant Sci. 1998, 3, 195–199. [Google Scholar] [CrossRef]
- Underwood, C.J.; Henderson, I.R.; Martienssen, R.A. Genetic and epigenetic variation of transposable elements in Arabidopsis. Curr. Opin. Plant Biol. 2017, 36, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Lysak, M.A.; Berr, A.; Pecinka, A.; Schmidt, R.; McBreen, K.; Schubert, I. Mechanisms of chromosome number reduction in Arabidopsis thaliana and related Brassicaceae species. Proc. Natl. Acad. Sci. USA 2006, 103, 5224–5229. [Google Scholar] [CrossRef] [PubMed]
- Yogeeswaran, K.; Frary, A.; York, T.L.; Amenta, A.; Lesser, A.H.; Nasrallah, J.B.; Tanksley, S.D.; Nasrallah, M.E. Comparative genome analyses of Arabidopsis spp.: Inferring chromosomal rearrangement events in the evolutionary history of A. thaliana. Genome Res. 2005, 15, 505–515. [Google Scholar] [CrossRef] [PubMed]
- Fransz, P.; Linc, G.; Lee, C.R.; Aflitos, S.A.; Lasky, J.R.; Toomajian, C.; Ali, H.; Peters, J.; van Dam, P.; Ji, X.; et al. Molecular, genetic and evolutionary analysis of a paracentric inversion in Arabidopsis thaliana. Plant J. 2016, 88, 159–178. [Google Scholar] [CrossRef] [PubMed]
- Lou, Q.; Iovene, M.; Spooner, D.M.; Buell, C.R.; Jiang, J. Evolution of chromosome 6 of Solanum species revealed by comparative fluorescence in situ hybridization mapping. Chromosoma 2010, 119, 435–442. [Google Scholar] [CrossRef] [PubMed]
- Febrer, M.; Goicoechea, J.L.; Wright, J.; McKenzie, N.; Xong, X.; Lin, J.; Collura, K.; Wissotski, M.; Yu, Y.; Ammiraju, J.S.S.; et al. An integrated physical, genetic and cytogenetic map of Brachypodium distachyon, a model system for grass research. PLoS ONE 2010, 5, e13461. [Google Scholar] [CrossRef] [PubMed]
- Betekhtin, A.; Jenkins, G.; Hasterok, R. Reconstructing the evolution of Brachypodium genomes using comparative chromosome painting. PLoS ONE 2014, 9, e115108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Da Silva dos Santos, M.; Kretschmer, R.; Frankl-Vilches, C.; Bakker, A.; Gahr, M.; O’Brien, P.C.M.; Ferguson-Smith, M.; de Oliveira, E.H.C. Comparative cytogenetics between two important songbird models: the zebra finch and the canary. PLoS ONE 2017, 12, e0170997. [Google Scholar] [CrossRef] [PubMed]
- The International Brachypodium Initiative. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 2010, 463, 763–768. [Google Scholar]
- Degrandi, T.M.; del Valle Garnero, A.; O’Brien, P.C.M.; Ferguson-Smith, M.A.; Kretschmer, R.; de Oliveira, E.H.C.; Gunski, R.J. Chromosome painting in Trogon s. surrucura (Aves, Trogoniformes) reveals a karyotype derived by chromosomal fissions, fusion, and inversions. Cytogenet. Genome Res. 2017, 151, 208–215. [Google Scholar] [CrossRef] [PubMed]
- Idziak, D.; Hazuka, I.; Poliwczak, B.; Wiszynska, A.; Wolny, E.; Hasterok, R. Insight into the karyotype evolution of Brachypodium species using comparative chromosome barcoding. PLoS ONE 2014, 9, e93503. [Google Scholar] [CrossRef] [PubMed]
- Chuzhanova, N.; Abeysinghe, S.S.; Krawczak, M.; Cooper, D.N. Translocation and gross deletion breakpoints in human inherited disease and cancer II: Potential involvement of repetitive sequences elements in secondary structure formation between DNA ends. Hum. Mutat. 2003, 22, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Badaeva, E.D.; Dedkova, O.S.; Gay, G.; Pukhalskyi, V.A.; Zelenin, A.V.; Bernard, S.; Bernard, M. Chromosome rearrangements in wheat: Their types and distribution. Genome 2007, 50, 907–926. [Google Scholar] [CrossRef] [PubMed]
- Siljak-Yakovlev, S.; Godelle, B.; Zoldos, V.; Vallès, J.; Garnatje, T.; Hidalgo, O. Evolutionary implications of heterochromatin and rDNA in chromosome number and genome size changes during dysploidy: A case study in Reichardia genus. PLoS ONE 2017, 12, e0182318. [Google Scholar] [CrossRef] [PubMed]
