Amphibian and Avian Karyotype Evolution: Insights from Lampbrush Chromosome Studies
Abstract
:1. Peculiarities of Amphibian and Avian Genomes and Karyotypes
2. Lampbrush Chromosomes as a Tool to Study Amphibian Chromosomal Evolution
2.1. Interspecies Differences
2.2. Interpopulation Differences
2.3. Sex Chromosomes
2.4. Interspecies Hybrids
3. Lampbrush Chromosomes as a Tool to Study Avian Chromosomal Evolution
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Gregory, T.R. Genome size and evolution in animals. In The Evolution of the Genome; Gregory, T.R., Ed.; Elsevier: Burlington, NJ, USA, 2005; pp. 3–87. [Google Scholar]
- Morescalchi, A.; Olmo, E. Single-copy DNA and vertebrate phylogeny. Cytogenet. Cell Genet. 1982, 34, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Shepard, D.B.; Chong, R.A.; Lopez Arriaza, J.; Hall, K.; Castoe, T.A.; Feschotte, C.; Pollock, D.D.; Mueller, R.L. LTR transposons contribute to genomic gigantism in plethodontid salamanders. Genome Biol. Evol. 2012, 4, 168–183. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.J.; Putta, S.; Zhu, W.; Pao, G.M.; Verma, I.M.; Hunter, T.; Bryant, S.V.; Gardiner, D.M.; Harkins, T.T.; Voss, S.R. Genic regions of a large salamander genome contain long introns and novel genes. BMC Genom. 2009, 10, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Hellsten, U.; Harland, R.M.; Gilchrist, M.J.; Hendrix, D.; Jurka, J.; Kapitonov, V.; Ovcharenko, I.; Putnam, N.H.; Shu, S.; Taher, L.; et al. The genome of the Western clawed frog Xenopus tropicalis. Science 2010, 328, 633–636. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.B.; Xiong, Z.J.; Xiang, X.Y.; Liu, S.P.; Zhou, W.W.; Tu, X.L.; Zhong, L.; Wang, L.; Wu, D.D.; Zhang, B.L.; et al. Whole-genome sequence of the Tibetan frog Nanorana parkeri and the comparative evolution of tetrapod genome. Proc. Natl. Acad. Sci. USA 2015, 112, 1257–1262. [Google Scholar] [CrossRef] [PubMed]
- Session, A.M.; Uno, Y.; Kwon, T.; Chapman, J.A.; Toyoda, A.; Takahashi, S.; Fukui, A.; Hikosaka, A.; Suzuki, A.; Kondo, M.; et al. Genome evolution in the allotetraploid frog Xenopus laevis. Nature 2016, 538, 336–343. [Google Scholar] [CrossRef] [PubMed]
- Schmid, M. Chromosome banding in Amphibia. I. Constitutive heterochromatin and nucleolus organizer regions in Bufo and Hyla. Chromosoma 1978, 66, 361–388. [Google Scholar] [CrossRef]
- Schempp, W.; Schmid, M. Chromosome banding in amphibia. VI. BrdU-replication patterns in anura and demonstration of XX/XY sex chromosomes in Rana esculenta. Chromosoma 1981, 83, 697–710. [Google Scholar] [CrossRef] [PubMed]
- Morescalchi, A. Evolution and karyology of the amphibians. Boll. Zool. 1980, 47, 113–126. [Google Scholar] [CrossRef]
- Schmid, M.; Steinlein, C.; Bogart, J.P.; Feichtinger, W.; León, P.; La Marca, E.; Díaz, L.M.; Sanz, A.; Chen, S.H.; Hedges, S.B. The chromosomes of terraranan frogs. Insights into vertebrate cytogenetics. Cytogenet. Genome Res. 2010, 130–131, 1–568. [Google Scholar] [CrossRef] [PubMed]
- Wallace, H.; Wallace, B.M.; Badawy, G.M. Lampbrush chromosomes and chiasmata of sex-reversed crested newts. Chromosoma 1997, 106, 526–533. [Google Scholar] [CrossRef] [PubMed]
- Wallace, H.; Badawy, G.M.; Wallace, B.M. Amphibian sex determination and sex reversal. Cell. Mol. Life Sci. 1999, 55, 901–909. [Google Scholar] [CrossRef] [PubMed]
- Lacroix, J.C.; Azzouz, R.; Simon, F.; Bellini, M.; Charlemagne, J.; Dournon, C. Lampbrush, W and Z heterochromosome characterization with a monoclonal antibody and heat-induced chromosomal markers in the newt Pleurodeles waltl: W chromosome plays a role in female sex determination. Chromosoma 1990, 99, 307–314. [Google Scholar] [CrossRef] [PubMed]
- Mable, B.K.; Alexandrou, M.A.; Taylor, M.I. Genome duplication in amphibians and fish: An extended synthesis. J. Zool. 2011, 284, 151–182. [Google Scholar] [CrossRef]
- Schmid, M.; Evans, B.J.; Bogart, J.P. Polyploidy in amphibia. Cytogenet. Genome Res. 2015, 145, 315–330. [Google Scholar] [CrossRef] [PubMed]
- Tegelström, H.; Ryttman, H. Chromosomes in birds (Aves): Evolutionary implications of macro-and microchromosome numbers and lengths. Hereditas 1981, 94, 225–233. [Google Scholar] [CrossRef]
- Christidis, L. Aves. In Animal Cytogenetics, Chordata 3 B; John, B., Kayano, H., Levan, A., Eds.; Gebrüder Borntraeger: Berlin, Germany, 1990; Volume 4. [Google Scholar]
- Rodionov, A.V. Evolution of avian chromosomes and linkage groups. Russ. J. Genet. 1997, 33, 605–617. [Google Scholar]
- Burt, D.W. Origin and evolution of avian microchromosomes. Cytogenet. Genome Res. 2002, 96, 97–112. [Google Scholar] [CrossRef] [PubMed]
- International Chicken Genome Sequencing Consortium (ICGSC). Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 2004, 432, 695–716. [Google Scholar]
- Masabanda, J.S.; Burt, D.W.; O’Brien, P.C.M. Molecular cytogenetic definition of the chicken genome: The first complete avian karyotype. Genetics 2004, 166, 1367–1373. [Google Scholar] [CrossRef] [PubMed]
- Griffin, D.K.; Robertson, L.B.W.; Tempest, H.G.; Skinner, B.M. The evolution of the avian genome as revealed by comparative molecular cytogenetics. Cytogenet. Genome Res. 2007, 117, 64–77. [Google Scholar] [CrossRef] [PubMed]
- McQueen, H.A.; Siriaco, G.; Bird, A.P. Chicken microchromosomes are hyperacetylated, early replicating, and gene rich. Genome Res. 1998, 8, 621–630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, J.; Bruley, C.K.; Paton, I.R.; Dunn, I.; Jones, C.T.; Windsor, D.; Morrice, D.R.; Law, A.S.; Masabanda, J.; Sazanov, A.; et al. Differences in gene density on chicken macrochromosomes and microchromosomes. Anim. Genet. 2000, 31, 96–103. [Google Scholar] [CrossRef] [PubMed]
- The National Center for Biotechnology Information (NCBI). Genomes. Available online: http://www.ncbi.nlm.nih.gov/genome/111 (accessed on 27 August 2017).
