A Strategy for Identifying Quantitative Trait Genes Using Gene Expression Analysis and Causal Analysis
Abstract
:1. Introduction
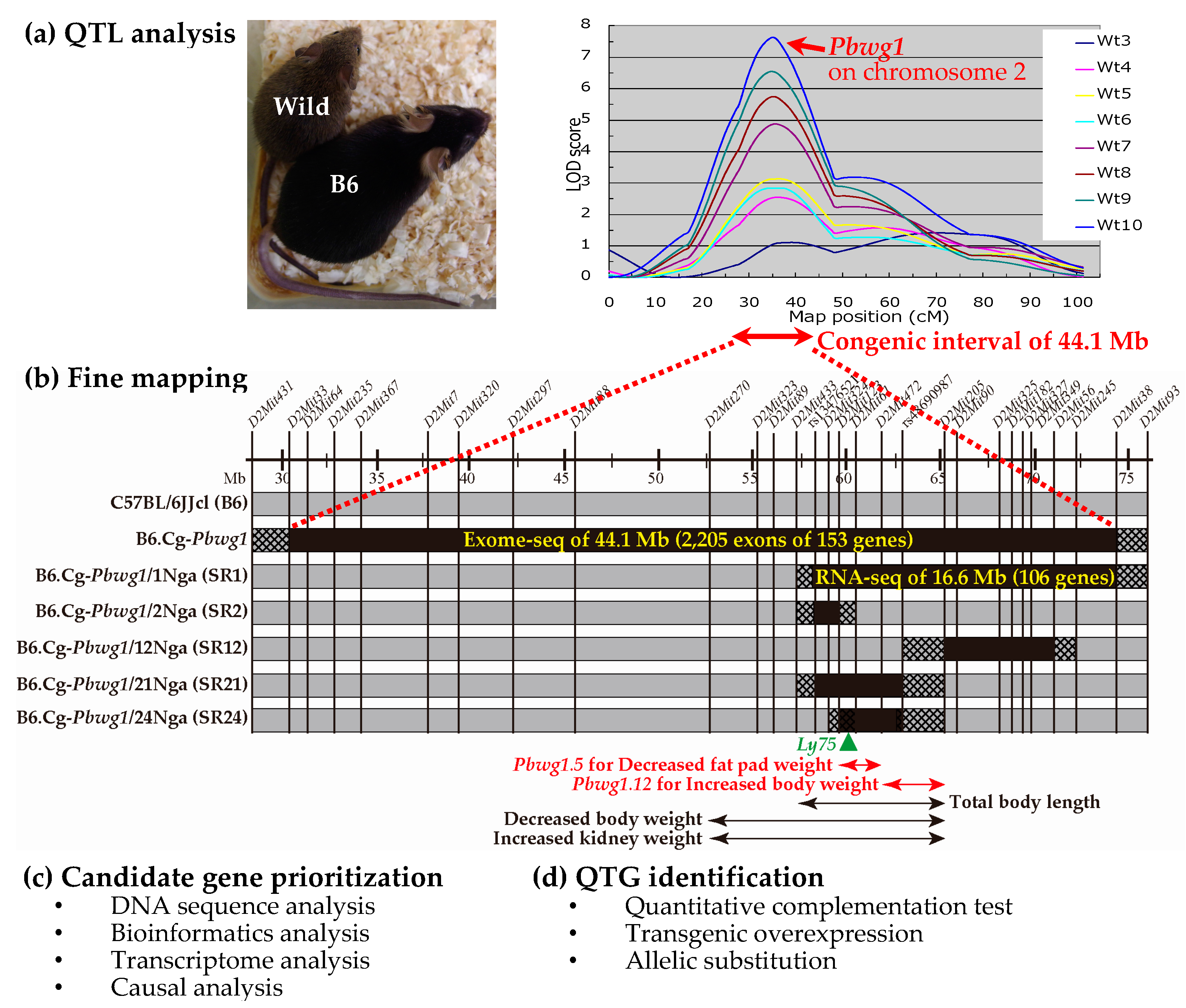
2. Quantitative Trait Loci Analysis
3. Fine Mapping
4. Candidate Gene Prioritization
4.1. Strategy
4.2. Limitations
5. Quantitative Trait Genes Identification
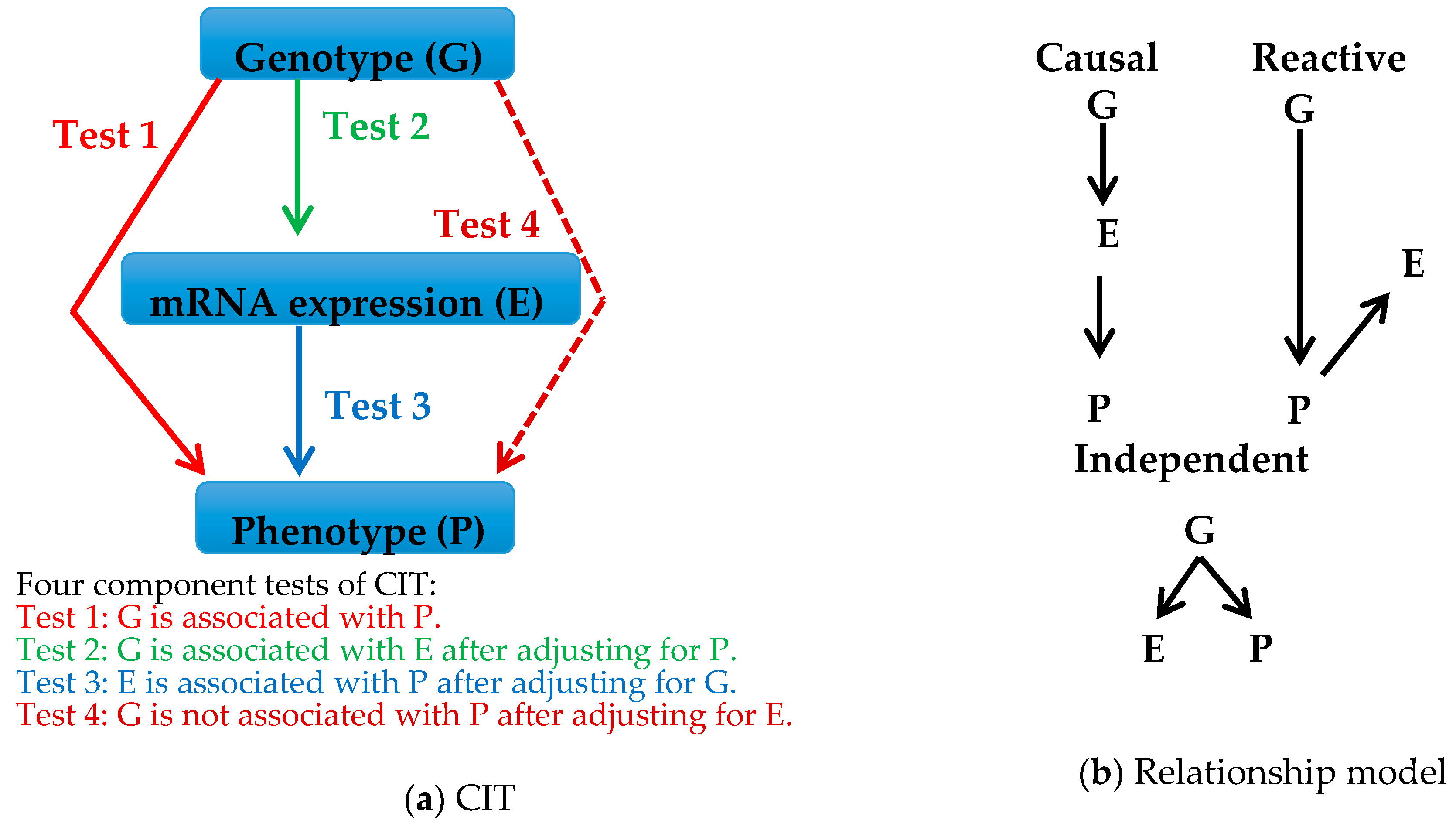
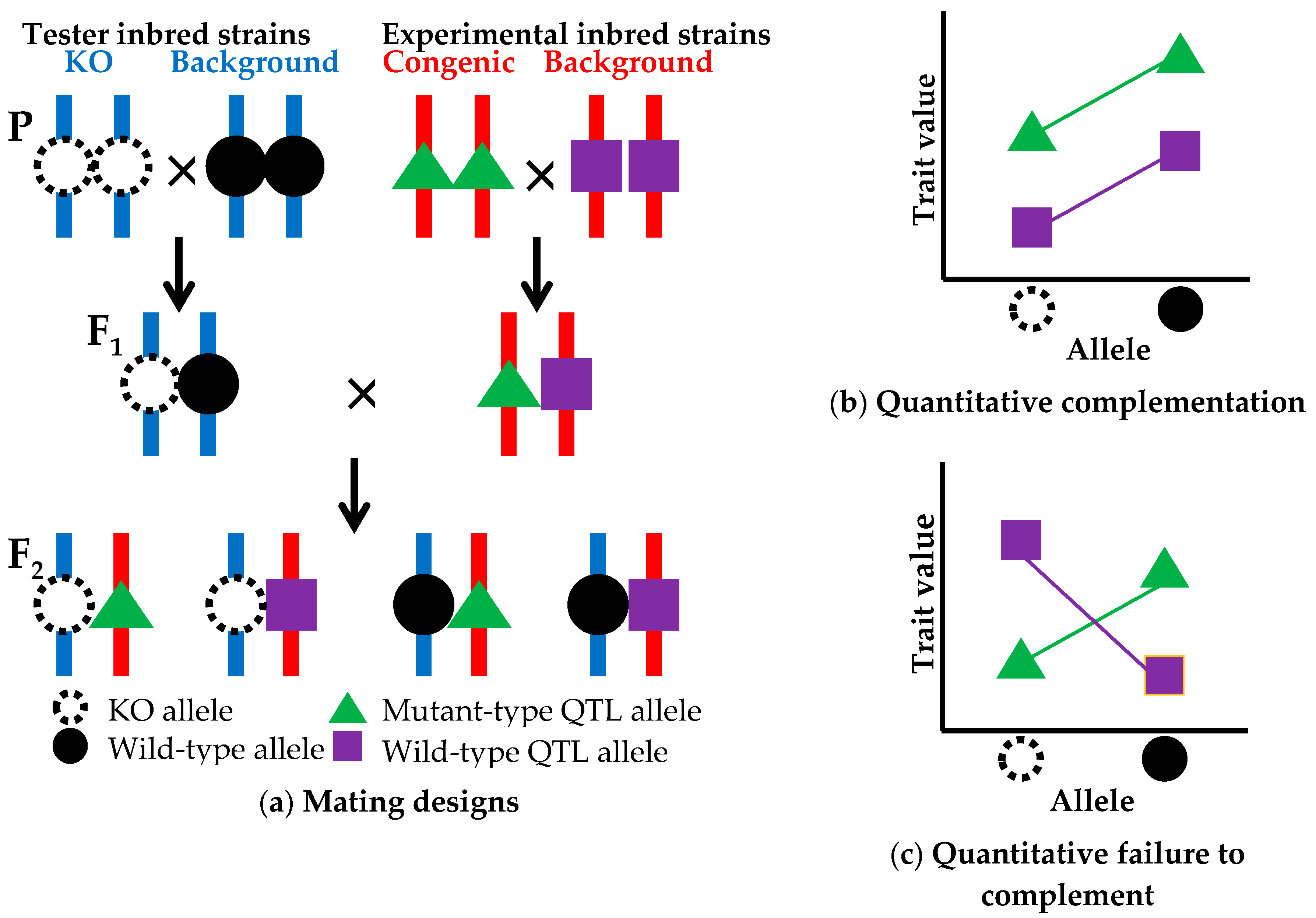
5.1. Strategy
5.2. Limitations
6. Future Perspective
Acknowledgments
Conflicts of Interest
Appendix A
References
- Miles, C.; Wayne, M. Quantitative trait locus (QTL) analysis. Nat. Educ. 2008, 1, 208. [Google Scholar]
- Welter, D.; MacArthur, J.; Morales, J.; Burdett, T.; Hall, P.; Junkins, H.; Klemm, A.; Flicek, P.; Manolio, T.; Hindorff, L.; Parkinson, H. The NHGRI-EBI Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014, 42, D1001–D1006. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.-L.; Park, C.A.; Reecy, J.M. Developmental progress and current status of the Animal QTLdb. Nucleic Acids Res. 2016, 44, D827–D833. [Google Scholar] [CrossRef] [PubMed]
- Eppig, J.T.; Blake, J.A.; Bult, C.J.; Kadin, J.A.; Richardson, J.E. Mouse Genome Database Group. The Mouse Genome Database (MGD): Facilitating mouse as a model for human biology and disease. Nucleic Acids Res. 2015, 43, D726–D736. [Google Scholar] [CrossRef] [PubMed]
- Darvasi, A.; Soller, M. A simple method to calculate resolving power and confidence interval of QTL map location. Behav. Genet. 1997, 27, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Darvasi, A. Interval-specific congenic strains (ISCS): An experimental design for mapping a QTL into a 1-centimorgan interval. Mamm. Genome 1997, 8, 163–167. [Google Scholar] [CrossRef] [PubMed]
- Markel, P.; Shu, P.; Ebeling, C.; Carlson, G.A.; Nagle, D.L.; Smutko, J.S.; Moore, K.J. Theoretical and empirical issues for marker-assisted breeding of congenic mouse strains. Nat. Genet. 1997, 17, 280–284. [Google Scholar] [CrossRef] [PubMed]
