Connections between Transcription Downstream of Genes and cis-SAGe Chimeric RNA
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture and Osmotic Stress Onduction
2.2. RNA Extraction
2.3. Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
2.4. Statistics
2.5. Bioinformatics
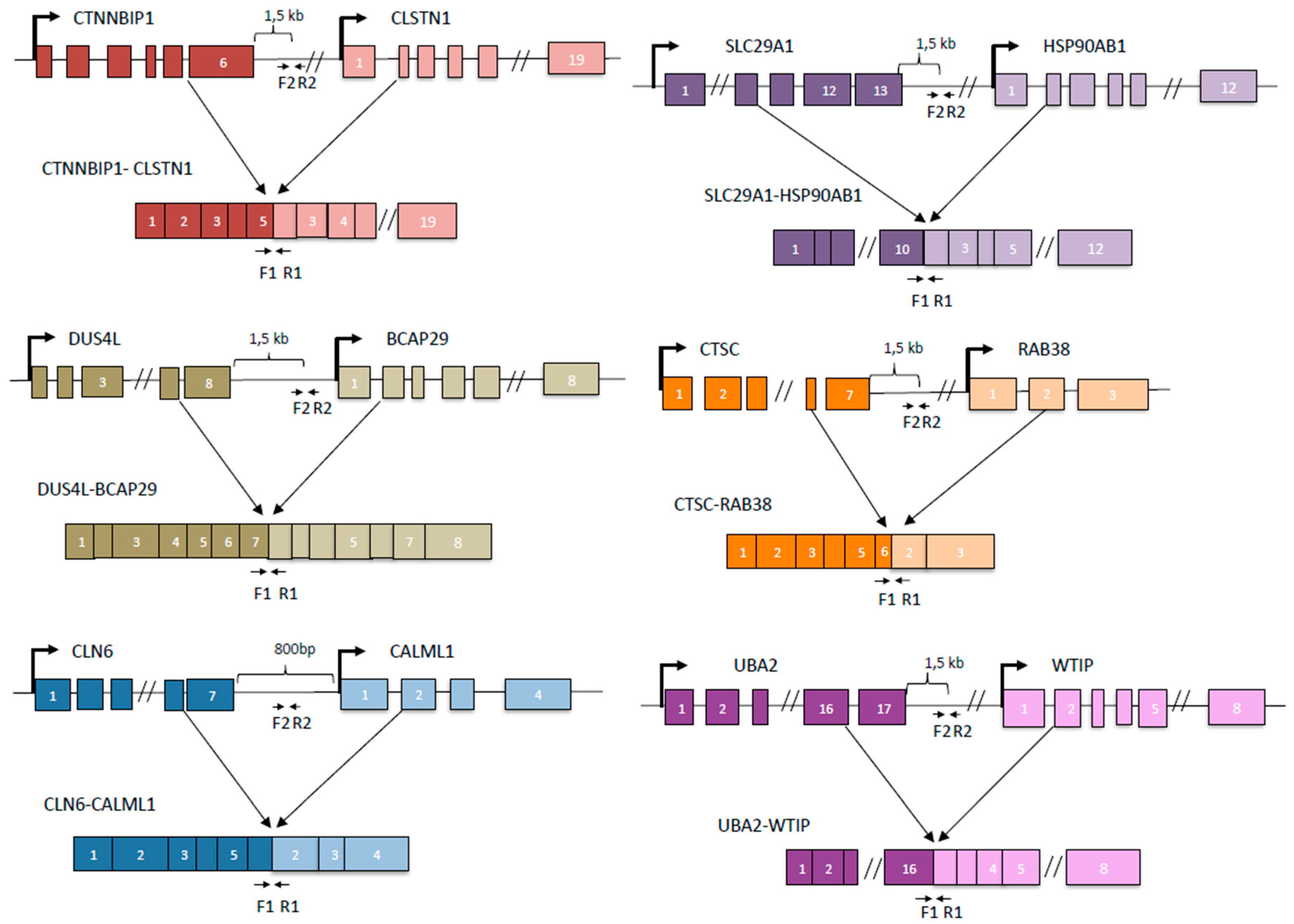
3. Results
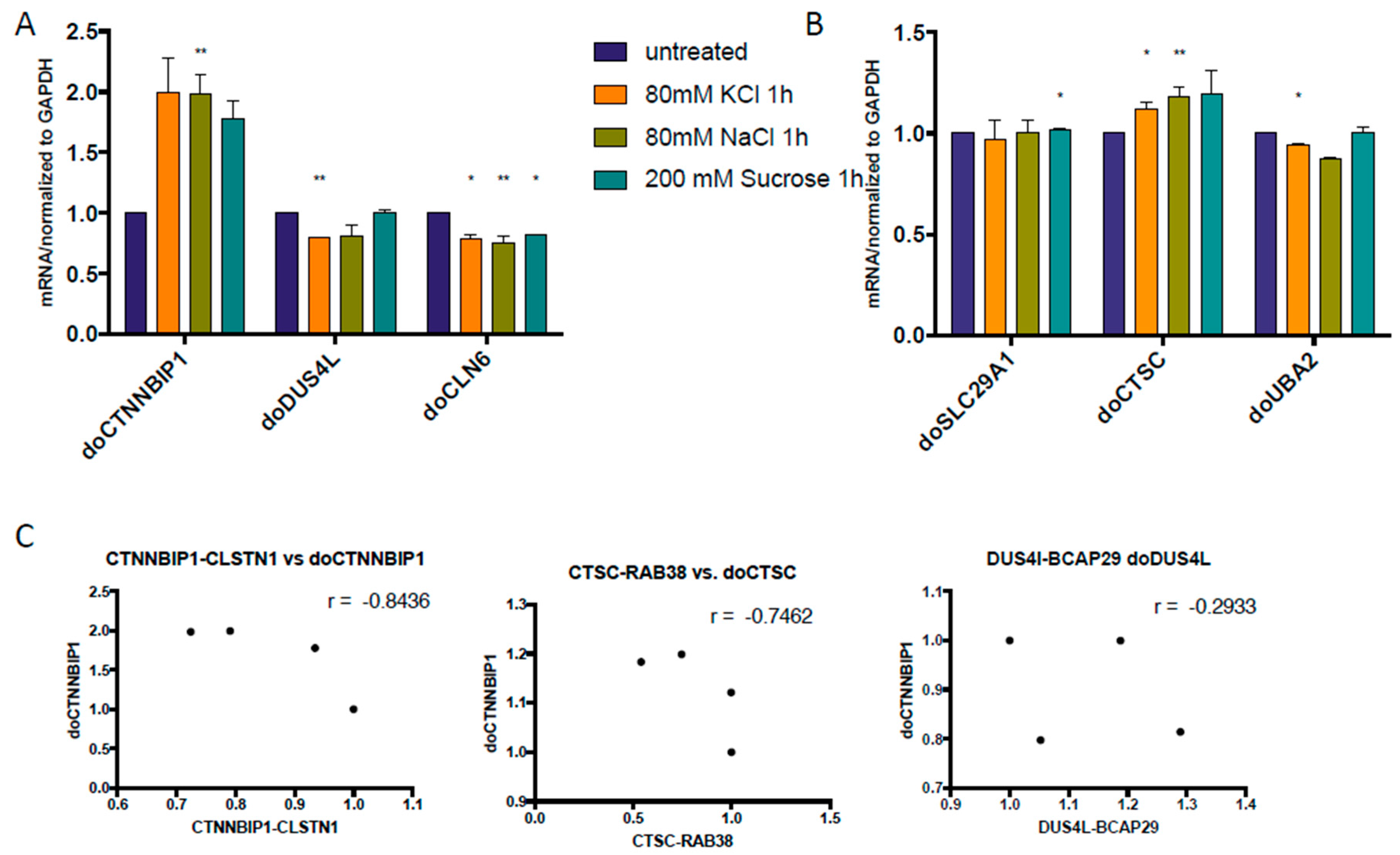
3.1. cis-SAGe Chimeric RNAs and Corresponding DoGs Are not All Induced after 1 h Treatment
3.2. Some DoGs and Their Corresponding Chimeric RNAs Correlate Negatively
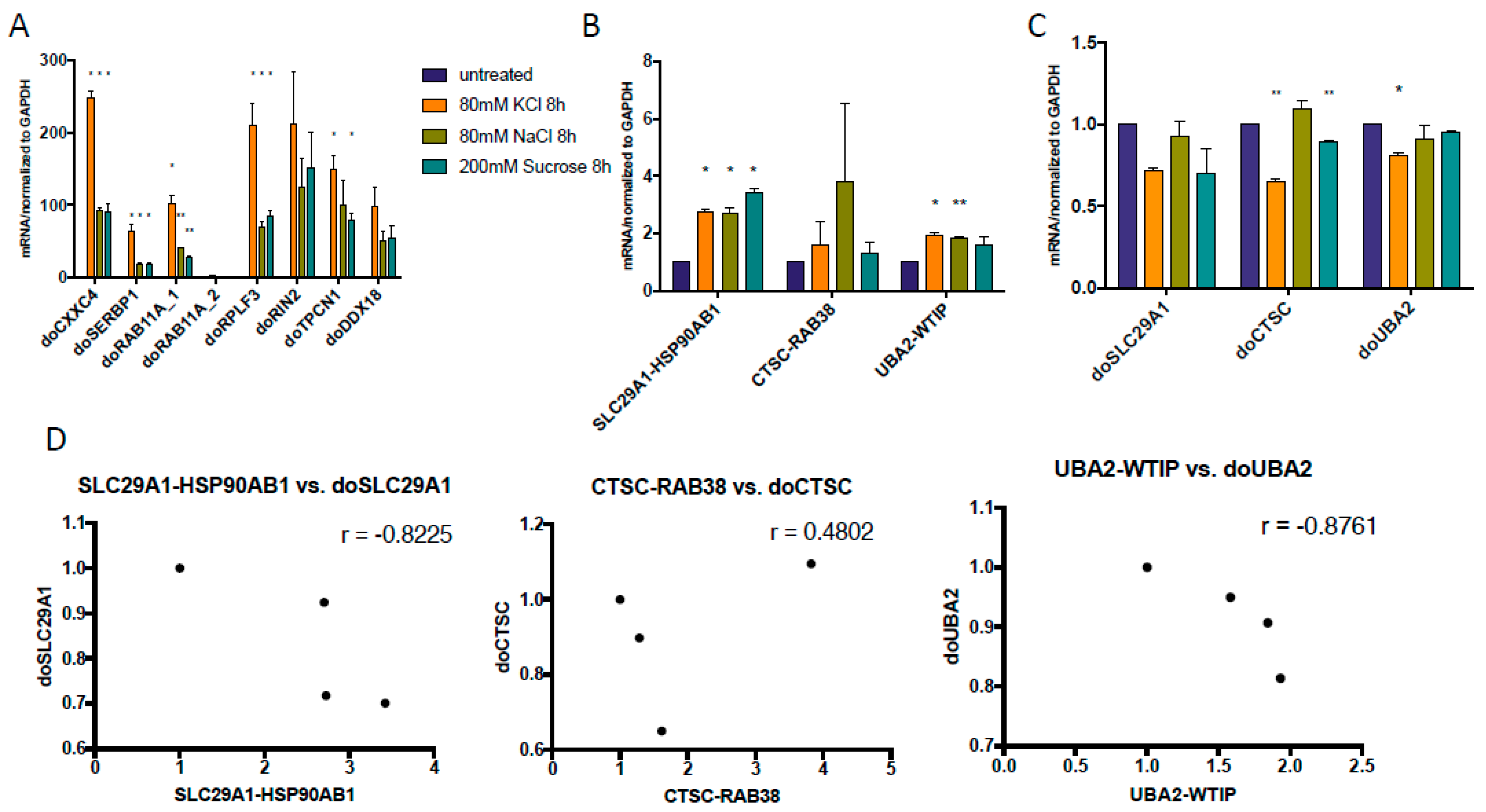
3.3. Both DoGs and Corresponding cis-SAGe Fusions Are Induced in Response to Prolonged Treatment
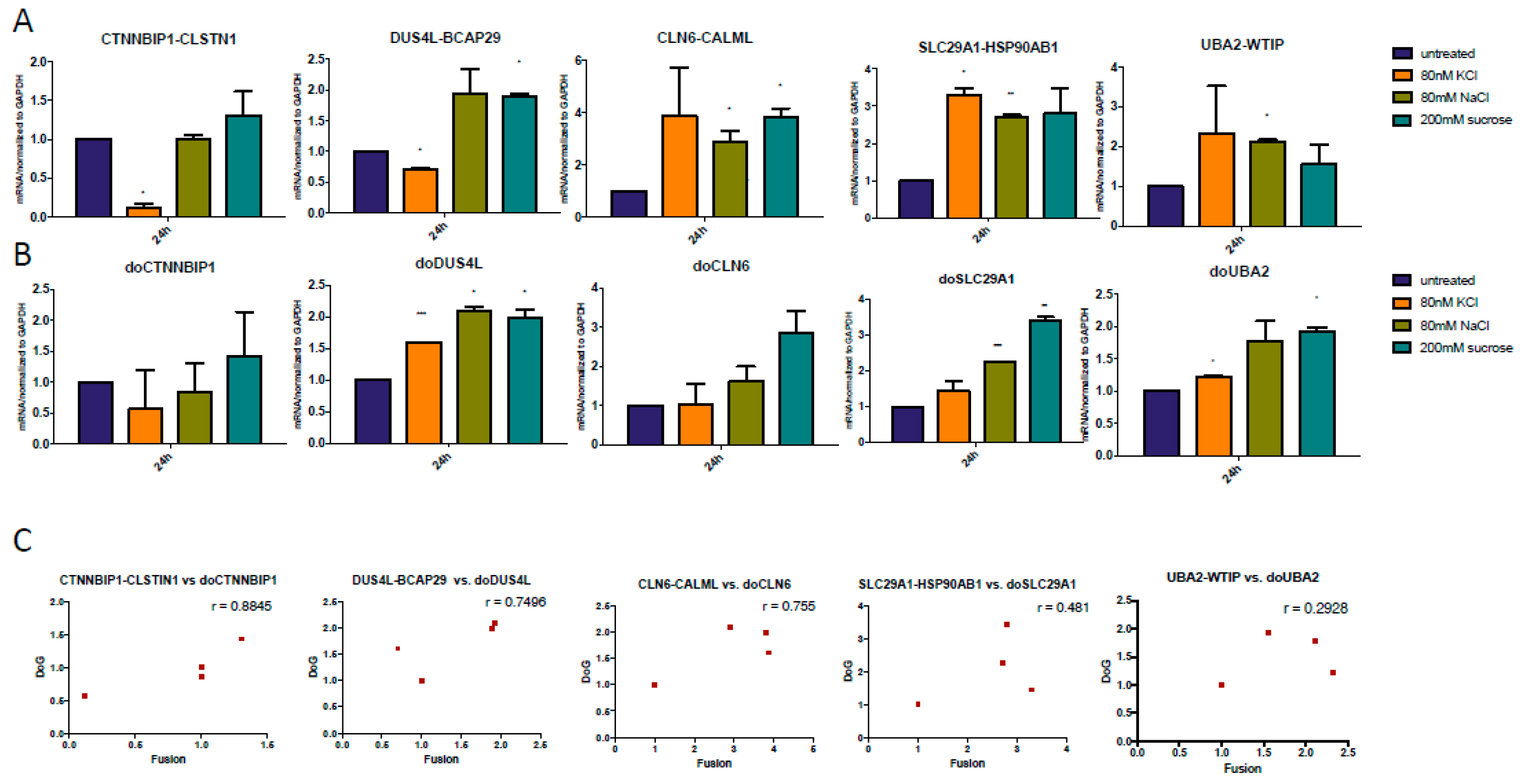
3.4. The Upregulation of DoGs and cis-SAGe Fusions Persists after Osmotic Stress
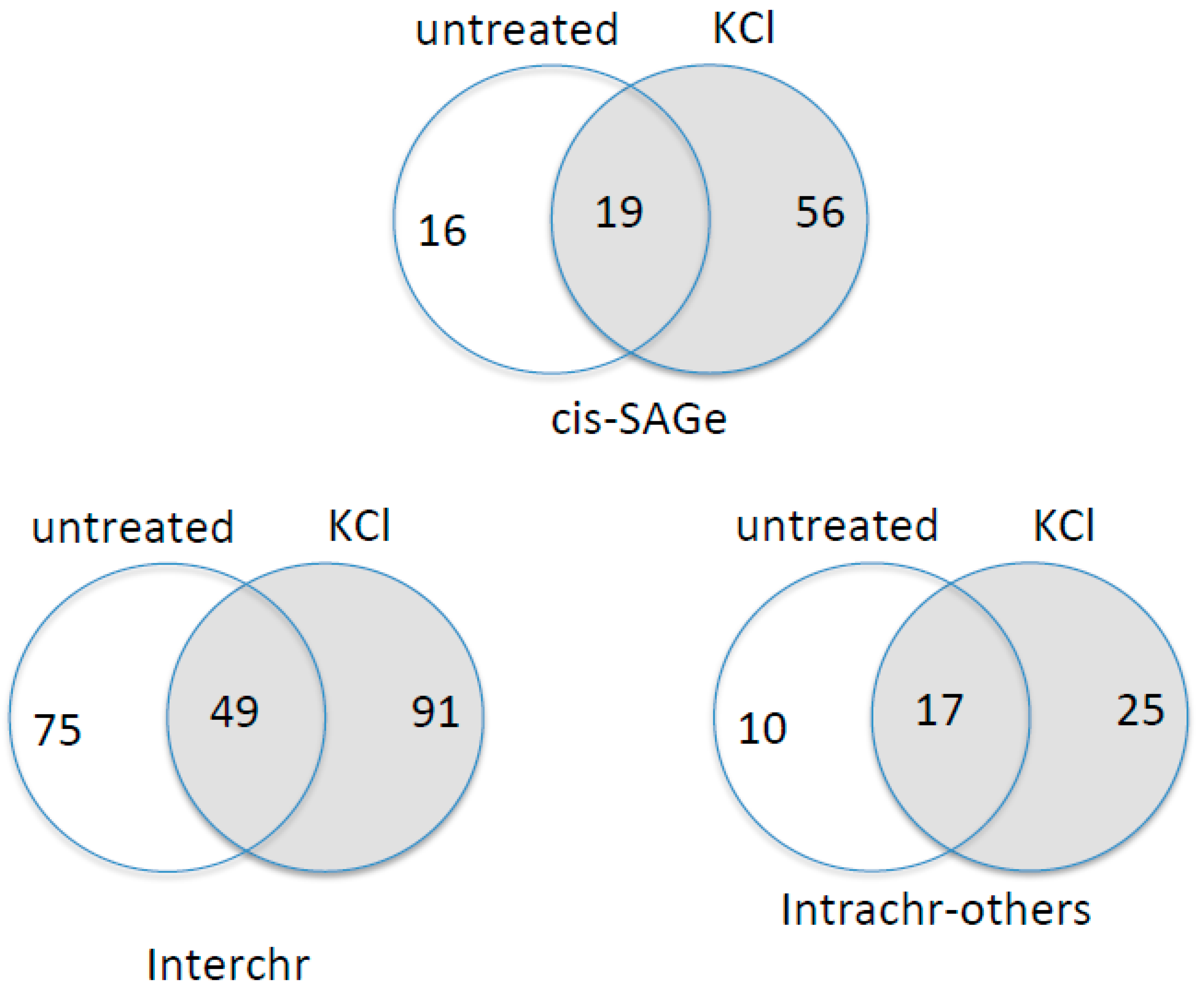
3.5. KCl-Induced Osmostress Increases the Global Occurrence of cis-SAGe Chimeric RNAs
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
References
- Parra, G.; Reymond, A.; Dabbouseh, N.; Dermitzakis, E.T.; Castelo, R.; Thomson, T.M.; Antonarakis, S.E.; Guigó, R. Tandem chimerism as a means to increase protein complexity in the human genome. Genome Res. 2006, 16, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Akiva, P.; Toporik, A.; Edelheit, S.; Peretz, Y.; Diber, A.; Shemesh, R.; Novik, A.; Sorek, R. Transcription-mediated