- Zoorob, R.; Billault, A.; Severac, V.; Fillon, V.; Vignal, A.; Auffray, C. Two chicken genomic libraries in the PAC and BAC cloning systems: Organization and characterization. Anim. Genet. 1996, 27, 2–69. [Google Scholar]
- Griffin, D.K.; Haberman, F.; Masabanda, J. Micro- and macrochromosome paints generated by flow cytometry and microdissection: Tools for mapping the chicken genome. Cytogenet. Cell Genet. 1999, 87, 278–281. [Google Scholar] [CrossRef] [PubMed]
- Crooijmans, R.P.M.A.; Vrebalov, J.; Dijkhof, R.J.; van der Poel, J.J.; Nicholls, R.D.; Bovenhuis, H.; Groenen, M.A. Two-dimensional screening of the Wageningen chicken BAC library. Mamm. Genome 2000, 11, 360–363. [Google Scholar] [CrossRef] [PubMed]
- BACPAC Resources Center (BPRC). Available online: https://bacpacresources.org (accessed on 20 August 2017).
- Schmid, M.; Nanda, I.; Guttenbach, M.; Steinlein, C.; Hoehn, M.; Schartl, M.; Haaf, T.; Weigend, S.; Fries, R.; Buerstedde, J.M.; et al. First report on chicken genes and chromosomes. Cytogenet. Cell Genet. 2000, 90, 169–218. [Google Scholar] [CrossRef] [PubMed]
- Schmid, M.; Nanda, I.; Hoehn, H.; Schartl, M.; Haaf, T.; Buerstedde, J.M.; Arakawa, H.; Caldwell, R.B.; Weigend, S.; Burt, D.W.; et al. Second report on chicken genes and chromosomes. Cytogenet. Genome Res. 2005, 109, 415–479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmid, M.; Smith, J.; Burt, D.W.; Aken, B.L.; Antin, P.B.; Archibald, A.L.; Ashwell, C.; Blackshear, P.J.; Boschiero, C.; Brown, C.T.; et al. Third report on chicken genes and chromosomes 2015. Cytogenet. Genome Res. 2015, 145, 78–179. [Google Scholar] [CrossRef] [PubMed]
- Larkin, D.M.; Pape, G.; Donthu, R.; Auvil, L.; Welge, M.; Lewin, H.A. Breakpoint regions and homologous synteny blocks in chromosomes have different evolutionary histories. Genome Res. 2009, 19, 770–777. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Li, C.; Li, Q.; Li, B.; Larkin, D.M.; Lee, C.; Storz, J.F.; Antunes, A.; Greenwold, M.J.; Meredith, R.W.; et al. Comparative genomics reveals insights into avian genome evolution and adaptation. Science 2014, 346, 1311–1320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farré, M.; Narayan, J.; Slavov, G.T.; Damas, J.; Auvil, L.; Li, C.; Jarvis, E.D.; Burt, D.W.; Griffin, D.K.; Larkin, D.M. Novel insights into chromosome evolution in birds, archosaurs, and reptiles. Genome Biol. Evol. 2016, 8, 2442–2451. [Google Scholar] [CrossRef] [PubMed]
- Damas, J.; O’Connor, R.; Farré, M.; Lenis, V.P.E.; Martell, H.J.; Mandawala, A.; Fowler, K.; Joseph, S.; Swain, M.T.; Griffin, D.K.; et al. Upgrading short-read animal genome assemblies to chromosome level using comparative genomics and a universal probe set. Genome Res. 2017, 27, 875–884. [Google Scholar] [CrossRef] [PubMed]
- Callan, H.G. Lampbrush Chromosomes. Mol. Biol. Biochem. Biophys. 1986, 36, 1–254. [Google Scholar] [PubMed]
- Morgan, G.T. Lampbrush chromosomes and associated bodies: New insights into principles of nuclear structure and function. Chromosom. Res. 2002, 10, 177–200. [Google Scholar] [CrossRef]
- Gaginskaya, E.; Kulikova, T.; Krasikova, A. Avian lampbrush chromosomes: A powerful tool for exploration of genome expression. Cytogenet. Genome Res. 2009, 124, 251–267. [Google Scholar] [CrossRef] [PubMed]
- Macgregor, H.C. Chromomeres revisited. Chromosom. Res. 2012, 20, 911–924. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chelysheva, L.A.; Solovei, I.V.; Rodionov, A.V.; Yakovlev, A.F.; Gaginskaya, E.R. The lampbrush chromosomes of the chicken. The cytological map of macrobivalents. Tsitologiia 1990, 32, 303–316. [Google Scholar] [PubMed]
- Callan, H.G.; Gall, J.G.; Berg, A. The lampbrush chromosomes of Xenopus laevis: Preparation, identification, and distribution of 5s DNA sequences. Chromosoma 1987, 95, 236–250. [Google Scholar] [CrossRef] [PubMed]
- Galkina, S.; Deryusheva, S.; Fillon, V.; Vignal, A.; Crooijmans, R.; Groenen, M.; Rodionov, A.; Gaginskaya, E. FISH on avian lampbrush chromosomes produces higher resolution gene mapping. Genetica 2006, 128, 241–251. [Google Scholar] [CrossRef] [PubMed]
- Daks, A.; Deryusheva, S.; Krasikova, A.; Zlotina, A.; Gaginskaya, E.; Galkina, S. Lampbrush Chromosomes of the Japanese Quail (Coturnix coturnix japonica): A New Version of Cytogenetic Maps. Genetika 2010, 46, 1335–1338. [Google Scholar] [CrossRef] [PubMed]
- Rodionov, A.V.; Chechik, M.S. Cytogenetic map of the Japanese quail lampbrush chromosomes. Genetika 2002, 38, 1246–1251. [Google Scholar] [PubMed]
- Penrad-Mobayed, M.; El Jamil, A.; Kanhoush, R.; Perrin, C. Working map of the lampbrush chromosomes of Xenopus tropicalis: A new tool for cytogenetic analyses. Dev. Dyn. 2009, 238, 1492–1501. [Google Scholar] [CrossRef] [PubMed]