- Visscher, P.M.; Wray, N.R.; Zhang, Q.; Sklar, P.; McCarthy, M.I.; Brown, M.A.; Yang, J. 10 years of GWAS discovery: Biology, function, and translation. Am. J. Hum. Genet. 2017, 101, 5–22. [Google Scholar] [CrossRef] [PubMed]
- Albert, F.W.; Kruglyak, L. The role of regulatory variation in complex traits and disease. Nat. Rev. Genet. 2015, 16, 197–212. [Google Scholar] [CrossRef] [PubMed]
- Keane, T.M.; Goodstadt, L.; Danecek, P.; White, M.A.; Wong, K.; Yalcin, B.; Heger, A.; Agam, A.; Slater, G.; Goodson, M.; et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 2011, 477, 289–294. [Google Scholar] [CrossRef] [PubMed]
- Eisen, E.J. The Mouse in Animal Genetics and Breeding Research; Imperial College Press: London, UK, 2005; pp. 1–364. ISBN 1-86094-565-1. [Google Scholar]
- Ishikawa, A.; Matsuda, Y.; Namikawa, T. Detection of quantitative trait loci for body weight at 10 weeks from Philippine wild mice. Mamm. Genome 2000, 11, 824–830. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, A.; Namikawa, T. Mapping major quantitative trait loci for postnatal growth in an intersubspecific backcross between C57BL/6J and Philippine wild mice by using principal component analysis. Genes Genet. Syst. 2004, 79, 27–39. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, A.; Hatada, S.; Nagamine, Y.; Namikawa, T. Further mapping of quantitative trait loci for postnatal growth in an intersubspecific backcross of wild Mus musculus castaneus and C57BL/6J mice. Genet. Res. 2005, 85, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Mollah, M.B.R.; Ishikawa, A. Intersubspecific subcongenic mouse strain analysis reveals closely linked QTLs with opposite effects on body weight. Mamm. Genome 2011, 22, 282–289. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, A.; Kim, E.-H.; Bolor, H.; Mollah, M.B.R.; Namikawa, T. A growth QTL (Pbwg1) region of mouse chromosome 2 contains closely linked loci affecting growth and body composition. Mamm. Genome 2007, 18, 229–239. [Google Scholar] [CrossRef] [PubMed]
