5-Fluorouracil Treatment Alters the Efficiency of Translational Recoding
Abstract
:1. Introduction
2. Materials and Methods
2.1. Genetic Methods and Plasmid Construction
2.2. Cells and Cell Culture
2.3. Dual-Luciferase Assays
2.4. Real-Time Reverse Transcriptase -PCR for Detecting Dual Luciferase mRNA
2.5. siRNAs and Transfection
2.6. Western Immunoblot Analysis
2.7. [32P]-Orthophosphate Labeling of rRNA in SW-480 Cells
2.8. Two-Dimensional-Thin Layer Chromatography Pseudouridylation Assay (2D-TLC)
2.9. Analysis of rRNA and U2 snRNA Levels
3. Results
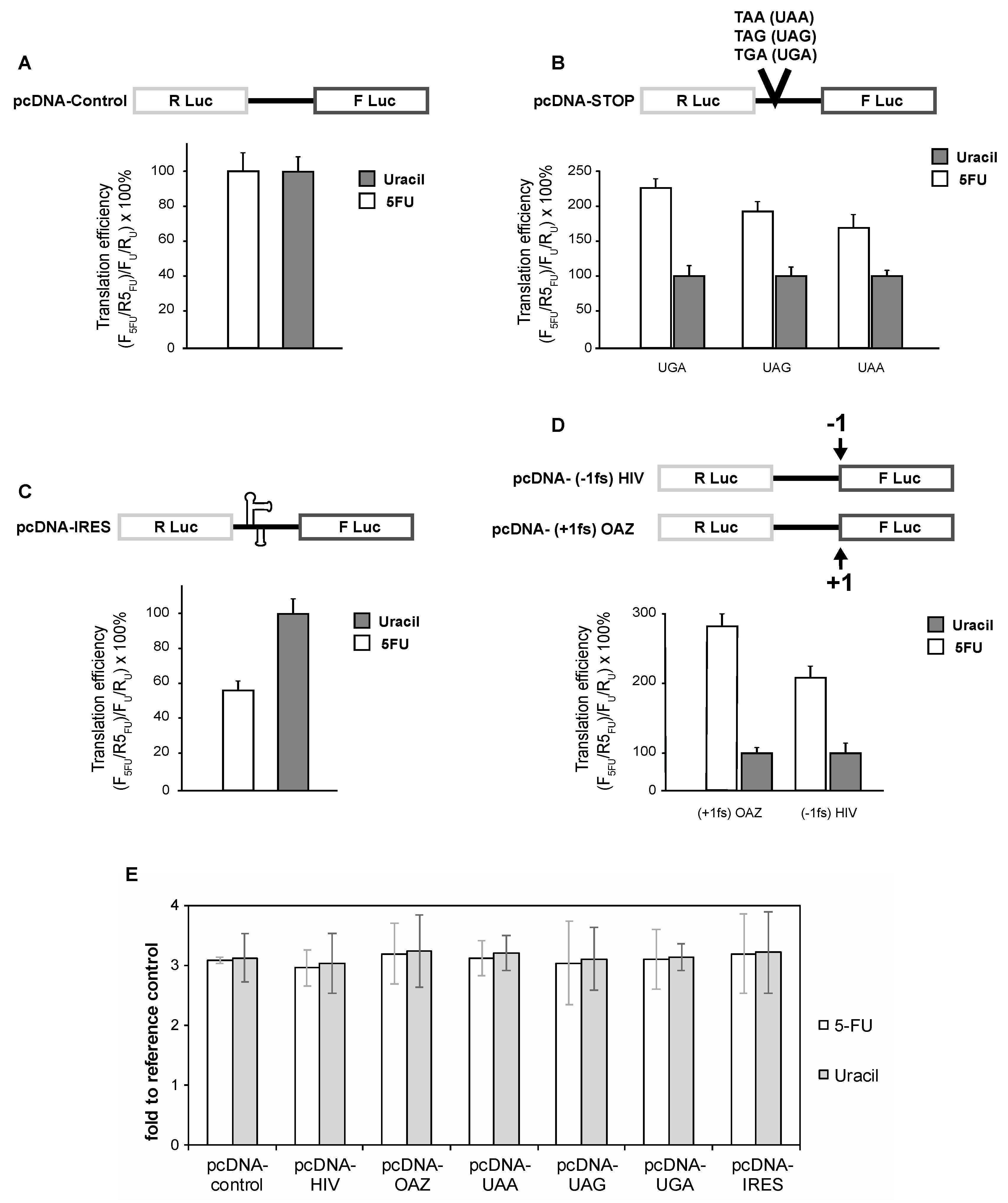
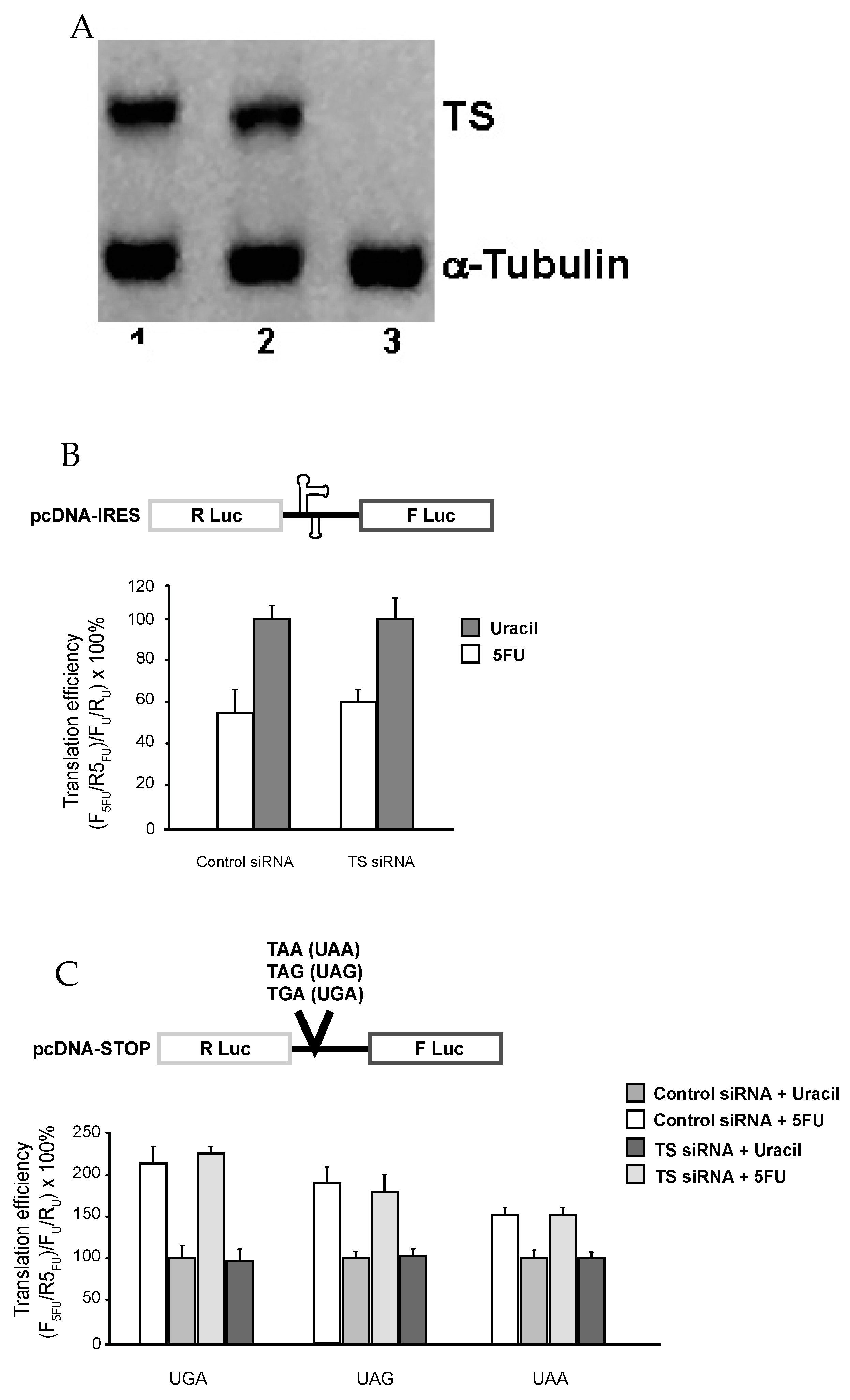
3.1. 5-FU Treated Cells Have Defects in Translational Recoding
3.2. The Effect of 5-FU on Translational Recoding Does Not Involve DNA-Directed Process
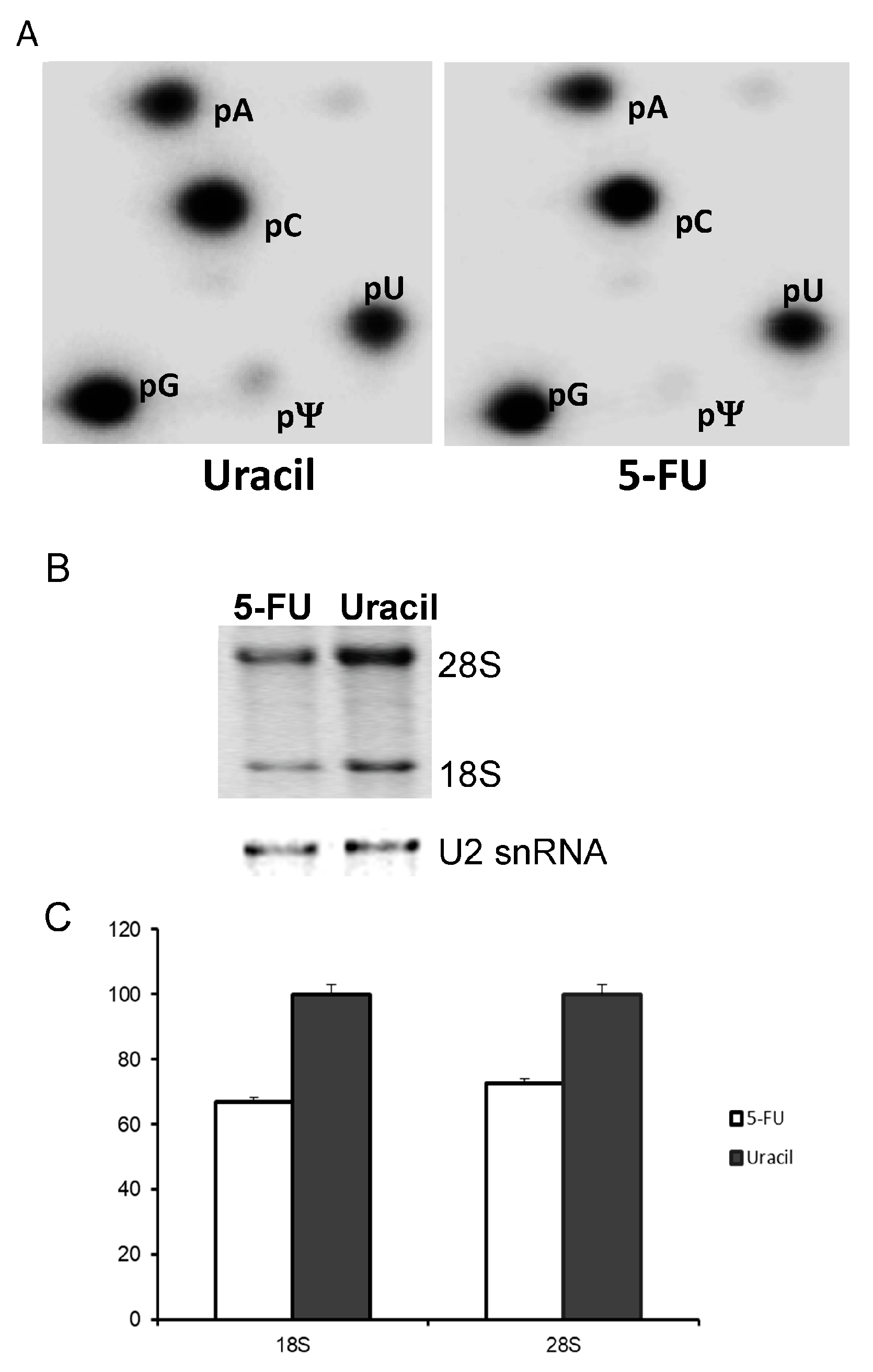
3.3. 5-FU Inhibits rRNA Pseudouridylation
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Rutman, R.J.; Cantarow, A.; Paschkis, K.E. Studies in 2-acetylaminofluorene carcinogenesis. III. The utilization of uracil-2-c14 by preneoplastic rat liver and rat hepatoma. Cancer Res. 1954, 14, 119–123. [Google Scholar] [PubMed]
- Tsimberidou, A.M.; Leick, M.B.; Lim, J.; Fu, S.; Wheler, J.; Piha-Paul, S.A.; Hong, D.; Falchook, G.S.; Naing, A.; Subbiah, I.M.; et al. Dose-finding study of hepatic arterial infusion of oxaliplatin-based treatment in patients with advanced solid tumors metastatic to the liver. Cancer Chemother. Pharmacol. 2013, 71, 389–397. [Google Scholar] [CrossRef] [PubMed]
- Longley, D.B.; Harkin, D.P.; Johnston, P.G. 5-fluorouracil: Mechanisms of action and clinical strategies. Nat. Rev. Cancer 2003, 3, 330–338. [Google Scholar] [CrossRef] [PubMed]
- Wohlhueter, R.M.; McIvor, R.S.; Plagemann, P.G. Facilitated transport of uracil and 5-fluorouracil, and permeation of orotic acid into cultured mammalian cells. J. Cell. Physiol. 1980, 104, 309–319. [Google Scholar] [CrossRef] [PubMed]