- Solinhac, R.; Leroux, S.; Galkina, S.; Chazara, O.; Feve, K.; Vignoles, F.; Morisson, M.; Derjusheva, S.; Bed’hom, B.; Vignal, A.; et al. Integrative mapping analysis of chicken microchromosome 16 organization. BMC Genom. 2010, 11, 616. [Google Scholar] [CrossRef] [PubMed]
- Zlotina, A.; Galkina, S.; Krasikova, A.; Crooijmans, R.P.; Groenen, M.A.; Gaginskaya, E.; Deryusheva, S. Centromere positions in chicken and Japanese quail chromosomes: De novo centromere formation versus pericentric inversions. Chromosom. Res. 2012, 20, 1017–1032. [Google Scholar] [CrossRef] [PubMed]
- Dedukh, D.; Mazepa, G.; Shabanov, D.; Rosanov, J.; Litvinchuk, S.; Borkin, L.; Saifitdinova, A.; Krasikova, A. Cytological maps of lampbrush chromosomes of European water frogs (Pelophylax esculentus complex) from the Eastern Ukraine. BMC Genet. 2013, 14, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Macgregor, H.C.; Sessions, S.K.; Arntzen, J.W. An integrative analysis of phylogenetic relationships among newts of the genus Triturus (family Salamandridae), using comparative biochemistry, cytogenetics, and reproductive interactions. J. Evol. Biol. 1990, 3, 329–373. [Google Scholar] [CrossRef]
- Lampbrush chromosome protocols. Available online: http://projects.exeter.ac.uk/lampbrush/protocols.htm (accessed on 7 September 2017).
- Zlotina, A.; Krasikova, A. FISH in Lampbrush Chromosomes. In Fluorescence in situ Hybridization (FISH)—Application Guide, 2nd ed.; Liehr, T., Ed.; Springer: Berlin, Germany, 2017; pp. 445–457. ISBN 978-3662529577. [Google Scholar]
- Zlotina, A.; Galkina, S.; Krasikova, A.; Crooijmans, R.P.; Groenen, M.A.; Gaginskaya, E.; Deryusheva, S. Precise centromere positioning on chicken chromosome 3. Cytogenet. Genome Res. 2010, 129, 310–313. [Google Scholar] [CrossRef] [PubMed]
- Guraya, S.S. The cell and molecular biology of fish oogenesis. Monogr. Dev. Biol. 1986, 18, 1–223. [Google Scholar] [PubMed]
- Callebaut, M.; Van Nassauw, L.; Harrisson, F. Comparison between oogenesis and related ovarian structures in a reptile, Pseudemys scripta elegans (turtle) and in a bird Coturnix coturnix japonica (quail). Reprod. Nutr. Dev. EDP Sci. 1997, 37, 233–252. [Google Scholar] [CrossRef]
- Moore, B.C.; Uribe-Aranabal, M.C.; Boggs, A.S.P.; Guillette, L.J.J. Developmental morphology of the neonatal alligator (Alligator mississippiensis) ovary. J. Morphol. 2008, 269, 302–312. [Google Scholar] [CrossRef] [PubMed]
- Lutes, A.A.; Neaves, W.B.; Baumann, D.P.; Wiegraebe, W.; Baumann, P. Sister chromosome pairing maintains heterozygosity in parthenogenetic lizards. Nature 2010, 464, 283–286. [Google Scholar] [CrossRef] [PubMed]
- Lukina, N.A. Characterization of meiotic chromosomes in the oocytes of some Lacertidae (Reptilia). Tsitologiia 1994, 36, 323–329. [Google Scholar]
- Kupriyanova, L. Cytogenetic and genetic trends in the evolution of unisexual lizards. Cytogenet. Genome Res. 2009, 127, 273–279. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Miyake, T.; Edwards, S.V.; Amemiya, C.T. Tuatara (Sphenodon) genomics: BAC library construction, sequence survey, and application to the DMRT gene family. J. Hered. 2006, 97, 541–548. [Google Scholar] [CrossRef] [PubMed]
- Olmo, E. Trends in the evolution of reptilian chromosomes. Integr. Comp. Biol. 2008, 48, 486–493. [Google Scholar] [CrossRef] [PubMed]
- Chapus, C.; Edwards, S.V. Genome evolution in Reptilia: In silico chicken mapping of 12,000 BAC-end sequences from two reptiles and a basal bird. BMC Genom. 2009, 10, S8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pokorna, M.; Giovannotti, M.; Kratochvil, L.; Caputo, V.; Olmo, E.; Ferguson-Smith, M.A.; Rens, W. Conservation of chromosomes syntenic with avian autosomes in squamate reptiles revealed by comparative chromosome painting. Chromosoma 2012, 121, 409–418. [Google Scholar] [CrossRef] [PubMed]
- Muller, W.P. The lampbrush chromosomes of Xenopus laevis. Chromosoma 1974, 47, 283–296. [Google Scholar] [CrossRef] [PubMed]
- Bucci, S.; Ragghianti, M.; Mancino, G.; Berger, L.; Hotz, H.; Uzzell, T. Lampbrush and mitotic chromosomes of the hemiclonally reproducing hybrid Rana esculenta and its parental species. J. Exp. Zool. Suppl. 1990, 255, 37–56. [Google Scholar] [CrossRef] [PubMed]
- Guerrini, F.; Bucci, S.; Ragghianti, M.; Mancino, G.; Hotz, H.; Uzzell, T.; Berger, L. Genomes of two water frog species resist germ line exclusion in interspecies hybrids. J. Exp. Zool. 1997, 279, 163–176. [Google Scholar] [CrossRef]
- Vishnyakova, N.M.; Lacroix, J.-C.; Rodionov, A.V. Cytogenetic maps of lampbrush chromosomes of newts of the genus Pleurodeles: An algorithm of lampbrush chromosome identification in Pleurodeles waltl by immunocytochemical staining of landmark loops with polyclonal anti-Ro52 antisera. Russ. J. Genet. 2004, 40, 491–499. [Google Scholar]