- Mollah, M.B.R.; Ishikawa, A. A wild derived quantitative trait locus on mouse chromosome 2 prevents obesity. BMC Genet. 2010, 11, 84. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, A.; Okuno, S. Fine mapping and candidate gene search of quantitative trait loci for growth and obesity using mouse intersubspecific subcongenic intercrosses and exome sequencing. PLoS ONE 2014, 9, e113233. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, A. Identification of a putative quantitative trait gene for resistance to obesity in mice using transcriptome analysis and causal inference tests. PLoS ONE 2017, 12, e0170652. [Google Scholar]
- Mollah, M.B.R.; Ishikawa, A. Fine mapping of quantitative trait loci affecting organ weights by mouse intersubspecific subcongenic strain analysis. Anim. Sci. J. 2013, 84, 296–302. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, A. Mapping an overdominant quantitative trait locus for heterosis of body weight in mice. J. Hered. 2009, 100, 501–504. [Google Scholar] [CrossRef] [PubMed]
- Darvasi, A.; Soller, M. Advanced intercross lines, an experimental population for fine genetic mapping. Genetics 1995, 141, 1199–1207. [Google Scholar] [PubMed]
- Mott, R.; Talbot, C.J.; Turri, M.G.; Collins, A.C.; Flint, J. A method for fine mapping quantitative trait loci in outbred animal stocks. Proc. Natl. Acad. Sci. USA 2000, 97, 12649–12654. [Google Scholar] [CrossRef] [PubMed]
- Hansen, C.; Spuhler, K. Development of the National Institutes of Health genetically heterogeneous rat stock. Alcohol. Clin. Exp. Res. 1984, 8, 477–479. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A.; Morgan, A.P.; Najarian, M.L.; Sarsani, V.K.; Sigmon, J.S.; Shorter, J.R.; Kashfeen, A.; McMullan, R.C.; Williams, L.H.; Giusti-Rodríguez, P.; et al. Genomes of the mouse collaborative cross. Genetics 2017, 206, 537–556. [Google Scholar] [CrossRef] [PubMed]
