Impact of Repetitive Elements on the Y Chromosome Formation in Plants
Abstract
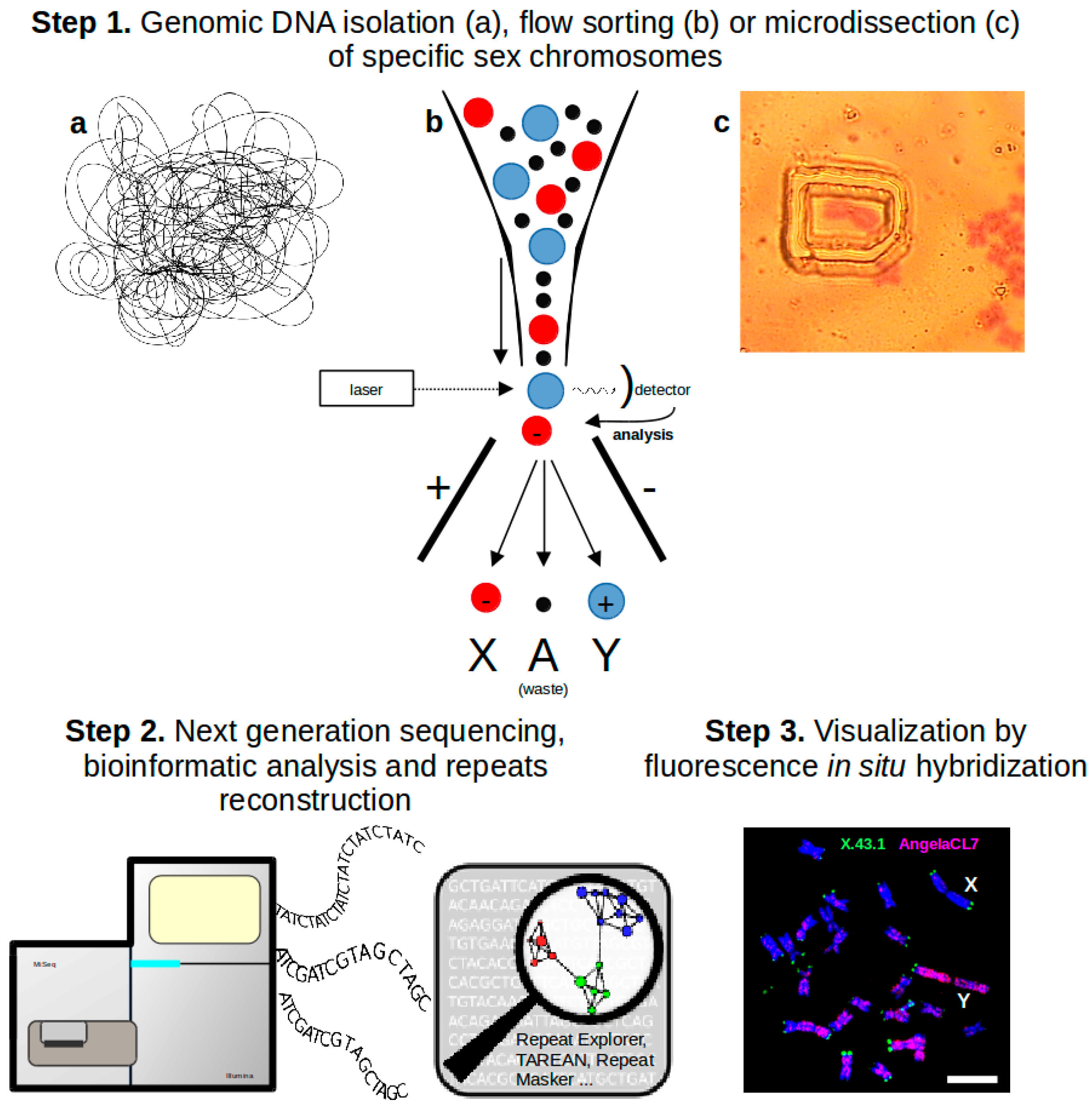
:1. Introduction
2. Promiscuous DNA—Frequent Passengers Colonizing Non-Recombining Regions
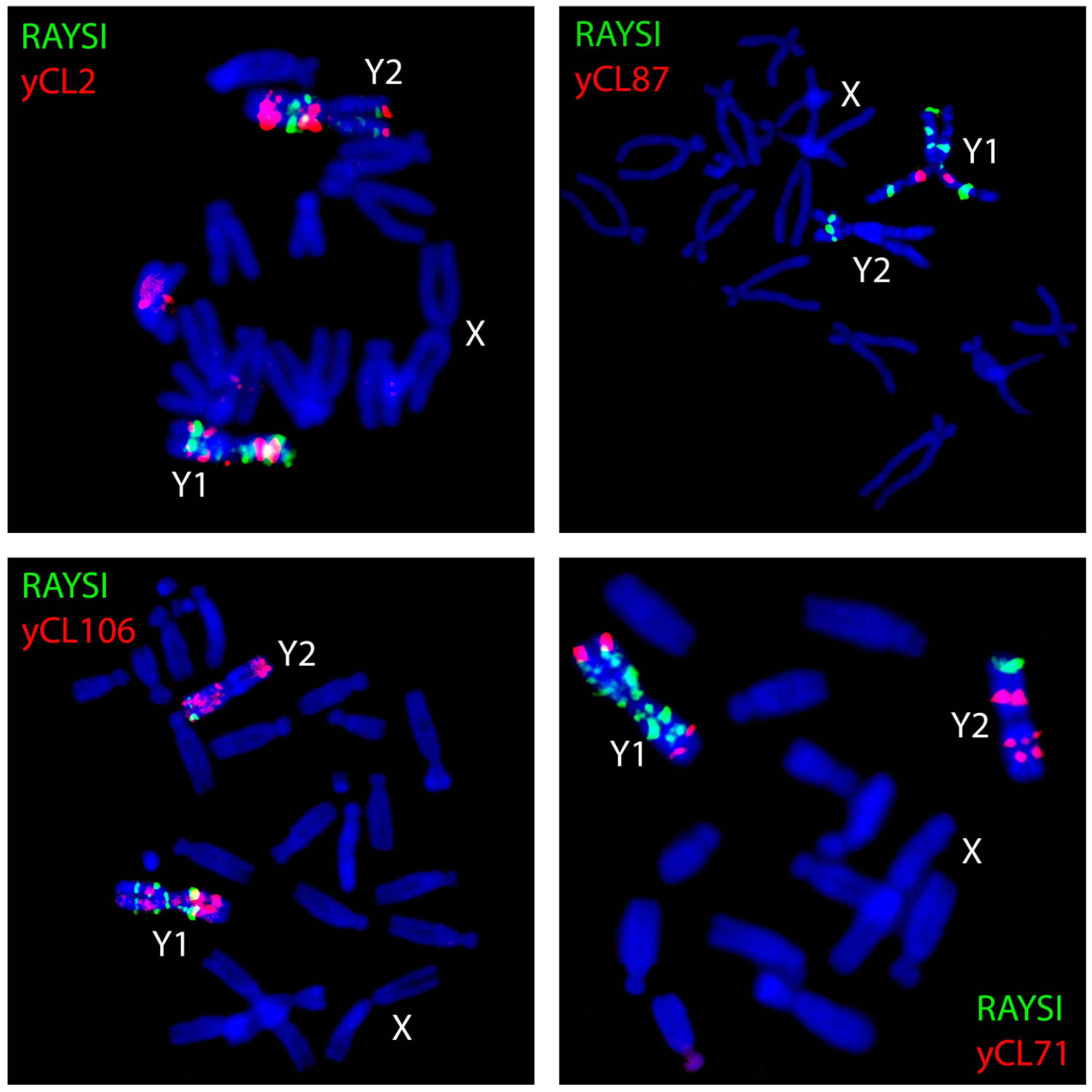
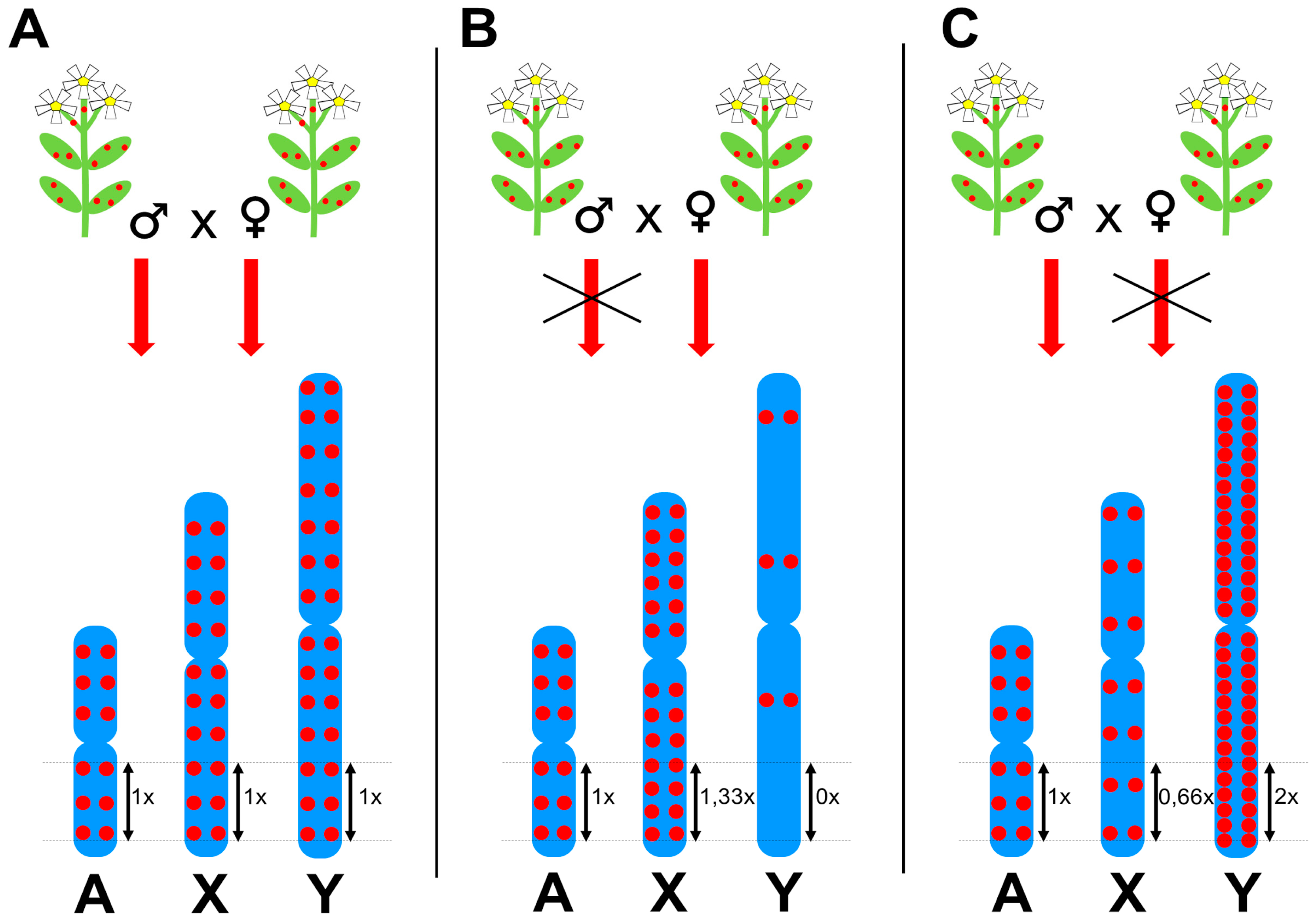
3. Satellites and Chromatin Changes
4. Transposable Elements—Major Force in Sex Chromosome Dynamics
5. Unusual DNA Structures on the Y Chromosome and Their Possible Function
6. Conclusions
Acknowledgments
Conflicts of Interest
References
- Bachtrog, D. Y-chromosome evolution: Emerging insights into processes of Y-chromosome degeneration. Nat. Rev. Genet. 2013, 14, 113–124. [Google Scholar] [CrossRef] [PubMed]
- Vyskot, B.; Hobza, R. The genomics of plant sex chromosomes. Plant Sci. 2015, 236, 126–315. [Google Scholar] [CrossRef] [PubMed]
- Akagi, T.; Henry, I.M.; Tao, R.; Comai, L. Plant genetics. A Y-chromosome-encoded small RNA acts as a sex determinant in persimmons. Science 2014, 346, 646–650. [Google Scholar] [CrossRef] [PubMed]
- Mohanty, J.N.; Nayak, S.; Jha, S.; Joshi, R.K. Transcriptome profiling of the floral buds and discovery of genes related to sex-differentiation in the dioecious cucurbit Coccinia grandis (L.) Voigt. Gene 2017, 626, 395–406. [Google Scholar] [CrossRef] [PubMed]
- Murase, K.; Shigenobu, S.; Fujii, S.; Ueda, K.; Murata, T.; Sakamoto, A.; Wada, Y.; Yamaguchi, K.; Osakabe, Y.; Osakabe, K.; et al. MYB transcription factor gene involved in sex determination in Asparagus officinalis. Genes Cells 2017, 22, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Tennessen, J.A.; Govindarajulu, R.; Liston, A.; Ashman, T.-L. Homomorphic ZW chromosomes in a wild strawberry show distinctive recombination heterogeneity but a small sex-determining region. New Phytol. 2016, 211, 1412–1423. [Google Scholar] [CrossRef] [PubMed]
- Slancarova, V.; Zdanska, J.; Janousek, B.; Talianova, M.; Zschach, C.; Zluvova, J.; Siroky, J.; Kovacova, V.; Blavet, H.; Danihelka, J.; et al. Evolution of sex determination systems with heterogametic males and females in silene. Evolution 2013, 67, 3669–3677. [Google Scholar] [CrossRef] [PubMed]
- Ming, R.; Bendahmane, A.; Renner, S.S. Sex chromosomes in land plants. Annu. Rev. Plant Biol. 2011, 62, 485–514. [Google Scholar] [CrossRef] [PubMed]