- Grogan, B.C.; Parker, J.B.; Guminski, A.F.; Stivers, J.T. Effect of the thymidylate synthase inhibitors on dUTP and TTP pool levels and the activities of DNA repair glycosylases on uracil and 5-fluorouracil in DNA. Biochemistry 2011, 50, 618–627. [Google Scholar] [CrossRef] [PubMed]
- Peters, G.J. Therapeutic potential of TAS-102 in the treatment of gastrointestinal malignancies. Ther. Adv. Med. Oncol. 2015, 7, 340–356. [Google Scholar] [CrossRef] [PubMed]
- Mitrovski, B.; Pressacco, J.; Mandelbaum, S.; Erlichman, C. Biochemical effects of folate-based inhibitors of thymidylate synthase in MGH-U1 cells. Cancer Chemother. Pharmacol. 1994, 35, 109–114. [Google Scholar] [CrossRef] [PubMed]
- Sommer, H.; Santi, D.V. Purification and amino acid analysis of an active site peptide from thymidylate synthetase containing covalently bound 5-fluoro-2′-deoxyuridylate and methylenetetrahydrofolate. Biochem. Biophys. Res. Commun. 1974, 57, 689–695. [Google Scholar] [CrossRef]
- Santi, D.V.; McHenry, C.S.; Sommer, H. Mechanism of interaction of thymidylate synthetase with 5-fluorodeoxyuridylate. Biochemistry 1974, 13, 471–481. [Google Scholar] [CrossRef] [PubMed]
- Aherne, G.W.; Hardcastle, A.; Raynaud, F.; Jackman, A.L. Immunoreactive dUMP and TTP pools as an index of thymidylate synthase inhibition; effect of tomudex (zd1694) and a nonpolyglutamated quinazoline antifolate (cb30900) in l1210 mouse leukaemia cells. Biochem. Pharmacol. 1996, 51, 1293–1301. [Google Scholar] [CrossRef]
- Bijnsdorp, I.V.; Comijn, E.M.; Padron, J.M.; Gmeiner, W.H.; Peters, G.J. Mechanisms of action of FdUMP[10]: Metabolite activation and thymidylate synthase inhibition. Oncol. Rep. 2007, 18, 287–291. [Google Scholar] [CrossRef] [PubMed]