- McClintock, B. Maize Genetics. In Washington Year Book; Carnegie Institution of Washington: Washington, DC, USA, 1946; Volume 45, pp. 176–186. [Google Scholar]
- Raskina, O.; Belyayev, A.; Nevo, E. Activity of the En/Spm-like transposons in meiosis as a base for chromosome repatterning in a small, isolated, peripheral population of Aegilops speltoides Tausch. Chromosome Res. 2004, 12, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Schubert, I.; Rieger, R.; Fuchs, J.; Pich, U. Sequence organization and the mechanism of interstitial deletion clustering in a plant genome (Vicia faba). Mutat. Res. 1994, 325, 1–5. [Google Scholar] [CrossRef]
- Kejnovský, E.; Michalovova, M.; Steflova, P.; Kejnovska, I.; Manzano, S.; Hobza, R.; Kubat, Z.; Kovarik, J.; Jamilena, M.; Vyskot, B. Expansion of microsatellites on evolutionary young Y chromosome. PLoS ONE 2013, 8, e45519. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Zhang, Z.; Liu, C.; Liu, J.; Huang, S.; Jiang, J.; Jin, W. Centromere repositioning in cucurbit species: Implication of the genomic impact from centromere activation and inactivation. Proc. Natl. Acad. Sci. USA 2009, 106, 14937–14941. [Google Scholar] [CrossRef] [PubMed]
- Cáceres, M.; Ranz, J.M.; Barbadilla, A.; Long, M.; Ruiz, A. Generation of a widespread Drosophia inversion by a transposable element. Science 1999, 285, 415–418. [Google Scholar] [PubMed]
- Rachidi, N.; Barre, P.; Blondin, B. Multiple Ty-mediated chromosomal translocations lead to karyotype changes in a wine strain of Saccharomyces cerevisiae. Mol. Gen. Genet. 1999, 261, 841–850. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yu, C.; Pulletikurti, V.; Lamb, J.; Danilova, T.; Weber, D.F.; Birchler, J.; Peterson, T. Alternative Ac/Ds transposition induces major chromosomal rearrangements in maize. Gene. Dev. 2009, 23, 755–765. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.C.; Deal, K.R.; Akhunov, E.D.; Akhunova, A.R.; Anderson, O.D.; Anderson, J.A.; Blake, N.; Clegg, M.T.; Coleman-Derr, D.; Conley, E.J.; et al. Genome comparisons reveal a dominant mechanism of chromosome number reduction in grasses and accelerated genome evolution in Triticeae. Proc. Natl. Acad. Sci. USA 2009, 106, 15780–15785. [Google Scholar] [CrossRef] [PubMed]
- Fishman, L.; Willis, J.H.; Wu, C.A.; Lee, Y.W. Comparative linkage maps suggest that fission, not polyploidy, underlies near-doubling of chromosome number within monkeyflowers (Mimulus; Phrymaceae). Heredity 2014, 112, 562–568. [Google Scholar] [CrossRef] [PubMed]
- Murat, F.; Xu, J.H.; Tannier, E.; Abrouk, M.; Guilhot, N.; Pont, C.; Messing, J.; Salse, J. Ancestral grass karyotype reconstruction unravels new mechanisms of genome shuffling as a source of plant evolution. Genome Res. 2010, 20, 1545–1557. [Google Scholar] [CrossRef] [PubMed]
- Bennetzen, J.L. Transposable elements, gene creation and genome rearrangement in flowering plants. Curr. Opin. Genet. Dev. 2005, 15, 621–627. [Google Scholar] [CrossRef] [PubMed]
- Chuong, E.B.; Elde, N.C.; Feschotte, C. Regulatory activities of transposable elements: From conflicts to benefits. Nat. Rev. Genet. 2017, 18, 71–86. [Google Scholar] [CrossRef] [PubMed]
- Lippman, Z.; Gendrel, A.V.; Black, M.; Vaughn, M.W.; Dedhia, N.; McCombie, W.R.; Lavine, K.; Mittal, V.; May, B.; Kasschau, K.D.; et al. Role of transposable elements in heterochromatin and epigenetic control. Nature 2004, 430, 471–476. [Google Scholar] [CrossRef] [PubMed]
- Li, S.F.; Zhang, G.J.; Yuan, J.H.; Deng, C.L.; Lu, L.D.; Gao, W.J. Effect of 5-azaC on the growth, flowering time and sexual phenotype in spinach. Russ. J. Plant Physiol. 2015, 62, 670–675. [Google Scholar] [CrossRef]