- Gatti, D.M.; Svenson, K.L.; Shabalin, A.; Wu, L.-Y.; Valdar, W.; Simecek, P.; Goodwin, N.; Cheng, R.; Pomp, D.; Palmer, A.; et al. Quantitative trait locus mapping methods for diversity outbred mice. Genes Genomes Genet. 2014, 4, 1623–1633. [Google Scholar] [CrossRef] [PubMed]
- Simon, M.M.; Greenaway, S.; White, J.K.; Fuchs, H.; Gailus-Durner, V.; Wells, S.; Sorg, T.; Wong, K.; Bedu, E.; Cartwright, E.J.; et al. A comparative phenotypic and genomic analysis of C57BL/6J and C57BL/6N mouse strains. Genome Biol. 2013, 14, R82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frazer, K.A.; Eskin, E.; Kang, H.M.; Bogue, M.A.; Hinds, D.A.; Beilharz, E.J.; Gupta, R.V.; Montgomery, J.; Morenzoni, M.M.; Nilsen, G.B.; et al. A sequence-based variation map of 8.27 million SNPs in inbred mouse strains. Nature 2007, 448, 1050–1053. [Google Scholar] [CrossRef] [PubMed]
- Tranchevent, L.-C.; Barriot, R.; Yu, S.; Vooren, S.V.; Loo, P.V.; Coessens, B.; Moor, B.D.; Aerts, S.; Moreau, Y. Endeavour update: A web resource for gene prioritization in multiple species. Nucleic Acids Res. 2008, 36, W377–W384. [Google Scholar] [CrossRef] [PubMed]
- Adzhubei, I.A.; Schmidt, S.; Peshkin, L.; Ramensky, V.E.; Gerasimova, A.; Bork, P.; Kondrashov, A.S.; Sunyaev, S.R. A method and server for predicting damaging missense mutations. Nat. Methods 2010, 7, 248–249. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Henikoff, S.; Ng, P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009, 4, 1073–1082. [Google Scholar] [CrossRef] [PubMed]
- Millstein, J.; Zhang, B.; Zhu, J.; Schadt, E.E. Disentangling molecular relationships with a causal inference test. BMC Genet. 2009, 10, 23. [Google Scholar] [CrossRef] [PubMed]
- Schadt, E.E.; Lamb, J.; Yang, X.Y.; Zhu, J.; Edwards, S.; GuhaThakurta, D.G.; Sieberts, S.K.; Monks, S.; Reitman, M.; Zhang, C.; et al. An integrative genomics approach to infer causal associations between gene expression and disease. Nat. Genet. 2005, 37, 710–717. [Google Scholar] [CrossRef] [PubMed]