- Lindahl, T. An N-Glycosidase from Escherichia coli that releases free uracil from DNA containing deaminated cytosine residues. Proc. Natl. Acad. Sci. USA 1974, 71, 3649–3653. [Google Scholar] [CrossRef] [PubMed]
- Ghoshal, K.; Jacob, S.T. An alternative molecular mechanism of action of 5-fluorouracil, a potent anticancer drug. Biochem. Pharmacol. 1997, 53, 1569–1575. [Google Scholar] [CrossRef]
- Parker, W.B.; Cheng, Y.C. Metabolism and mechanism of action of 5-fluorouracil. Pharmacol. Ther. 1990, 48, 381–395. [Google Scholar] [CrossRef]
- Engelbrecht, C.; Ljungquist, I.; Lewan, L.; Yngner, T. Modulation of 5-fluorouracil metabolism by thymidine. In vivo and in vitro studies on RNA-directed effects in rat liver and hepatoma. Biochem. Pharmacol. 1984, 33, 745–750. [Google Scholar] [CrossRef]
- Pritchard, D.M.; Watson, A.J.; Potten, C.S.; Jackman, A.L.; Hickman, J.A. Inhibition by uridine but not thymidine of p53-dependent intestinal apoptosis initiated by 5-fluorouracil: Evidence for the involvement of RNA perturbation. Proc. Natl. Acad. Sci. USA 1997, 94, 1795–1799. [Google Scholar] [CrossRef] [PubMed]
- Kufe, D.W.; Major, P.P. 5-fluorouracil incorporation into human breast carcinoma RNA correlates with cytotoxicity. J. Biol. Chem. 1981, 256, 9802–9805. [Google Scholar] [PubMed]
- Glazer, R.I.; Lloyd, L.S. Association of cell lethality with incorporation of 5-fluorouracil and 5-fluorouridine into nuclear RNA in human colon carcinoma cells in culture. Mol. Pharmacol. 1982, 21, 468–473. [Google Scholar] [PubMed]
- Ghoshal, K.; Jacob, S.T. Specific inhibition of pre-ribosomal RNA processing in extracts from the lymphosarcoma cells treated with 5-fluorouracil. Cancer Res. 1994, 54, 632–636. [Google Scholar] [PubMed]
- Herrick, D.; Kufe, D.W. Lethality associated with incorporation of 5-fluorouracil into pre-ribosomal RNA. Mol. Pharmacol. 1984, 26, 135–140. [Google Scholar] [PubMed]