- Smith, J.J.; Voss, S.R. Gene order data from a model amphibian (Ambystoma): New perspectives on vertebrate genome structure and evolution. BMC Genom. 2006, 7, 219. [Google Scholar] [CrossRef]
- Voss, S.R.; Kump, D.K.; Putta, S.; Pauly, N.; Reynolds, A.; Henry, R.J.; Basa, S.; Walker, J.A.; Smith, J.J. Origin of amphibian and avian chromosomes by fission, fusion, and retention of ancestral chromosomes. Genome Res. 2011, 21, 1306–1312. [Google Scholar] [CrossRef] [PubMed]
- Miura, I.; Ohtani, H.; Kashiwagi, A.; Hanada, H.; Nakamura, M. Structural differences between XX and ZW sex lampbrush chromosomes in Rana rugosa females (Anura: Ranidae). Chromosoma 1996, 105, 237–241. [Google Scholar] [CrossRef] [PubMed]
- Miura, I.; Ohtani, H.; Hanada, H.; Ichikawa, Y.; Kashiwagi, A.; Nakamura, M. Evidence for two successive pericentric inversions in sex lampbrush chromosomes of Rana rugosa (Anura: Ranidae). Chromosoma 1997, 106, 178–182. [Google Scholar] [CrossRef] [PubMed]
- Mancino, G. Evolutionary cytogenetics of newts of the genus Triturus as revealed using monoclonal antibodies. In Amphibian Cytogenetics and Evolution; Green, D.M., Sessions, S.K., Eds.; Academic Press: New York, NY, USA, 1991; pp. 197–215. [Google Scholar]
- Ohtani, H. Lampbrush chromosomes of Rana nigromaculata, R. brevipoda, R. plancyi chosenica, R. p. fukienensis and their reciprocal hybrids. Sci. Rep. Lab. Amphib. Biol. Hiroshima Univ. 1990, 10, 165–221. [Google Scholar]
- Wielstra, B.; Baird, A.B.; Arntzen, J.W. A multimarker phylogeography of crested newts (Triturus cristatus superspecies) reveals cryptic species. Mol. Phylogenet. Evol. 2013, 67, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Ohtani, H. Polymorphism of lampbrush chromosomes in Japanese populations of Rana nigromaculata. Zool. Sci. 1994, 11, 337–342. [Google Scholar]
- Ohtani, H. Local variations in the lampbrush chromosomes of the Japanese pond frog, Rana porosa. Caryologia 1995, 48, 189–199. [Google Scholar] [CrossRef]
- Coyne, J.A.; Orr, H.A. The evolutionary genetics of speciation. Philos. Trans. R. Soc. Lond. Ser. B 1998, 353, 287–305. [Google Scholar] [CrossRef] [PubMed]
- Miura, I. An evolutionary witness: The frog Rana rugosa underwent change of heterogametic sex from XY male to ZW female. Sex. Dev. 2007, 1, 323–331. [Google Scholar] [CrossRef] [PubMed]
- Ohta, S. Sex-determining mechanism in Buergeria buergeri (Schlegel): I. Heterozygosity of chromosome pair No. 7 in the female. Sci. Rep. Lab. Amphib. Biol. Hiroshima Univ. 1986, 8, 29–43. [Google Scholar]
- Callan, H.G.; Lloyd, L. Lampbrush chromosomes of crested newts Triturus cristatus (Laurenti). Philos. Trans. R. Soc. B 1960, 243, 135–219. [Google Scholar] [CrossRef]
- Sims, S.H.; Macgregor, H.C.; Pellatt, P.S.; Horner, H.A. Chromosome 1 in crested and marbled newts (Triturus). An extraordinary case of heteromorphism and independent chromosome evolution. Chromosoma 1984, 89, 169–185. [Google Scholar] [CrossRef]
- Sessions, S.K.; Macgregor, H.C.; Schmid, M.; Haaf, T. Cytology, embryology, and evolution of the development arrest syndrome in newts of the genus Triturus (Caudata: Salamandridae). J. Exp. Zool. 1988, 248, 321–334. [Google Scholar] [CrossRef] [PubMed]
- Hillis, D.M.; Green, D.M. Evolutionary changes of heterogametic sex in the phylogenetic history of amphibians. J. Evol. Biol. 1990, 3, 49–64. [Google Scholar] [CrossRef]
- Green, D.M.; Kezer, J.; Nussbaum, R.A. Supernumerary chromosome variation and heterochromatin distribution in the endemic New Zealand frog Leopelma hochstetteri. Chromosoma 1987, 95, 339–344. [Google Scholar] [CrossRef] [PubMed]
- Green, D.M. Cytogenetics of the endemic New Zealand frog, Leopelma hochstetteri: Extraordinary supernumerary chromosome variation and a unique sex chromosome system. Chromosoma 1988, 97, 55–70. [Google Scholar] [CrossRef]
- Green, D.M. Heteromorphic sex chromosomes in the rare and primitive frog Leopelma hamiltonii from New Zealand. J. Heredity 1988, 79, 165–169. [Google Scholar] [CrossRef]
- Zeul, C. Genome Evolution in the Primitive Frog Leopelma hochstetteri. Ph.D. Thesis, McGill University, Montreal, QC, Canada, 1991. [Google Scholar]
- Green, D.M.; Zeyl, C.W.; Sharbel, T.F. The evolution of hypervariable sex and supernumerary (B) chromosomes in the relict New Zealand frog, Leiopelma hochstetteri. J. Evol. Biol. 1993, 6, 417–441. [Google Scholar] [CrossRef]
- Bogart, J.P. The influence of life history on karyotypic evolution in frogs. In Amphibian Cytogencttcs and Evolution; Green, D.M., Sessions, S.K., Eds.; Academic Press: San Diego, CA, USA, 1991; pp. 233–258. [Google Scholar]
- Dawley, R.M.; Bogart, J.P. Evolution and Ecology of Unisexual Vertebrates; New York State Museum Bulletin 466; New York State Museum: Albany, NY, USA, 1989; p. 302.