- Aten, J.; Fuller, T.; Lusis, A.; Horvath, S. Using genetic markers to orient the edges in quantitative trait networks: The NEO software. BMC Syst. Biol. 2008, 2, 34. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Tsaih, S.W.; Shockley, K.; Stylianou, I.M.; Wergedal, J.; Paigen, B.; Churchill, G.A. Structural model analysis of multiple quantitative traits. PLoS Genet. 2006, 2, e114. [Google Scholar] [CrossRef] [PubMed]
- Relton, C.; Smith, G.D. Two-step epigenetic Mendelian randomization: A strategy for establishing the causal role of epigenetic processes in pathways to disease. Int. J. Epidemiol. 2012, 41, 161–176. [Google Scholar] [CrossRef] [PubMed]
- Rockman, M.V. Reverse engineering the genotype–phenotype map with natural genetic variation. Nature 2008, 456, 738–744. [Google Scholar] [CrossRef] [PubMed]
- Johnsson, M.; Williams, M.J.; Jensen, P.; Wright, D. Genetical genomics of behavior: A novel chicken genomic model for anxiety behavior. Genetics 2016, 202, 327–340. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Aryee, M.J.; Padyukov, L.; Fallin, M.D.; Hesselberg, E.; Runarsson, A.; Reinius, L.; Acevedo, N.; Taub, M.; Ronninger, M.; et al. Epigenome-wide association data implicate DNA methylation as an intermediary of genetic risk in rheumatoid arthritis. Nat. Biotechnol. 2013, 31, 142–147. [Google Scholar] [CrossRef] [PubMed]
- Olsson, A.H.; Volkov, P.; Bacos, K.; Dayeh, T.; Hall, E.; Nilsson, E.A.; Ladenvall, C.; Rönn, T.; Ling, C. Genome-wide associations between genetic and epigenetic variation influence mRNA expression and insulin secretion in human pancreatic islets. PLoS Genet. 2014, 10, e1004735. [Google Scholar] [CrossRef] [PubMed]
- Wahl, S.; Drong, A.; Lehne, B.; Loh, M.; Scott, W.R.; Kunze, S.; Tsai, P.-C.; Ried, J.S.; Zhang, W.; Yang, Y.; et al. Epigenome-wide association study of body mass index, and the adverse outcomes of adiposity. Nature 2017, 541, 81–86. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, Y. Metabolic impact of glucagon deficiency. Diabetes Obes. Metab. 2011, 13 (Suppl. S1), 151–157. [Google Scholar] [CrossRef] [PubMed]