- Renner, S.S. The relative and absolute frequencies of angiosperm sexual systems: Dioecy, monoecy, gynodioecy, and an updated online database. Am. J. Bot. 2014, 101, 1588–1596. [Google Scholar] [CrossRef] [PubMed]
- Na, J.-K.; Wang, J.; Ming, R. Accumulation of interspersed and sex-specific repeats in the non-recombining region of papaya sex chromosomes. BMC Genomics 2014, 15, 335. [Google Scholar] [CrossRef] [PubMed]
- Gschwend, A.R.; Yu, Q.; Tong, E.J.; Zeng, F.; Han, J.; VanBuren, R.; Aryal, R.; Charlesworth, D.; Moore, P.H.; Paterson, A.H.; et al. Rapid divergence and expansion of the X chromosome in papaya. Proc. Natl. Acad. Sci. USA 2012, 109, 13716–13721. [Google Scholar] [CrossRef] [PubMed]
- Kejnovsky, E.; Hobza, R.; Cermak, T.; Kubat, Z.; Vyskot, B. The role of repetitive DNA in structure and evolution of sex chromosomes in plants. Heredity 2009, 102, 533–541. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Wang, X.; Yu, Q.; Ming, R.; Jiang, J. DNA methylation and heterochromatinization in the male-specific region of the primitive Y chromosome of papaya. Genome Res. 2008, 18, 1938–1943. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, K.; Ohmido, N.; Fukui, K.; Kamada, H.; Satoh, S. Site-specific accumulation of a line-like retrotransposon in a sex chromosome of the dioecious plant Cannabis sativa. Plant Mol. Biol. 2000, 44, 723–732. [Google Scholar] [CrossRef] [PubMed]
- Puterova, J.; Razumova, O.; Martinek, T.; Alexandrov, O.; Divashuk, M.; Kubat, Z.; Hobza, R.; Karlov, G.; Kejnovsky, E. Satellite DNA and Transposable Elements in Seabuckthorn (Hippophae rhamnoides), a Dioecious Plant with Small Y and Large X Chromosomes. Genome Biol. Evol. 2017, 9, 197–212. [Google Scholar] [CrossRef] [PubMed]
- Sousa, A.; Bellot, S.; Fuchs, J.; Houben, A.; Renner, S.S. Analysis of transposable elements and organellar DNA in male and female genomes of a species with a huge Y chromosome reveals distinct Y centromeres. Plant J. 2016, 88, 387–396. [Google Scholar] [CrossRef] [PubMed]
- Vyskot, B.; Hobza, R. Gender in plants: Sex chromosomes are emerging from the fog. Trends Genet. 2004, 20, 432–438. [Google Scholar] [CrossRef] [PubMed]
- Kubat, Z.; Zluvova, J.; Vogel, I.; Kovacova, V.; Cermak, T.; Cegan, R.; Hobza, R.; Vyskot, B.; Kejnovsky, E. Possible mechanisms responsible for absence of a retrotransposon family on a plant Y chromosome. New Phytol. 2014, 202, 662–678. [Google Scholar] [CrossRef] [PubMed]