- Muller, W.P. Diplotene chromosomes of Xenopus hybrid oocytes. Chromosoma 1977, 59, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Neaves, W.B.; Baumann, P. Unisexual reproduction among vertebrates. Trends Genet. 2011, 27, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Stenberg, P.; Saura, A. Meiosis and its deviations in polyploid animals. Cytogenet. Genome Res. 2013, 140, 185–203. [Google Scholar] [CrossRef] [PubMed]
- Macgregor, H.C.; Uzzell, T.M. Gynogenesis in salamanders related to Ambystoma jeffersonianum. Science 1964, 143, 1043–1045. [Google Scholar] [CrossRef] [PubMed]
- Dedukh, D.; Litvinchuk, S.; Rosanov, J.; Mazepa, G.; Saifitdinova, A.; Shabanov, D.; Krasikova, A. Optional endoreplication and selective elimination of parental genomes during oogenesis in diploid and triploid hybrid European water frogs. PLoS ONE 2015, 10, e0123304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stöck, M.; Lamatsch, K.; Steinlein, C.; Epplen, J.T.; Grosse, W.-R.; Klapperstück, T.; Lampert, K.P.; Scheer, U.; Schmid, M.; Schartl, M. A bisexually reproducing all-triploid vertebrate. Nat. Genet. 2002, 30, 325–328. [Google Scholar] [CrossRef] [PubMed]
- Bi, K.; Bogart, J.P.; Fu, J. Intergenomic translocations in unisexual salamanders of the genus Ambystoma (Amphibia, Caudata). Cytogenet. Genome Res. 2007, 116, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Bi, K.; Bogart, J.P. Probing the meiotic mechanism of intergenomic exchanges by genomic in situ hybridization on lampbrush chromosomes of unisexual Ambystoma (Amphibia: Caudata). Chromosom. Res. 2010, 18, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Ohno, S. Evolution by Gene Duplication; Springer: New York, NY, USA, 1970; p. 160. [Google Scholar]
- Edwards, N.S.; Murray, A.W. Identification of Xenopus CENP-A and an associated centromeric DNA repeat. Mol. Biol. Cell 2005, 16, 1800–1810. [Google Scholar] [CrossRef] [PubMed]
- Guttenbach, M.; Nanda, I.; Feichtinger, W.; Masabanda, J.S.; Griffin, D.K.; Schmid, M. Comparative chromosome painting of chicken autosomal paints 1-9 in nine different bird Species. Cytogenet. Genome Res. 2003, 103, 173–184. [Google Scholar] [CrossRef] [PubMed]