- Hollister, J.D.; Gaut, B.S. Epigenetic silencing of transposable elements: A trade-off between reduced transposition and deleterious effects on neighboring gene expression. Genome Res. 2009, 19, 1419–1428. [Google Scholar] [CrossRef] [PubMed]
- Slotkin, R.K.; Vaughn, M.; Borges, F.; Tanurdžić, M.; Becker, J.D.; Feijó, J.A.; Martienssen, R.A. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 2009, 136, 461–472. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, K.; Katakami, H.; Takata, R. Mobilization of a retrotransposon in 5-azacytidine-treated fungus Fusarium oxysporum. Plant Biotechnol. 2007, 24, 345–348. [Google Scholar] [CrossRef]
- Ohama, N.; Sato, H.; Shinozaki, K.; Yamaguchi-Shinozaji, K. Transcriptional regulatory network of plant heat stress response. Trends Plant Sci. 2017, 22, 53–65. [Google Scholar] [CrossRef] [PubMed]
- Martinez, G.; Köhler, C. Role of small RNAs in epigenetic reprogramming during plant sexual reproduction. Curr. Opin. Plant Biol. 2017, 36, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Lämke, J.; Bäurle, I. Epigenetic and chromatin-based mechanisms in environmental stress adaptation and stress memory in plants. Genome Biol. 2017, 18, 124. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.C.; Karpen, G.H. Epigenetic regulation of heterochromatic DNA stability. Curr. Opin. Genet. Dev. 2008, 18, 204–211. [Google Scholar] [CrossRef] [PubMed]
- Iwata-Otsubo, A.; Lin, J.Y.; Gill, N.; Jackson, S.A. Highly distinct chromosomal structures in cowpea (Vigna unguiculata), as revealed by molecular cytogenetic analysis. Chromosome Res. 2016, 24, 197–216. [Google Scholar] [CrossRef] [PubMed]
- Kowar, T.; Zakrzewski, F.; Macas, J.; Kobližková, A.; Viehoever, P.; Weisshaar, B.; Schmidt, T. Repeat composition of CenH3-chromomatin and H3K9me2-marked heterochromatin in Sugar Beet (Beta vulgaris). BMC Plant Biol. 2016, 16, 20. [Google Scholar] [CrossRef] [PubMed]
- Soppe, W.J.J.; Jasencakova, Z.; Houben, A.; Kakutani, T.; Meister, A.; Huang, M.S.; Jacobsen, S.E.; Schubert, I.; Fransz, P.F. DNA methylation controls histone H3 lysine 9 methylation and heterochromatin assembly in Arabidopsis. EMBO J. 2002, 21, 6549–6559. [Google Scholar] [CrossRef] [PubMed]
- Zemach, A.; Kim, M.Y.; Hsieh, P.H.; Coleman-Derr, D.; Eshed-Williams, L.; Thao, K.; Harmer, S.L.; Zilberman, D. The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell 2013, 153, 193–205. [Google Scholar] [CrossRef] [PubMed]
- Dernburg, A.F.; Sedat, J.W.; Hawley, R.S. Direct evidence of a role for heterochromatin in meiotic chromosome segregation. Cell 1996, 86, 135–146. [Google Scholar] [CrossRef]
- Bernard, P.; Maure, J.F.; Partridge, J.F.; Genier, S.; Javerzat, J.P.; Allshire, R.C. Requirement of heterochromatin for cohesion at centromeres. Science 2001, 294, 2539–2542. [Google Scholar] [CrossRef] [PubMed]
- Jezek, M.; Gast, A.; Choi, G.; Kulkarni, R.; Quijote, J.; Graham-Yooll, A.; Park, D.; Green, E.M. The histone methyltransferases Set5 and Set1 have overlapping functions in gene silencing and telomere maintenance. Epigenetics 2017, 12. [Google Scholar] [CrossRef] [PubMed]
- Nishibuchi, G.; Déjardin, J. The molecular basis of the organization of repetitive DNA-containing constitutive heterochromatin in mammals. Chromosome Res. 2017, 25, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhu, J.K. RNA-directed DNA methylation. Curr. Opin. Plant Biol. 2011, 14, 142–147. [Google Scholar] [CrossRef] [PubMed]