- Navajas-Pérez, R.; de la Herrán, R.; López González, G.; Jamilena, M.; Lozano, R.; Ruiz Rejón, C.; Ruiz Rejón, M.; Garrido-Ramos, M.A. The evolution of reproductive systems and sex-determining mechanisms within rumex (polygonaceae) inferred from nuclear and chloroplastidial sequence data. Mol. Biol. Evol. 2005, 22, 1929–1939. [Google Scholar] [CrossRef] [PubMed]
- Sousa, A.; Fuchs, J.; Renner, S.S. Molecular cytogenetics (FISH, GISH) of Coccinia grandis: A ca. 3 myr-old species of cucurbitaceae with the largest Y/autosome divergence in flowering plants. Cytogenet. Genome Res. 2013, 139, 107–118. [Google Scholar] [CrossRef] [PubMed]
- Novák, P.; Neumann, P.; Macas, J. Graph-based clustering and characterization of repetitive sequences in next-generation sequencing data. BMC Bioinformatics 2010, 11, 378. [Google Scholar] [CrossRef] [PubMed]
- Novak, P.; Neumann, P.; Pech, J.; Steinhaisl, J.; Macas, J. RepeatExplorer: A Galaxy-based web server for genome-wide characterization of eukaryotic repetitive elements from next-generation sequence reads. Bioinformatics 2013, 29, 792–793. [Google Scholar] [CrossRef] [PubMed]
- Novák, P.; Ávila Robledillo, L.; Koblížková, A.; Vrbová, I.; Neumann, P.; Macas, J. TAREAN: A computational tool for identification and characterization of satellite DNA from unassembled short reads. Nucleic Acids Res. 2017, 45, e111. [Google Scholar] [CrossRef] [PubMed]
- Kejnovsky, E.; Kubat, Z.; Hobza, R.; Lengerova, M.; Sato, S.; Tabata, S.; Fukui, K.; Matsunaga, S.; Vyskot, B. Accumulation of chloroplast DNA sequences on the Y chromosome of Silene latifolia. Genetica 2006, 128, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Steflova, P.; Hobza, R.; Vyskot, B.; Kejnovsky, E. Strong accumulation of chloroplast DNA in the Y chromosomes of Rumex acetosa and Silene latifolia. Cytogenet. Genome Res. 2014, 142, 59–65. [Google Scholar] [CrossRef] [PubMed]
- VanBuren, R.; Ming, R. Organelle DNA accumulation in the recently evolved papaya sex chromosomes. Mol. Genet. Genomics 2013, 288, 277–284. [Google Scholar] [CrossRef] [PubMed]
- Shibata, F.; Hizume, M.; Kuroki, Y. Chromosome painting of Y chromosomes and isolation of a Y chromosome-specific repetitive sequence in the dioecious plant Rumex acetosa. Chromosoma 1999, 108, 266–270. [Google Scholar] [CrossRef] [PubMed]