- Cory, J.G.; Breland, J.C.; Carter, G.L. Effect of 5-fluorouracil on RNA metabolism in Novikoff hepatoma cells. Cancer Res. 1979, 39, 4905–4913. [Google Scholar] [PubMed]
- Greenhalgh, D.A.; Parish, J.H. Effects of 5-fluorouracil on cytotoxicity and RNA metabolism in human colonic carcinoma cells. Cancer Chemother. Pharmacol. 1989, 25, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Greenhalgh, D.A.; Parish, J.H. Effect of 5-fluorouracil combination therapy on RNA processing in human colonic carcinoma cells. Br. J. Cancer 1990, 61, 415–419. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Yu, Y.T. Incorporation of 5-fluorouracil into U2 snRNA blocks pseudouridylation and pre-mRNA splicing in vivo. Nucleic Acids Res. 2007, 35, 550–558. [Google Scholar] [CrossRef] [PubMed]
- Doong, S.L.; Dolnick, B.J. 5-fluorouracil substitution alters pre-mRNA splicing in vitro. J. Biol. Chem. 1988, 263, 4467–4473. [Google Scholar] [PubMed]
- Gustavsson, M.; Ronne, H. Evidence that tRNA modifying enzymes are important in vivo targets for 5-fluorouracil in yeast. RNA 2008, 14, 666–674. [Google Scholar] [CrossRef] [PubMed]
- Nakada, D.; Magasanik, B. The roles of inducer and catabolite repressor in the synthesis of β-galactosidase by Escherichia coli. J. Mol. Biol. 1964, 8, 105–127. [Google Scholar] [CrossRef]
- Rosen, B.; Rothman, F.; Weigert, M.G. Miscoding caused by 5-fluorouracil. J. Mol. Biol. 1969, 44, 363–375. [Google Scholar] [CrossRef]
- Glazer, R.I.; Hartman, K.D. In vitro translation of messenger RNA following exposure of human colon carcinoma cells in culture to 5-fluorouracil and 5-fluorouridine. Mol. Pharmacol. 1983, 23, 540–546. [Google Scholar] [PubMed]
- Glazer, R.I.; Hartman, K.D. Analysis of the effect of 5-fluorouracil on the synthesis and translation of polysomal poly(A) RNA from Ehrlich ascites cells. Mol. Pharmacol. 1981, 19, 117–121. [Google Scholar] [PubMed]