- Burgess, S.; Thompson, D.J.; Rees, J.M.B.; Day, F.R.; Perry, J.R.; Ong, K.K. Dissecting causal pathways using Mendelian randomization with summarized genetic data: Application to age at menarche and risk of breast cancer. Genetics 2017, 207, 481–487. [Google Scholar] [CrossRef] [PubMed]
- Rikke, B.A.; Johnson, T.E. Towards the cloning of genes underlying murine QTLs. Mamm. Genome. 1998, 9, 963–968. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Gong, S.; Maric, S.; Misulovin, Z.; Pack, M.; Mahnke, K.; Nussenzweig, M.C.; Steinman, R.M. A monoclonal antibody to the DEC-205 endocytosis receptor on human dendritic cells. Hum. Immunol. 2000, 61, 729–738. [Google Scholar] [CrossRef]
- Christians, J.K.; de Zwaan, D.R.; Fung, S.H.Y. Pregnancy associated plasma protein A2 (PAPP-A2) affects bone size and shape and contributes to natural variation in postnatal growth in mice. PLoS ONE 2013, 8, e56260. [Google Scholar] [CrossRef] [PubMed]
- Tomida, S.; Mamiya, T.; Sakamaki, H.; Miura, M.; Aosaki, T.; Masuda, M.; Niwa, M.; Kameyama, T.; Kobayashi, J.; Iwaki, Y.; et al. Usp46 is a quantitative trait gene regulating mouse immobile behavior in the tail suspension and forced swimming tests. Nat. Genet. 2009, 41, 688–695. [Google Scholar] [CrossRef] [PubMed]
- Morton, N.M.; Beltram, J.; Carter, R.N.; Michailidou, Z.; Gorjanc, G.; McFadden, C.; Barrios-Llerena, M.E.; Rodriguez-Cuenca, S.; Gibbins, M.T.G.; Aird, R.E.; et al. Genetic identification of thiosulfate sulfurtransferase as an adipocyte-expressed antidiabetic target in mice selected for leanness. Nat. Med. 2016, 22, 771–779. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Waghulde, H.; Mell, B.; Morgan, E.E.; Pruett-Miller, S.M.; Joe, B. Positional cloning of quantitative trait nucleotides for blood pressure and cardiac QT-interval by targeted CRISPR/Cas9 editing of a novel long non-coding RNA. PLoS Genet. 2017, 13, e1006961. [Google Scholar] [CrossRef] [PubMed]