- McMurchy, A.N.; Stempor, P.; Gaarenstroom, T.; Wysolmerski, B.; Dong, Y.; Aussianikava, D.; Appert, A.; Huang, N.; Kolasinska-Zwierz, P.; Sapetschnig, A.; et al. A team of heterochromatin factors collaborates with small RNA pathways to combat repetitive elements and germline stress. eLife 2017, 6, e21666. [Google Scholar] [PubMed]
- Fultz, D.; Choudury, S.G.; Slotkin, P.K. Silencing of active transposable elements in plants. Curr. Opin. Plant Biol. 2015, 27, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Cao, X. Epigenetic regulation and functional exaptation of transposable elements in higher plants. Curr. Opin. Plant Biol. 2014, 21, 83–88. [Google Scholar] [CrossRef] [PubMed]
- Eichten, S.R.; Ellis, N.A.; Makarevitch, I.; Yeh, C.T.; Gent, J.I.; Guo, L.; McGinnis, K.M.; Zhang, X.; Schnable, P.S.; Vaughn, M.W.; et al. Spreading of heterochromatin is limited to specific families of maize retrotransposons. PLoS Genet. 2012, 8, e1003127. [Google Scholar] [CrossRef] [PubMed]
- Gent, J.I.; Ellis, N.A.; Guo, L.; Harkess, A.E.; Yao, Y.; Zhang, X.; Dawe, R.K. CHH island: De novo DNA methylation in near-gene chromatin regulation in maize. Genome Res. 2013, 23, 628–637. [Google Scholar] [CrossRef] [PubMed]
- Comai, L.; Maheshwari, S.; Marimuthu, M.P.A. Plant centromeres. Curr. Opin. Plant Biol. 2017, 36, 158–167. [Google Scholar] [CrossRef] [PubMed]
- Heslop-Harrison, J.S.; Brandes, A.; Schwarzacher, T. Tandemly repeated DNAs sequences and centromeric chromosomal regions of Arabidopsis species. Chromosome Res. 2003, 11, 241–253. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Masonbrink, R.E.; Shan, W.; Song, F.; Zhang, J.; Yu, W.; Wang, K.; Wu, Y.; Tang, H.; Wendel, J.F.; et al. Rapid proliferation and nucleolar organizer targeting centromeric retrotransposons in cotton. Plant J. 2016, 88, 992–1005. [Google Scholar] [CrossRef] [PubMed]
- Allshire, R.C.; Karpen, G.H. Epigenetic regulation of centromeric chromatin: Old dogs, new tricks? Nat. Rev. Genet. 2008, 9, 923–937. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, F.; Gao, Z.; Birchler, J.A. Reactivation of an inactive centromere reveals epigenetic and structural components for centromere specification in maize. Plant Cell 2009, 21, 1929–1939. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Friebe, B.; Gill, B.S.; Jiang, J. Centromere inactivation and epigenetic modifications of a plant chromosome with three functional centromeres. Chromosoma 2010, 119, 553–563. [Google Scholar] [CrossRef] [PubMed]
- Han, F.; Lamb, J.C.; Birchler, J.A. High frequency of centromere inactivation resulting in stable dicentric chromosomes of maize. Proc. Natl. Acad. Sci. USA 2006, 103, 3238–3243. [Google Scholar] [CrossRef] [PubMed]
- Koo, D.H.; Han, F.; Birchler, J.A.; Jiang, J. Distinct DNA methylation patterns associated with active and inactive centromeres of the maize B chromosome. Genome Res. 2011, 21, 908–914. [Google Scholar] [CrossRef] [PubMed]
- Su, H.; Liu, Y.; Liu, Y.; Lv, Z.; Li, H.; Xie, S.; Gao, Z.; Pang, J.; Wang, X.; Lai, J.; et al. Dynamic chromatin changes associated with de novo centromere formation in maize euchromatin. Plant J. 2016, 88, 854–866. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Lv, Z.; Pang, J.; Liu, Y.; Guo, X.; Fu, S.; Li, J.; Dong, Q.; Wu, H.; Gao, Z.; et al. Formation of a functional maize centromere after loss of centromeric sequences and gain of ectopic sequences. Plant Cell 2013, 25, 1979–1989. [Google Scholar] [CrossRef] [PubMed]
- Schneider, K.L.; Xie, Z.; Wolfgruber, T.K.; Presting, G.G. Inbreeding drives maize centromere evolution. Proc. Natl. Acad. Sci. USA 2016, 113, E987–E996. [Google Scholar] [CrossRef] [PubMed]