gene fusion in the human genome. Genome Res. 2006, 16, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Kannan, K.; Wang, L.; Wang, J.; Ittmann, M.M.; Li, W.; Yen, L. Recurrent chimeric RNAs enriched in human prostate cancer identified by deep sequencing. Proc. Natl. Acad. Sci. USA 2011, 108, 9172–9177. [Google Scholar] [CrossRef] [PubMed]
- Nacu, S.; Yuan, W.; Kan, Z.; Bhatt, D.; Rivers, C.S.; Stinson, J.; Peters, B.A.; Modrusan, Z.; Jung, K.; Seshagiri, S.; et al. Deep RNA sequencing analysis of readthrough gene fusions in human prostate adenocarcinoma and reference samples. BMC Med. Genomics 2011, 4, 11. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.; Wu, J.; Zhao, G.; Wang, Z.; Chen, W.; Mu, S. Abundant and broad expression of transcription-induced chimeras and protein products in mammalian genomes. Biochem. Biophys. Res. Commun. 2016, 470, 759–765. [Google Scholar] [CrossRef] [PubMed]
- Greger, L.; Su, J.; Rung, J.; Ferreira, P.G.; Lappalainen, T.; Dermitzakis, E.T.; Brazma, A. Tandem RNA chimeras contribute to transcriptome diversity in human population and are associated with intronic genetic variants. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Prakash, T.; Sharma, V.K.; Adati, N.; Ozawa, R.; Kumar, N.; Nishida, Y.; Fujikake, T.; Takeda, T.; Taylor, T.D. Expression of conjoined genes: Another mechanism for gene regulation in eukaryotes. PLoS ONE 2010, 5. [Google Scholar] [CrossRef] [PubMed]