| QTL | Gene | sSNP | nsSNP | Gene Ranking 1 | Damage of Protein 2 | |
|---|---|---|---|---|---|---|
| PolyPhen-2 | SIFT | |||||
| Pbwg1.5 | Dapl1 | 1 | 0 | |||
| Tanc1 | 21 | 4 | ||||
| Wdsub1 | 6 | 1 | ||||
| Baz2b | 15 | 6 | ||||
| March7 | 6 | 0 | ||||
| Cd302 | 1 | 0 | ||||
| Ly75 | 27 | 9 | 1 | Benign | Tolerated | |
| Pla2r1 | 18 | 8 | ||||
| Itgb6 | 11 | 3 | 2 | Benign | Affected | |
| Rbms1 | 2 | 0 | ||||
| Tank | 1 | 5 | ||||
| Psmd14 | 1 | 0 | ||||
| Pbwg1.12 | Tbr1 | 2 | 1 | |||
| Slc4a10 | 6 | 0 | ||||
| Dpp4 | 6 | 0 | ||||
| Gcg | 0 | 1 | 1 | Benign | Tolerated | |
| Fap | 2 | 2 | ||||
| Ifih1 | 17 | 5 | ||||
| Gca | 3 | 1 | ||||
| Kcnh7 | 6 | 1 | ||||
| Fign | 4 | 1 | ||||
| Grb14 | 7 | 2 | 2 | Benign | Tolerated | |
| Cobll1 | 14 | 18 | ||||
| Organ | Gene | Relative Expression Level 1 | Differences 2 | ||
|---|---|---|---|---|---|
| B/B | B/C | C/C | |||
| Liver | Ly75 | 1.00 | 1.81 | 3.19 | C/C>B/C>B/B |
| Pla2r1 | 1.00 | −1.58 | 0.58 | B/B≥C/C≥B/C | |
| Fap | 1.00 | 5.89 | 8.03 | C/C>B/C>B/B | |
| Gca | 1.00 | 0.79 | 0.34 | B/B≥B/C≥C/C | |
| Gonadal fat | Fap | 1.00 | 1.43 | 2.11 | C/C>B/C>B/B |
| Ifih1 | 1.00 | −0.47 | −0.53 | B/B>B/C≥C/C | |
| Grb14 | 1.00 | 0.73 | 0.50 | B/B≥B/C≥C/C | |
© 2017 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ishikawa, A. A Strategy for Identifying Quantitative Trait Genes Using Gene Expression Analysis and Causal Analysis. Genes 2017, 8, 347. https://doi.org/10.3390/genes8120347
Ishikawa A. A Strategy for Identifying Quantitative Trait Genes Using Gene Expression Analysis and Causal Analysis. Genes. 2017; 8(12):347. https://doi.org/10.3390/genes8120347
Chicago/Turabian StyleIshikawa, Akira. 2017. "A Strategy for Identifying Quantitative Trait Genes Using Gene Expression Analysis and Causal Analysis" Genes 8, no. 12: 347. https://doi.org/10.3390/genes8120347