- Shibusawa, M.; Nishibori, M.; Nishida-Umehara, C.; Tsudzuki, M.; Masabanda, J.; Griffin, D.K.; Matsuda, Y. Karyotypic evolution in the Galliformes: An examination of the process of karyotypic evolution by comparison of the molecular cytogenetic findings with the molecular phylogeny. Cytogenet. Genome Res. 2004, 106, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Shibusawa, M.; Nishida-Umehara, C.; Tsudzuki, M.; Masabanda, J.; Griffin, D.K.; Matsuda, Y. A comparative karyological study of the blue-breasted quail (Coturnix chinensis, Phasianidae) and California quail (Callipepla californica, Odontophoridae). Cytogenet. Genome Res. 2004, 106, 82–90. [Google Scholar] [CrossRef] [PubMed]
- Griffin, D.K.; Robertson, L.B.; Tempest, H.G.; Vignal, A.; Fillon, V.; Crooijmans, R.P.; Groenen, M.A.; Deryusheva, S.; Gaginskaya, E.; Carré, W.; et al. Whole genome comparative studies between chicken and turkey and their implications for avian genome evolution. BMC Genom. 2008, 9, 168. [Google Scholar] [CrossRef] [PubMed]
- Shibusawa, M.; Minai, S.; Nishida-Umehara, C.; Suzuki, T.; Mano, T.; Yamada, K.; Namikawa, T.; Matsuda, Y. A comparative cytogenetic study of chromosome homology between chicken and Japanese quail. Cytogenet. Cell Genet. 2001, 95, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Kayang, B.B.; Fillon, V.; Inoue-Murayama, M.; Miwa, M.; Leroux, S.; Feve, K.; Monvoisin, J.; Pitel, F.; Vignoles, M.; Mouilhayrat, C.; et al. Integrated maps in quail (Coturnix japonica) confirm the high degree of synteny conservation with chicken (Gallus gallus) despite 35 million years of divergence. BMC Genom. 2006, 7, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Sasazaki, S.; Hinenoya, T.; Lin, B.; Fujiwara, A.; Mannen, H. A comparative map of macrochromosomes between chicken and Japanese quail based on orthologous genes. Anim. Genet. 2006, 37, 316–320. [Google Scholar] [CrossRef] [PubMed]
- Ryttman, H.; Tegelström, H. G-banded karyotypes of three Galliformes species, domestic fowl (Gallus domesticus), Quail (Coturnix coturnix japonica), and turkey (Meleagris gallopavo). Hereditas 1981, 94, 165–170. [Google Scholar] [CrossRef]
- Sasaki, M. High resolution G-band karyotypes of the domestic fowl and the Japanese quail. Chromosom. Inf. Sevice 1981, 31, 26–28. [Google Scholar]
- Stock, A.D.; Bunch, T.D. The evolutionary implications of chromosome banding pattern homologies in the bird order Galliformes. Cytogenet. Cell Genet. 1982, 34, 136–148. [Google Scholar] [CrossRef] [PubMed]