- Kim, R.N.; Kim, A.; Choi, S.H.; Kim, D.S.; Nam, S.H.; Kim, D.W.; Kim, D.W.; Kang, A.; Kim, M.Y.; Park, K.H.; et al. Novel mechanism of conjoined gene formation in the human genome. Funct. Integr. Genomics 2012, 12, 45–61. [Google Scholar] [CrossRef] [PubMed]
- Varley, K.E.; Gertz, J.; Roberts, B.S.; Davis, N.S.; Bowling, K.M.; Kirby, M.K.; Nesmith, A.S.; Oliver, P.G.; Grizzle, W.E.; Forero, A.; et al. Recurrent read-through fusion transcripts in breast cancer. Breast Cancer Res. Treat. 2014, 146, 287–297. [Google Scholar] [CrossRef] [PubMed]
- Qin, F.; Song, Z.; Chang, M.; Song, Y.; Frierson, H.; Li, H. Recurrent cis-SAGe chimeric RNA, D2HGDH-GAL3ST2, in prostate cancer. Cancer Lett. 2016, 380, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Qin, F.; Song, Z.; Babiceanu, M.; Song, Y.; Facemire, L.; Singh, R.; Adli, M.; Li, H. Discovery of CTCF-Sensitive Cis-Spliced Fusion RNAs between Adjacent Genes in Human Prostate Cells. PLoS Genet. 2015, 11. [Google Scholar] [CrossRef]
- Qin, F.; Song, Y.; Zhang, Y.; Facemire, L.; Frierson, H.; Li, H. Role of CTCF in regulating SLC45A3-ELK4 chimeric RNA. PLoS ONE 2016, 11. [Google Scholar] [CrossRef] [PubMed]