- Meyer, P. Epigenetic variation and environmental change. J. Exp. Bot. 2015, 66, 3541–3548. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wu, R.; Lin, X.; Bai, Y.; Song, C.; Yu, X.; Xu, C.; Zhao, N.; Dong, Y.; Liu, B. Tissue culture-induced genetic and epigenetic alterations in rice pure-lines, F1 hybrids and polyploids. BMC Plant Biol. 2013, 3, 77. [Google Scholar] [CrossRef] [PubMed]
- Jackson, S.A. Epigenomics: Dissecting hybridization and polyploidization. Genome Biol. 2017, 18, 117. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, R.J.W.; O’Neill, M.J.; Graves, J.A.M. Undermethylation associated with retroelement activation and chromosome remodeling in an interspecific mammalian hybrid. Nature 1998, 393, 68–72. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Jiang, J.; Chen, S.; Qi, X.; Fang, W.; Guan, Z.; Teng, N.; Liao, Y.; Chen, F. Rapid genetic and epigenetic alterations under intergeneric genomic shock in newly synthesized Chrysanthemum morifolium × Leucanthemum paludosum hybrids (Asteraceae). Genome Biol. Evol. 2014, 6, 247–259. [Google Scholar] [CrossRef] [PubMed]
- Baack, E.J.; Whitney, K.D.; Rieseberg, L.H. Hybridization and genome size evolution: Timing and magnitude of nuclear DNA content increases in Helianthus homoploid hybrid species. New Phytol. 2005, 167, 623–630. [Google Scholar] [CrossRef] [PubMed]
- Lai, Z.; Nakazato, T.; Salmaso, M.; Burke, J.M.; Tang, S.; Knapp, S.J.; Rieseberg, L.H. Extensive chromosomal repatterning and the evolution of sterility barriers in hybrid sunflower species. Genetics 2005, 171, 291–303. [Google Scholar] [CrossRef] [PubMed]
- Ungerer, M.C.; Strakosh, S.C.; Zhen, Y. Genome expansion in three hybrid sunflower species is associated with retrotransposon proliferation. Curr. Biol. 2006, 16, 872–873. [Google Scholar] [CrossRef] [PubMed]
- Kawakami, T.; Strakosh, S.C.; Zhen, Y.; Ungerer, M.C. Different scales of Ty1/copia-like retrotransposon proliferation in the genomes of three diploid hybrid sunflower species. Heredity 2010, 104, 341–350. [Google Scholar] [CrossRef] [PubMed]
- Kubat, Z.; Zluvova, J.; Vogel, I.; Kovacova, V.; Cermak, T.; Cegan, R.; Hobza, R.; Vyskot, B.; Kejnovsky, E. Possible mechanism responsible for absence of a retrotransposon family on a plant Y chromosome. New Phytol. 2014, 202, 662–678. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Wendel, J.F. Epigenetic phenomena and the evolution of plant allopolyploids. Mol. Phylogenet. Evol. 2003, 29, 365–379. [Google Scholar] [CrossRef]
- Chen, X.; Ge, X.; Wang, J.; Tan, C.; King, G.J.; Liu, K. Genome-wide DNA methylation profiling by modified reduced representation bisulfite sequencing in Brassica rapa suggests that epigenetic modifications play a key role in polyploidy genome evolution. Front. Plant Sci. 2015, 6, 836. [Google Scholar] [CrossRef] [PubMed]
- Novák, P.; Neumann, P.; Pech, J.; Steinhaisl, J.; Macas, J. RepeatExplorer: A Galaxy-based web server for genome-wide characterization of eukaryotic repetitive elements from next-generation sequence reads. Bioinformatics 2013, 29, 792–793. [Google Scholar] [CrossRef] [PubMed]

© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, S.-F.; Su, T.; Cheng, G.-Q.; Wang, B.-X.; Li, X.; Deng, C.-L.; Gao, W.-J. Chromosome Evolution in Connection with Repetitive Sequences and Epigenetics in Plants. Genes 2017, 8, 290. https://doi.org/10.3390/genes8100290
Li S-F, Su T, Cheng G-Q, Wang B-X, Li X, Deng C-L, Gao W-J. Chromosome Evolution in Connection with Repetitive Sequences and Epigenetics in Plants. Genes. 2017; 8(10):290. https://doi.org/10.3390/genes8100290
Chicago/Turabian StyleLi, Shu-Fen, Ting Su, Guang-Qian Cheng, Bing-Xiao Wang, Xu Li, Chuan-Liang Deng, and Wu-Jun Gao. 2017. "Chromosome Evolution in Connection with Repetitive Sequences and Epigenetics in Plants" Genes 8, no. 10: 290. https://doi.org/10.3390/genes8100290