- Shibata, F.; Hizume, M.; Kuroki, Y. Differentiation and the polymorphic nature of the Y chromosomes revealed by repetitive sequences in the dioecious plant, Rumex acetosa. Chromosome Res. 2000, 8, 229–236. [Google Scholar] [CrossRef] [PubMed]
- Mariotti, B.; Manzano, S.; Kejnovský, E.; Vyskot, B.; Jamilena, M. Accumulation of Y-specific satellite DNAs during the evolution of Rumex acetosa sex chromosomes. Mol. Genet. Genomics 2009, 281, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Steflova, P.; Tokan, V.; Vogel, I.; Lexa, M.; Macas, J.; Novak, P.; Hobza, R.; Vyskot, B.; Kejnovsky, E. Contrasting patterns of transposable element and satellite distribution on sex chromosomes (XY1Y2) in the dioecious plant Rumex acetosa. Genome Biol. Evol. 2013, 5, 769–782. [Google Scholar] [CrossRef] [PubMed]
- Michalovova, M.; Kubat, Z.; Hobza, R.; Vyskot, B.; Kejnovsky, E. Fully automated pipeline for detection of sex linked genes using RNA-Seq data. BMC Bioinformatics 2015, 16, 78. [Google Scholar] [CrossRef] [PubMed]
- Hobza, R.; Lengerova, M.; Svoboda, J.; Kubekova, H.; Kejnovsky, E.; Vyskot, B. An accumulation of tandem DNA repeats on the Y chromosome in Silene latifolia during early stages of sex chromosome evolution. Chromosoma 2006, 115, 376–382. [Google Scholar] [CrossRef] [PubMed]
- Hobza, R.; Kejnovsky, E.; Vyskot, B.; Widmer, A. The role of chromosomal rearrangements in the evolution of Silene latifolia sex chromosomes. Mol. Genet. Genomics 2007, 278, 633–638. [Google Scholar] [CrossRef] [PubMed]
- Mehrotra, S.; Goyal, V. Repetitive Sequences in Plant Nuclear DNA: Types, Distribution, Evolution and Function. Genom. Proteom. Bioinform. 2014, 12, 164–171. [Google Scholar] [CrossRef] [PubMed]
- Cermak, T.; Kubat, Z.; Hobza, R.; Koblizkova, A.; Widmer, A.; Macas, J.; Vyskot, B.; Kejnovsky, E. Survey of repetitive sequences in Silene latifolia with respect to their distribution on sex chromosomes. Chromosom. Res. 2008, 16, 961–976. [Google Scholar] [CrossRef] [PubMed]
- Filatov, D.A.; Howell, E.C.; Groutides, C.; Armstrong, S.J. Recent spread of a retrotransposon in the Silene latifolia genome, apart from the Y chromosome. Genetics 2009, 181, 811–817. [Google Scholar] [CrossRef] [PubMed]