- Chwalenia, K.; Facemire, L.; Li, H. Chimeric RNAs in cancer and normal physiology. Wiley Interdiscip. Rev. RNA 2017. [CrossRef] [PubMed]
- Kowalski, P.E.; Freeman, J.D.; Mager, D.L. Intergenic Splicing between a HERV-H Endogenous Retrovirus and Two Adjacent Human Genes. Genomics 1999, 57, 371–379. [Google Scholar] [CrossRef] [PubMed]
- Eul, J.; Graessmann, M.; Graessmann, A. Trans-splicing and alternative-tandem-cis-splicing: Two ways by which mammalian cells generate a truncated SV40 T-antigen. Nucleic Acids Res. 1996, 24, 1653–1661. [Google Scholar] [CrossRef] [PubMed]
- Batista, L.; Bourachot, B.; Mateescu, B.; Reyal, F.; Mechta-Grigoriou, F.; Hurteau, G.J.; Carlson, J.A.; Spivack, S.D.; Brock, G.J.; Iorio, M.V.; et al. Regulation of miR-200c/141 expression by intergenic DNA-looping and transcriptional read-through. Nat. Commun. 2016, 7, 8959. [Google Scholar] [CrossRef] [PubMed]
- Aksoy, S.; Squires, C.L.; Squires, C. Evidence for antitermination in Escherichia coli RRNA transcription. J. Bacteriol. 1984, 159, 260–264. [Google Scholar] [PubMed]
- Santangelo, T.J.; Artsimovitch, I. Termination and antitermination: RNA polymerase runs a stop sign. Nat. Rev. Microbiol. 2011, 9, 319–329. [Google Scholar] [CrossRef] [PubMed]
- Vilborg, A.; Passarelli, M.C.C.; Yario, T.A.A.; Tycowski, K.T.T.; Steitz, J.A.A. Widespread Inducible Transcription Downstream of Human Genes. Mol. Cell 2015, 59, 449–461. [Google Scholar] [CrossRef] [PubMed]