- Montefalcone, G.; Tempesta, S.; Rocchi, M.; Archidiacono, N. Centromere repositioning. Genome Res. 1999, 9, 184–1188. [Google Scholar] [CrossRef]
- Nagaki, K.; Cheng, Z.; Ouyang, S.; Talbert, P.B.; Kim, M.; Jones, K.M.; Henikoff, S.; Buell, C.R.; Jiang, J. Sequencing of a rice centromere uncovers active genes. Nat. Genet. 2004, 36, 138–145. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, R.J.; Eldridge, M.D.; Metcalfe, C.J. Centromere dynamics and chromosome evolution in marsupials. J. Hered. 2004, 95, 375–381. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Zhang, Z.; Liu, C.; Liu, J.; Huang, S.; Jiang, J.; Jina, W. Centromere repositioning in cucurbit species: Implication of the genomic impact from centromere activation and inactivation. Proc. Natl. Acad. Sci. USA 2009, 106, 14937–14941. [Google Scholar] [CrossRef] [PubMed]
- Piras, F.M.; Nergadze, S.G.; Magnani, E.; Bertoni, L.; Attolini, C.; Khoriauli, L.; Raimondi, E.; Giulotto, E. Uncoupling of satellite DNA and centromeric function in the genus Equus. PLoS Genet. 2010, 6, e1000845. [Google Scholar] [CrossRef] [PubMed]
- Rocchi, M.; Archidiacono, N.; Schempp, W.; Capozzi, O.; Stanyon, R. Centromere repositioning in mammals. Heredity 2012, 108, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Trifonov, V.A.; Musilova, P.; Kulemsina, A.I. Chromosome Evolution in Perissodactyla. Cytogenet. Genome Res. 2012, 137, 208–217. [Google Scholar] [CrossRef] [PubMed]
- Kasai, F.; Garcia, C.; Arruga, M.V.; Ferguson-Smith, M.A. Chromosome homology between chicken (Gallus gallus domesticus) and the red-legged partridge (Alectoris rufa); evidence of the occurrence of a neocentromere during evolution. Cytogenet. Genome Res. 2003, 102, 326–330. [Google Scholar] [CrossRef] [PubMed]
- Skinner, B.M.; Robertson, L.B.W.; Tempest, H.G.; Langley, E.J.; Ioannou, D.; Fowler, K.E.; Crooijmans, R.P.M.A.; Hall, A.D.; Griffin, D.K.; Völker, M. Comparative genomics in chicken and Pekin duck using FISH mapping and microarray analysis. BMC Genom. 2009, 10, 357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krasikova, A.; Deryusheva, S.; Galkina, S.; Kurganova, A.; Evteev, A.; Gaginskaya, E. On the positions of centromeres in chicken lampbrush chromosomes. Chromosom. Res. 2006, 14, 777–789. [Google Scholar] [CrossRef] [PubMed]