- Carrico, C.K.; Glazer, R.I. Effect of 5-fluorouracil on the synthesis and translation of polyadenylic acid-containing RNA from regenerating rat liver. Cancer Res. 1979, 39, 3694–3701. [Google Scholar] [PubMed]
- Takimoto, C.H.; Voeller, D.B.; Strong, J.M.; Anderson, L.; Chu, E.; Allegra, C.J. Effects of 5-fluorouracil substitution on the RNA conformation and in vitro translation of thymidylate synthase messenger RNA. J. Biol. Chem. 1993, 268, 21438–21442. [Google Scholar] [PubMed]
- Harger, J.W.; Dinman, J.D. An in vivo dual-luciferase assay system for studying translational recoding in the yeast Saccharomyces cerevisiae. RNA 2003, 9, 1019–1024. [Google Scholar] [CrossRef] [PubMed]
- Boulant, S.; Becchi, M.; Penin, F.; Lavergne, J.P. Unusual multiple recoding events leading to alternative forms of hepatitis C virus core protein from genotype 1b. J. Biol. Chem. 2003, 278, 45785–45792. [Google Scholar] [CrossRef] [PubMed]
- Ruggero, D. Translational control in cancer etiology. Cold Spring Harb. Perspect. Biol. 2013, 5, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Hellen, C.U.; Sarnow, P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 2001, 15, 1593–1612. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.H.; Liu, Q.; Fournier, M.J. Loss of rRNA modifications in the decoding center of the ribosome impairs translation and strongly delays pre-rRNA processing. RNA 2009, 15, 1716–1728. [Google Scholar] [CrossRef] [PubMed]
- Piekna-Przybylska, D.; Przybylski, P.; Baudin-Baillieu, A.; Rousset, J.P.; Fournier, M.J. Ribosome performance is enhanced by a rich cluster of pseudouridines in the a-site finger region of the large subunit. J. Biol. Chem. 2008, 283, 26026–26036. [Google Scholar] [CrossRef] [PubMed]
- King, T.H.; Liu, B.; McCully, R.R.; Fournier, M.J. Ribosome structure and activity are altered in cells lacking snoRNPs that form pseudouridines in the peptidyl transferase center. Mol. Cell 2003, 11, 425–435. [Google Scholar] [CrossRef]