- Babiceanu, M.; Qin, F.; Xie, Z.; Jia, Y.; Lopez, K.; Janus, N.; Facemire, L.; Kumar, S.; Pang, Y.; Qi, Y.; et al. Recurrent chimeric fusion RNAs in non-cancer tissues and cells. Nucleic Acids Res. 2016, 44, 2859–2872. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Qin, F.; Liu, A.; Li, H. Recurrent fusion RNA DUS4L-BCAP29 in non-cancer human tissues and cells. Oncotarget 2017, 8, 31415–31423. [Google Scholar] [CrossRef] [PubMed]
- Grosso, A.R.; Leite, A.P.; Carvalho, S.; Matos, M.R.; Martins, F.B.; Vítor, A.C.; Desterro, J.M.; Carmo-Fonseca, M.; de Almeida, S.F. Pervasive transcription read-through promotes aberrant expression of oncogenes and RNA chimeras in renal carcinoma. eLife 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Martín, B.; Palumbo, E.; Marco-Sola, S.; Griebel, T.; Ribeca, P.; Alonso, G.; Rastrojo, A.; Aguado, B.; Guigó, R.; Djebali, S. ChimPipe: Accurate detection of fusion genes and transcription-induced chimeras from RNA-seq data. BMC Genomics 2017, 18, 7. [Google Scholar] [CrossRef] [PubMed]
- Sheets, M.D.; Ogg, S.C.; Wickens, M.P. Point mutations in AAUAAA and the poly (A) addition site: effects on the accuracy and efficiency of cleavage and polyadenylation in vitro. Nucleic Acids Res. 1990, 18, 5799–5805. [Google Scholar] [CrossRef] [PubMed]