- Kralova, T.; Cegan, R.; Kubat, Z.; Vrana, J.; Vyskot, B.; Vogel, I.; Kejnovsky, E.; Hobza, R. Identification of a novel retrotransposon with sex chromosome-specific distribution in Silene latifolia. Cytogenet. Genome Res. 2014, 143, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Cegan, R.; Vyskot, B.; Kejnovsky, E.; Kubat, Z.; Blavet, H.; Šafář, J.; Doležel, J.; Blavet, N.; Hobza, R. Genomic diversity in two related plant species with and without sex chromosomes—Silene latifolia and S. vulgaris. PLoS ONE 2012, 7, e31898. [Google Scholar] [CrossRef] [PubMed]
- Kuhl, J.C.; Havey, M.J.; Martin, W.J.; Cheung, F.; Yuan, Q.; Landherr, L.; Hu, Y.; Leebens-Mack, J.; Town, C.D.; Sink, K.C. Comparative genomic analyses in Asparagus. Genome 2005, 48, 1052–1060. [Google Scholar] [CrossRef] [PubMed]
- Harkess, A.; Mercati, F.; Abbate, L.; McKain, M.; Pires, J.C.; Sala, T.; Sunseri, F.; Falavigna, A.; Leebens-Mack, J. Retrotransposon proliferation coincident with the evolution of dioecy in Asparagus. G3 Genes, Genomes, Genet. 2016, 6, 2679–2685. [Google Scholar] [CrossRef] [PubMed]
- Akagi, H.; Yokozeki, Y.; Inagaki, A.; Mori, K.; Fujimura, T. Micron, a microsatellite-targeting transposable element in the rice genome. Mol. Genet. Genomics 2001, 266, 471–480. [Google Scholar] [CrossRef] [PubMed]
- Jiang, N.; Wessler, S.R. Insertion preference of maize and rice miniature inverted repeat transposable elements as revealed by the analysis of nested elements. Plant Cell 2001, 13, 2553–2564. [Google Scholar] [CrossRef] [PubMed]
- Naito, K.; Monden, Y.; Yasuda, K.; Saito, H.; Okumoto, Y. mPing: The bursting transposon. Breed. Sci. 2014, 64, 109–114. [Google Scholar] [CrossRef] [PubMed]
- Reinders, J.; Mirouze, M.; Nicolet, J.; Paszkowski, J. Parent-of-origin control of transgenerational retrotransposon proliferation in Arabidopsis. EMBO Rep 2013, 14, 823–828. [Google Scholar] [CrossRef] [PubMed]
- Gehring, M.; Henikoff, S. DNA methylation dynamics in plant genomes. Biochim. Biophys. Acta BBA Gene Struct. Expr. 2002, 1769, 276–286. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, T.-F.; Ibarra, C.A.; Silva, P.; Zemach, A.; Eshed-Williams, L.; Fischer, R.L.; Zilberman, D. Genome-wide demethylation of Arabidopsis endosperm. Science 2009, 324, 1451–1454. [Google Scholar] [CrossRef] [PubMed]
- Calarco, J.P.; Borges, F.; Donoghue, M.T.A.; Van Ex, F.; Jullien, P.E.; Lopes, T.; Gardner, R.; Berger, F.; Feijó, J.A.; Becker, J.D.; et al. Reprogramming of DNA methylation in pollen guides epigenetic inheritance via small RNA. Cell 2012, 151, 194–205. [Google Scholar] [CrossRef] [PubMed]