- Calderon, P.L.; Pigozzi, M.I. MLH1-focus mapping in birds shows equal recombination between sexes and diversity of crossover patterns. Chromosom. Res. 2006, 14, 605–612. [Google Scholar] [CrossRef] [PubMed]
- Solari, A.J. Synaptosomal complexes and associated structures in microspread human spermatocytes. Chromosoma 1980, 81, 315–337. [Google Scholar] [CrossRef] [PubMed]
- Kaelbling, M.; Fechheimer, N.S. Synaptonemal complexes and the chromosomes of the domestic fowl, Gallus domesticus. Cytogenet. Cell Genet. 1983, 35, 87–92. [Google Scholar] [CrossRef] [PubMed]
- Krasikova, A.; Daks, A.; Zlotina, A.; Gaginskaya, E. Polymorphic heterochromatic segments in Japanese quail microchromosomes. Cytogenet. Genome Res. 2009, 126, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Völker, M.; Backstrom, N.; Skinner, B.M.; Langley, E.J.; Bunzey, S.K.; Ellegren, H.; Griffin, D.K. Copy number variation, chromosome rearrangement, and their association with recombination during avian evolution. Genome Res. 2010, 20, 503–511. [Google Scholar] [CrossRef] [PubMed]
- Skinner, B.M.; Griffin, D.K. Intrachromosomal rearrangements in avian genome evolution: Evidence for regions prone to breakpoints. Heredity 2012, 108, 37–41. [Google Scholar] [CrossRef] [PubMed]
- Lithgow, P.E.; O’Connor, R.; Smith, D.; Fonseka, G.; Mutery, A.A.; Rathje, C.; Frodsham, R.; O’Brien, P.; Kasai, F.; Ferguson-Smith, M.A.; et al. Novel tools for characterising inter and intra chromosomal rearrangements in avian microchromosomes. Chromosom. Res. 2014, 22, 85–97. [Google Scholar] [CrossRef] [PubMed]
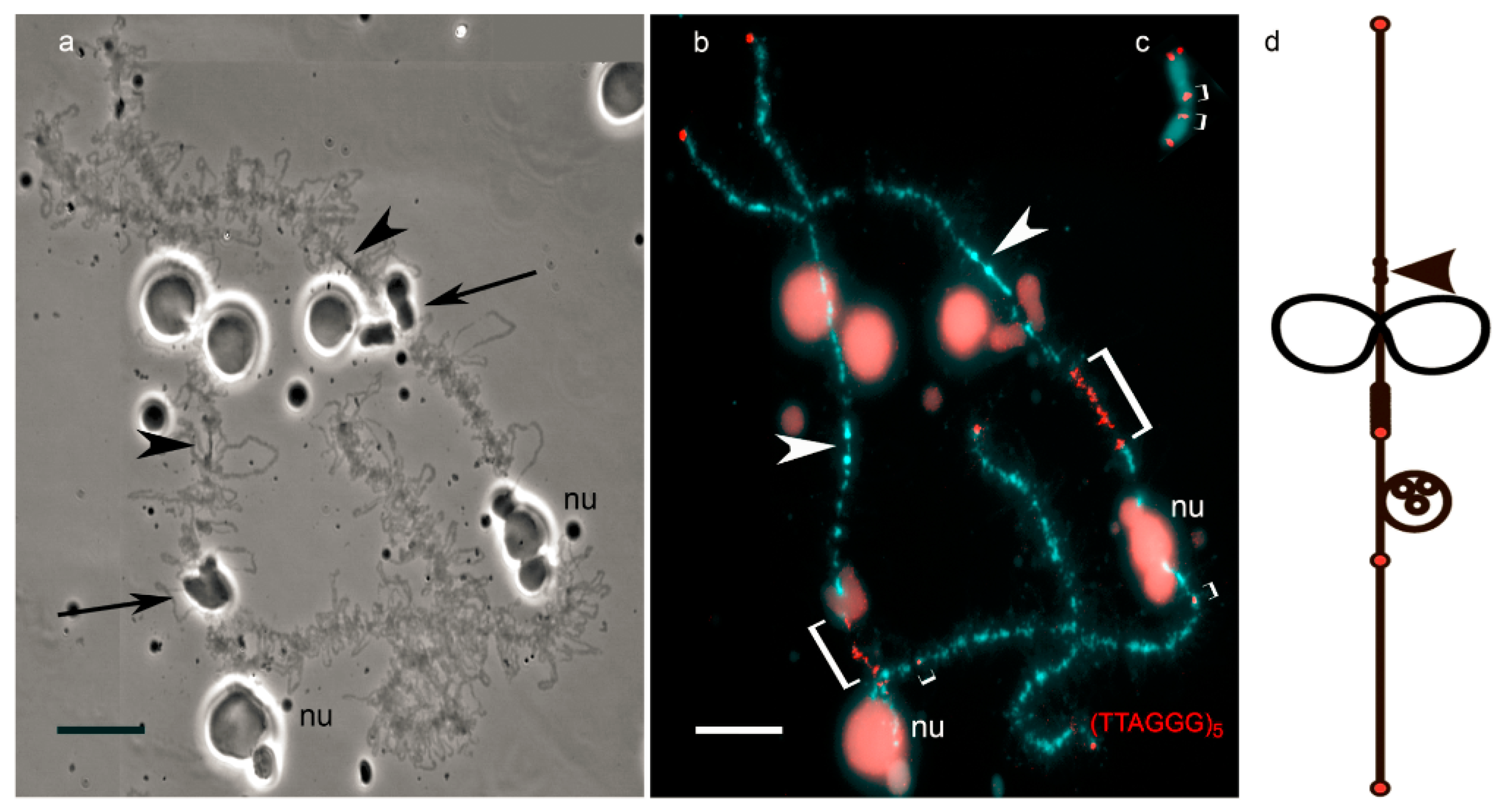
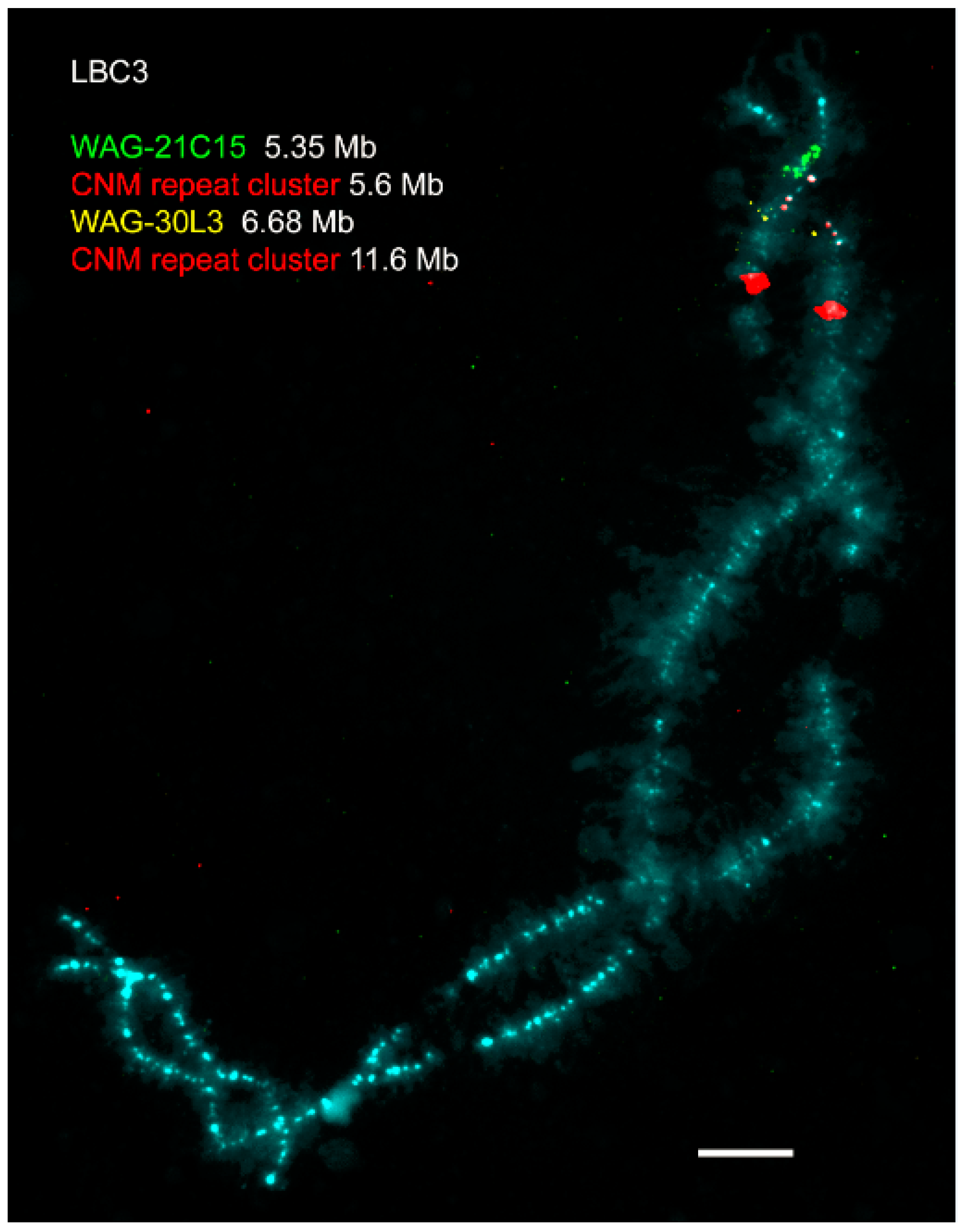
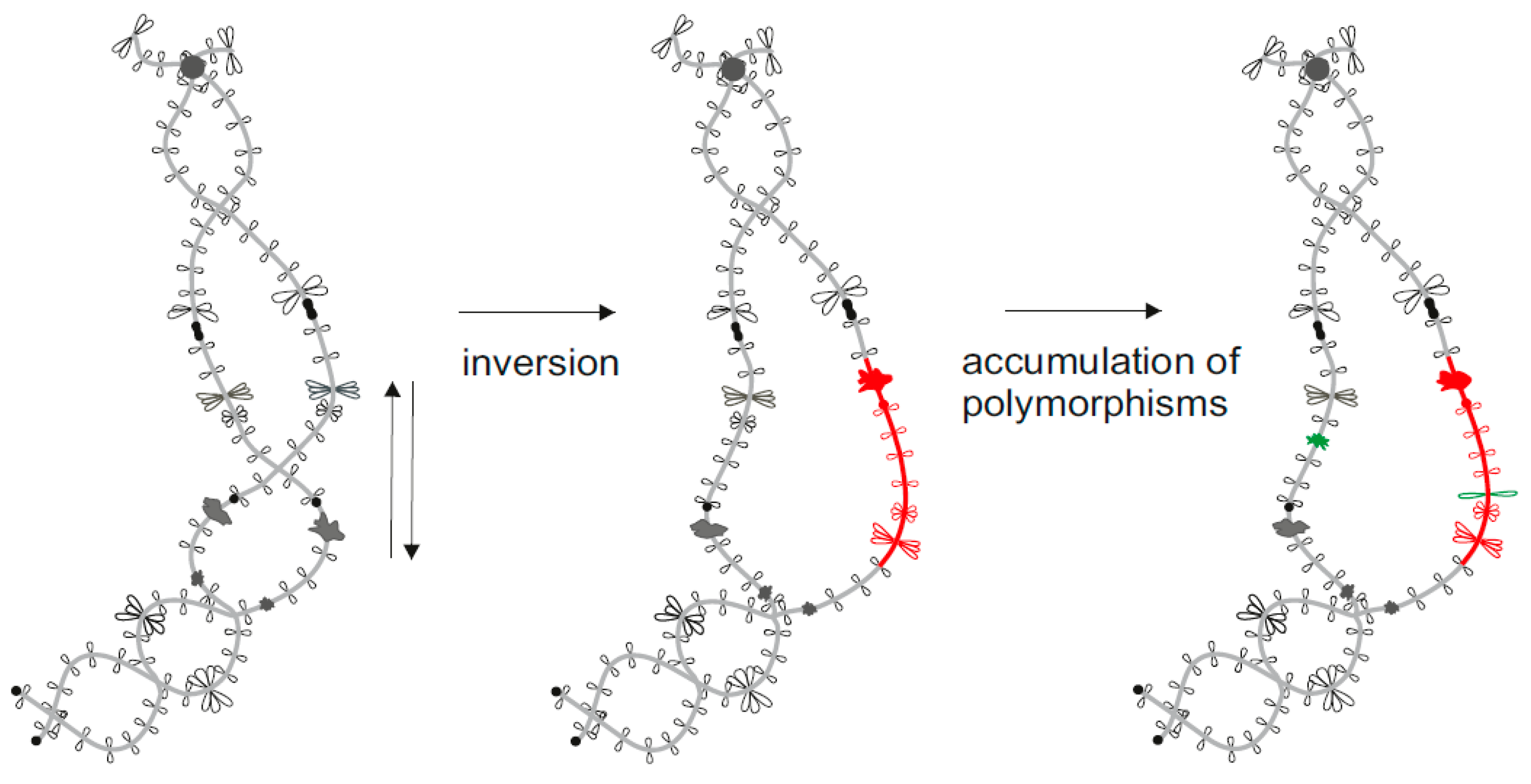
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zlotina, A.; Dedukh, D.; Krasikova, A. Amphibian and Avian Karyotype Evolution: Insights from Lampbrush Chromosome Studies. Genes 2017, 8, 311. https://doi.org/10.3390/genes8110311
Zlotina A, Dedukh D, Krasikova A. Amphibian and Avian Karyotype Evolution: Insights from Lampbrush Chromosome Studies. Genes. 2017; 8(11):311. https://doi.org/10.3390/genes8110311
Chicago/Turabian StyleZlotina, Anna, Dmitry Dedukh, and Alla Krasikova. 2017. "Amphibian and Avian Karyotype Evolution: Insights from Lampbrush Chromosome Studies" Genes 8, no. 11: 311. https://doi.org/10.3390/genes8110311