- Benelli, M.; Pescucci, C.; Marseglia, G.; Severgnini, M.; Torricelli, F.; Magi, A. Discovering chimeric transcripts in paired-end RNA-seq data by using EricScript. Bioinformatics 2012, 28, 3232–3239. [Google Scholar] [CrossRef] [PubMed]
- Sattler, M.; Griffin, J.D. Mechanisms of transformation by the BCR/ABL oncogene. Int. J. Hematol. 2001, 73, 278–291. [Google Scholar] [CrossRef] [PubMed]
- Linardic, C.M. PAX3-FOXO1 fusion gene in rhabdomyosarcoma. Cancer Lett. 2008, 270, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Schmitt-Ney, M.; Camussi, G. The PAX3-FOXO1 fusion protein present in rhabdomyosarcoma interferes with normal FOXO activity and the TGF-β pathway. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- Rabbitts, T.H. Chromosomal translocations in human cancer. Nature 1994, 372, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Qin, F.; Movassagh, M.; Park, H.; Golden, W.; Xie, Z.; Zhang, P.; Sklar, J.; Li, H. A chimeric RNA characteristic of rhabdomyosarcoma in normal myogenesis process. Cancer Discov. 2013, 3, 1394–1403. [Google Scholar] [CrossRef] [PubMed]
- Jividen, K.; Li, H. Chimeric RNAs generated by intergenic splicing in normal and cancer cells. Genes Chromosom. Cancer 2014, 53, 963–971. [Google Scholar] [CrossRef] [PubMed]