- Slotkin, R.K.; Vaughn, M.; Borges, F.; Tanurdžić, M.; Becker, J.D.; Feijó, J.A.; Martienssen, R.A. Epigenetic Reprogramming and Small RNA Silencing of Transposable Elements in Pollen. Cell 2009, 136, 461–472. [Google Scholar] [CrossRef] [PubMed]
- Ibarra, C.A.; Feng, X.; Schoft, V.K.; Hsieh, T.-F.; Uzawa, R.; Rodrigues, J.A.; Zemach, A.; Chumak, N.; Machlicova, A.; Nishimura, T.; et al. Active DNA Demethylation in Plant Companion Cells Reinforces Transposon Methylation in Gametes. Science 2012, 337, 1360–1364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martínez, G.; Panda, K.; Köhler, C.; Slotkin, R.K. Silencing in sperm cells is directed by RNA movement from the surrounding nurse cell. Nat. Plants 2016, 2, 16030. [Google Scholar] [CrossRef] [PubMed]
- Fultz, D.; Choudury, S.G.; Slotkin, R.K. Silencing of active transposable elements in plants. Curr. Opin. Plant Biol. 2015, 27, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Cuerda-Gil, D.; Slotkin, R.K. Non-canonical RNA-directed DNA methylation. Nat. Plants 2016, 2, 16163. [Google Scholar] [CrossRef] [PubMed]
- Chibalina, M.V.; Filatov, D.A. Plant Y chromosome degeneration is retarded by haploid purifying selection. Curr. Biol. 2011, 21, 1475–1479. [Google Scholar] [CrossRef] [PubMed]
- Creasey, K.M.; Zhai, J.; Borges, F.; Van Ex, F.; Regulski, M.; Meyers, B.C.; Martienssen, R.A. miRNAs trigger widespread epigenetically activated siRNAs from transposons in Arabidopsis. Nature 2014, 508, 411–415. [Google Scholar] [CrossRef] [PubMed]
- Kejnovsky, E.; Leitch, I.J.; Leitch, A.R. Contrasting evolutionary dynamics between angiosperm and mammalian genomes. Trends Ecol. Evol. 2009, 24, 572–582. [Google Scholar] [CrossRef] [PubMed]
- Ito, H. Small RNAs and transposon silencing in plants. Dev. Growth Differ. 2012, 54, 100–107. [Google Scholar] [CrossRef] [PubMed]
- Rozen, S.; Skaletsky, H.; Marszalek, J.D.; Minx, P.J.; Cordum, H.S.; Waterston, R.H.; Wilson, R.K.; Page, D.C. Abundant gene conversion between arms of palindromes in human and ape Y chromosomes. Nature 2003, 423, 873–876. [Google Scholar] [CrossRef] [PubMed]
- Trombetta, B.; Cruciani, F. Y chromosome palindromes and gene conversion. Hum. Genet. 2017, 136, 605–619. [Google Scholar] [CrossRef] [PubMed]
- Skaletsky, H.; Kuroda-Kawaguchi, T.; Minx, P.J.; Cordum, H.S.; Hillier, L.; Brown, L.G.; Repping, S.; Pyntikova, T.; Ali, J.; Bieri, T.; et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 2003, 423, 825–837. [Google Scholar] [CrossRef] [PubMed]