- Jack, K.; Bellodi, C.; Landry, D.M.; Niederer, R.O.; Meskauskas, A.; Musalgaonkar, S.; Kopmar, N.; Krasnykh, O.; Dean, A.M.; Thompson, S.R.; et al. rRNA pseudouridylation defects affect ribosomal ligand binding and translational fidelity from yeast to human cells. Mol. Cell 2011, 44, 660–666. [Google Scholar] [CrossRef] [PubMed]
- Artman, M.; Werthamer, S.; Gelb, P. Catabolie repression in inhibition of β-galactosidase synthesis by Escherichia coli in the presence of agents producing translation errors. Antimicrob. Agents Chemother. 1972, 2, 449–455. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Yu, Y.T. Pseudouridines in and near the branch site recognition region of U2 snRNA are required for snRNP biogenesis and pre-mRNA splicing in Xenopus oocytes. RNA 2004, 10, 681–690. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.T.; Shu, M.D.; Steitz, J.A. Modifications of U2 snRNA are required for snRNP assembly and pre-mRNA splicing. EMBO J. 1998, 17, 5783–5795. [Google Scholar] [CrossRef] [PubMed]
- Patton, J.R.; Jacobson, M.R.; Pederson, T. Pseudouridine formation in U2 small nuclear RNA. Proc. Natl. Acad. Sci. USA 1994, 91, 3324–3328. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ge, J.; Karijolich, J.; Zhai, Y.; Zheng, J.; Yu, Y.-T. 5-Fluorouracil Treatment Alters the Efficiency of Translational Recoding. Genes 2017, 8, 295. https://doi.org/10.3390/genes8110295
Ge J, Karijolich J, Zhai Y, Zheng J, Yu Y-T. 5-Fluorouracil Treatment Alters the Efficiency of Translational Recoding. Genes. 2017; 8(11):295. https://doi.org/10.3390/genes8110295
Chicago/Turabian StyleGe, Junhui, John Karijolich, Yingzhen Zhai, Jianming Zheng, and Yi-Tao Yu. 2017. "5-Fluorouracil Treatment Alters the Efficiency of Translational Recoding" Genes 8, no. 11: 295. https://doi.org/10.3390/genes8110295