- Magrangeas, F.; Pitiot, G.; Dubois, S.; Bragado-Nilsson, E.; Chérel, M.; Jobert, S.; Lebeau, B.; Boisteau, O.; Lethé, B.; Mallet, J.; et al. Cotranscription and intergenic splicing of human galactose-1-phosphate uridylyltransferase and interleukin-11 receptor alpha-chain genes generate a fusion mRNA in normal cells. Implication for the production of multidomain proteins during evolution. J. Biol. Chem. 1998, 273, 16005–16010. [Google Scholar]
- Li, H.; Wang, J.; Mor, G.; Sklar, J. A neoplastic gene fusion mimics trans-splicing of TRNAs in normal human cells. Science 2008, 321. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wang, J.; Ma, X.; Sklar, J. Gene fusions and RNA trans-splicing in normal and neoplastic human cells. Cell Cycle 2009, 8, 218–222. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Walsh, C.E. Spliceosome-mediated RNA trans-splicing. Mol. Ther. 2005, 12, 1006–1012. [Google Scholar] [CrossRef] [PubMed]
- Solnick, D. Trans splicing of mRNA precursors. Cell 1985, 42, 157–164. [Google Scholar] [CrossRef]
- Caudevilla, C.; Serra, D.; Miliar, A.; Codony, C.; Asins, G.; Bach, M.; Hegardt, F.G. Natural trans-splicing in carnitine octanoyltransferase pre-mRNAs in rat liver. Proc. Natl. Acad. Sci. USA 1998, 95, 12185–12190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akopian, A.N.; Okuse, K.; Souslova, V.; England, S.; Ogata, N.; Wood, J.N. Trans-splicing of a voltage-gated sodium channel is regulated by nerve growth factor. FEBS Lett. 1999, 445, 177–182. [Google Scholar] [CrossRef]