- Betrán, E.; Demuth, J.P.; Williford, A. Why chromosome palindromes? Int. J. Evol. Biol. 2012, 2012, 207958. [Google Scholar] [CrossRef] [PubMed]
- Warburton, P.E.; Giordano, J.; Cheung, F.; Gelfand, Y.; Benson, G. Inverted repeat structure of the human genome: The X-chromosome contains a preponderance of large, highly homologous inverted repeats that contain testes genes. Genome Res. 2004, 14, 1861–1869. [Google Scholar] [CrossRef] [PubMed]
- De Vries, M.; Vosters, S.; Merkx, G.; D’Hauwers, K.; Wansink, D.G.; Ramos, L.; de Boer, P. Human male meiotic sex chromosome inactivation. PLoS ONE 2012, 7, e31485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lexa, M.; Kejnovský, E.; Steflová, P.; Konvalinová, H.; Vorlícková, M.; Vyskot, B. Quadruplex-forming sequences occupy discrete regions inside plant LTR retrotransposons. Nucleic Acids Res. 2014, 42, 968–978. [Google Scholar] [CrossRef] [PubMed]
- Lexa, M.; Steflova, P.; Martinek, T.; Vorlickova, M.; Vyskot, B.; Kejnovsky, E. Guanine quadruplexes are formed by specific regions of human transposable elements. BMC Genomics 2014, 15, 1032. [Google Scholar] [CrossRef] [PubMed]
- Lisch, D. Epigenetic Regulation of Transposable Elements in Plants. Annu. Rev. Plant Biol. 2009, 60, 43–66. [Google Scholar] [CrossRef] [PubMed]
- Lengerova, M.; Vyskot, B. Sex chromatin and nucleolar analyses in Rumex acetosa L. Protoplasma 2001, 217, 147–153. [Google Scholar] [CrossRef] [PubMed]
- Sahakyan, A.B.; Murat, P.; Mayer, C.; Balasubramanian, S. G-quadruplex structures within the 3′ UTR of LINE-1 elements stimulate retrotransposition. Nat. Struct. Mol. Biol. 2017, 24, 243–247. [Google Scholar] [CrossRef] [PubMed]
- Kubat, Z.; Hobza, R.; Vyskot, B.; Kejnovsky, E. Microsatellite accumulation on the Y chromosome in Silene latifolia. Genome 2008, 51, 350–356. [Google Scholar] [CrossRef] [PubMed]
- Kejnovský, E.; Michalovova, M.; Steflova, P.; Kejnovska, I.; Manzano, S.; Hobza, R.; Kubat, Z.; Kovarik, J.; Jamilena, M.; Vyskot, B. Expansion of Microsatellites on Evolutionary Young Y Chromosome. PLoS ONE 2013, 8, e45519. [Google Scholar] [CrossRef] [PubMed]
- Huppert, J.L.; Balasubramanian, S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007, 35, 406–413. [Google Scholar] [CrossRef] [PubMed]
- Nadir, E.; Margalit, H.; Gallily, T.; Ben-Sasson, S.A. Microsatellite spreading in the human genome: Evolutionary mechanisms and structural implications. Proc. Natl. Acad. Sci. USA 1996, 93, 6470–6475. [Google Scholar] [CrossRef] [PubMed]
- Ellegren, H. Microsatellites: Simple sequences with complex evolution. Nat. Rev. Genet. 2004, 5, 435–445. [Google Scholar] [CrossRef] [PubMed]
- Kass, D.H.; Batzer, M.A.; Deininger, P.L. Gene conversion as a secondary mechanism of short interspersed element (SINE) evolution. Mol. Cell. Biol. 1995, 15, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, J.; Sangaré, D.; Besansky, N.J. Satellite DNA from the Y chromosome of the malaria vector Anopheles gambiae. Genetics 2005, 169, 185–196. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, J.S.; Proulx, S.R.; Rapp, R.A.; Wendel, J.F. Rapid DNA loss as a counterbalance to genome expansion through retrotransposon proliferation in plants. Proc. Natl. Acad. Sci. USA 2009, 106, 17811–17816. [Google Scholar] [CrossRef] [PubMed]
| Mechanism | References |
|---|---|
| Expansion phase | |
| Satellites expansion | Sousa et al. (2016) [16], Shibata et al. (1999) [26], |
| Kubat et al. (2008) [18], Puterova et al. (2017) [15], | |
| Retrotranspositions | Na et al. (2014) [10], Cermak et al. (2008) [34], |
| Sousa et al. (2016) [16] | |
| Promiscuous DNA insertions | VanBuren et al. (2013) [25], Kejnovsky et al. (2006) [23] |
| Shrinkage phase | |
| Ectopic recombination | Kejnovsky et al. (2009) [12] |
| Deletions | Hawkins et al. (2009) [55] |
| Both expansion and shrinkage phase | |
| Epigenetic regulation of TEs | Kubat et al. (2014) [18] |
| Chromatin modification | Zhang et al. (2008) [13], Lengerova et al. (2001) [56] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hobza, R.; Cegan, R.; Jesionek, W.; Kejnovsky, E.; Vyskot, B.; Kubat, Z. Impact of Repetitive Elements on the Y Chromosome Formation in Plants. Genes 2017, 8, 302. https://doi.org/10.3390/genes8110302
Hobza R, Cegan R, Jesionek W, Kejnovsky E, Vyskot B, Kubat Z. Impact of Repetitive Elements on the Y Chromosome Formation in Plants. Genes. 2017; 8(11):302. https://doi.org/10.3390/genes8110302
Chicago/Turabian StyleHobza, Roman, Radim Cegan, Wojciech Jesionek, Eduard Kejnovsky, Boris Vyskot, and Zdenek Kubat. 2017. "Impact of Repetitive Elements on the Y Chromosome Formation in Plants" Genes 8, no. 11: 302. https://doi.org/10.3390/genes8110302