- Eul, J.; Graessmann, M.; Graessmann, A. Experimental evidence for RNA trans-splicing in mammalian cells. EMBO J. 1995, 14, 3226–3235. [Google Scholar] [PubMed]
- Vilborg, A.; Steitz, J.A. Readthrough transcription: How are DoGs made and what do they do? RNA Biol. 2017, 14, 632–636. [Google Scholar] [CrossRef] [PubMed]
- Burg, M.B.; Ferraris, J.D.; Dmitrieva, N.I. Cellular Response to Hyperosmotic Stresses. Physiol. Rev. 2007, 87, 1441–1474. [Google Scholar] [CrossRef] [PubMed]


© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chwalenia, K.; Qin, F.; Singh, S.; Tangtrongstittikul, P.; Li, H. Connections between Transcription Downstream of Genes and cis-SAGe Chimeric RNA. Genes 2017, 8, 338. https://doi.org/10.3390/genes8110338
Chwalenia K, Qin F, Singh S, Tangtrongstittikul P, Li H. Connections between Transcription Downstream of Genes and cis-SAGe Chimeric RNA. Genes. 2017; 8(11):338. https://doi.org/10.3390/genes8110338
Chicago/Turabian StyleChwalenia, Katarzyna, Fujun Qin, Sandeep Singh, Panjapon Tangtrongstittikul, and Hui Li. 2017. "Connections between Transcription Downstream of Genes and cis-SAGe Chimeric RNA" Genes 8, no. 11: 338. https://doi.org/10.3390/genes8110338





