Evolution of Tandem Repeat Satellite Sequences in Two Closely Related Caenorhabditis Species. Diminution of Satellites in Hermaphrodites
Abstract
:1. Introduction
2. Materials and Methods
2.1. Genome Sequences and Alignments
2.2. Satellite Identification
2.3. Identification of Satellite Families
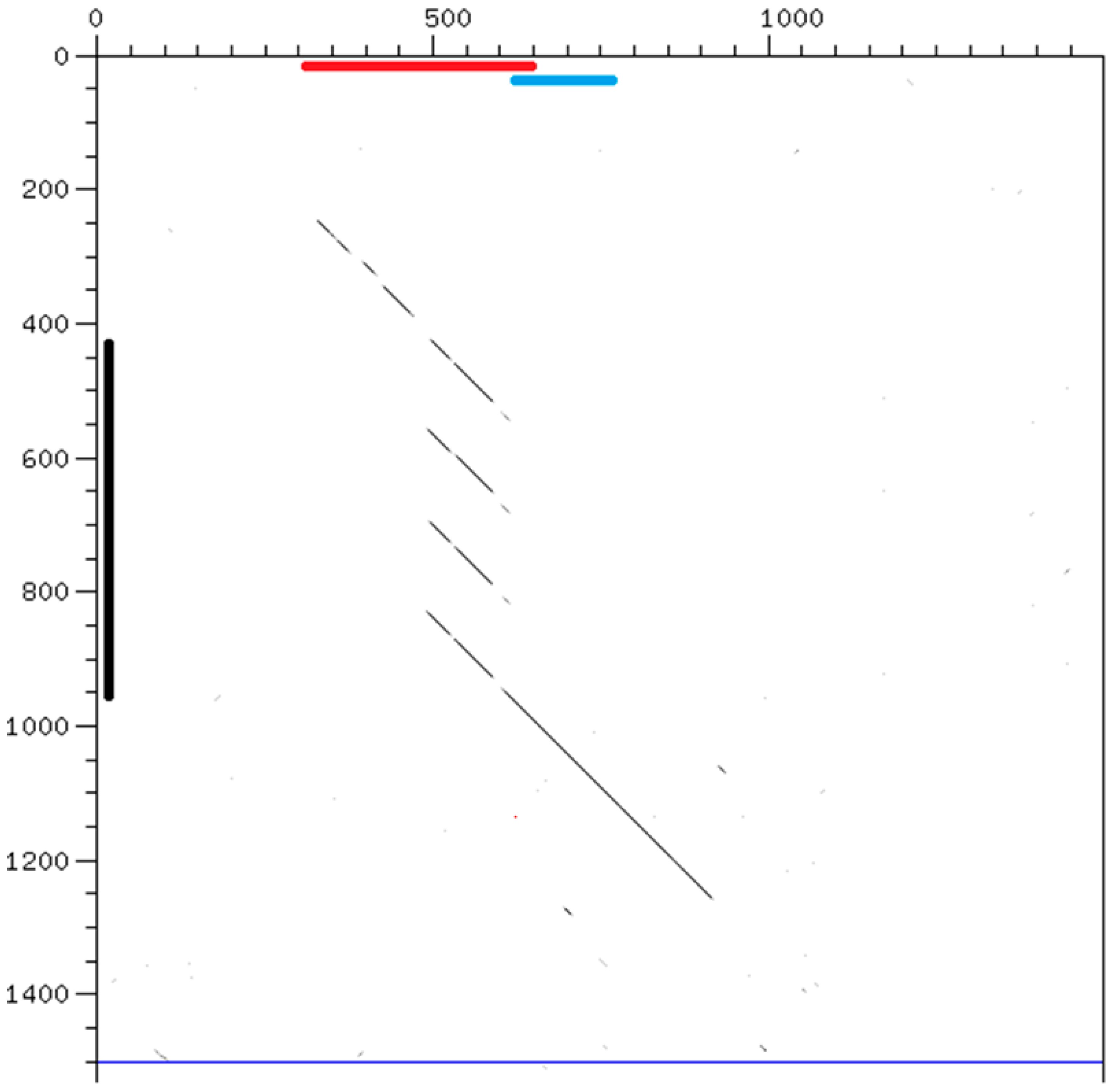
2.4. Comparison of Syntenic Satellites
3. Results and Discussion
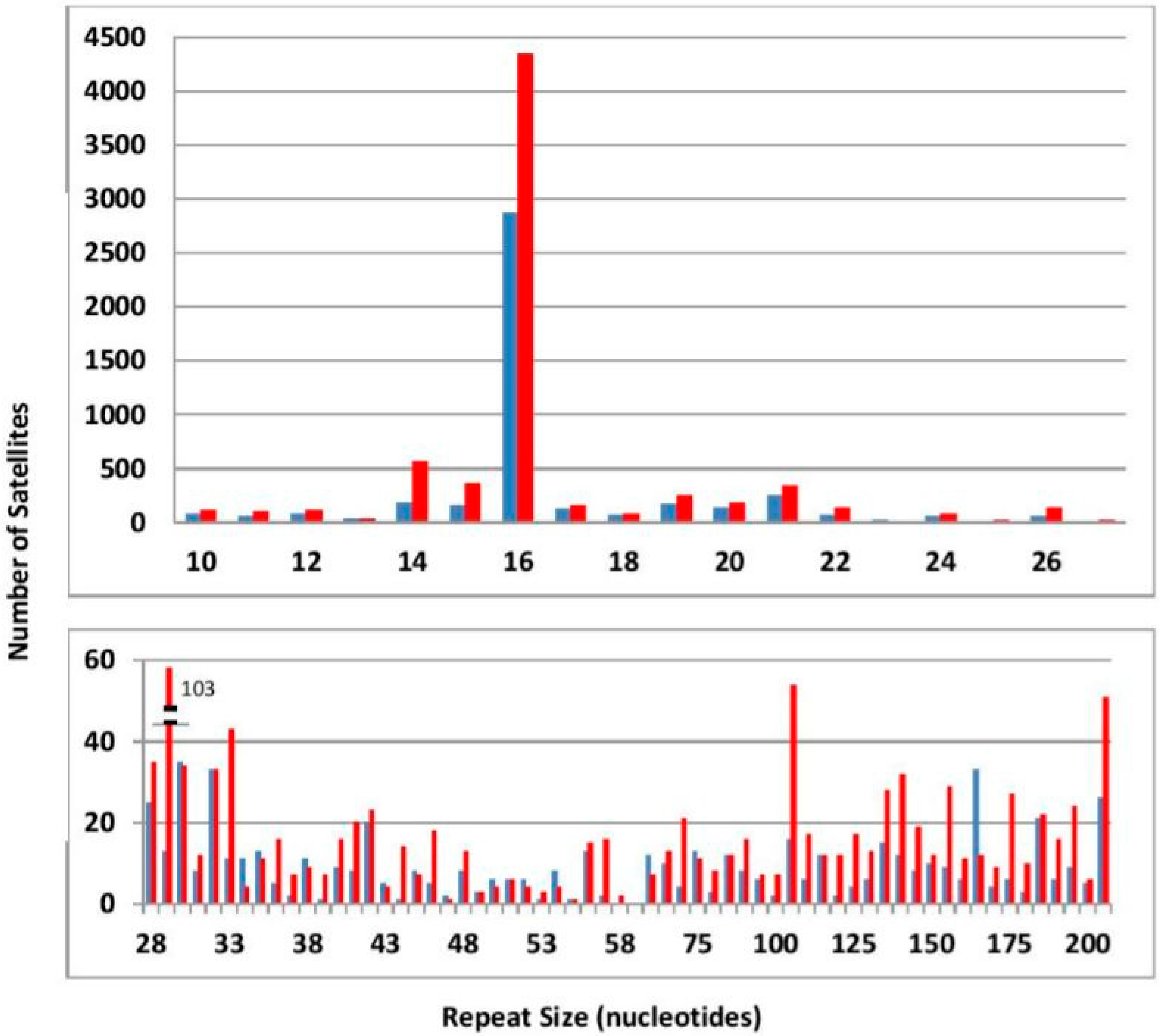
3.1. General Satellite Features
3.2. Satellite Families
3.3. Conserved Pairs of Satellites
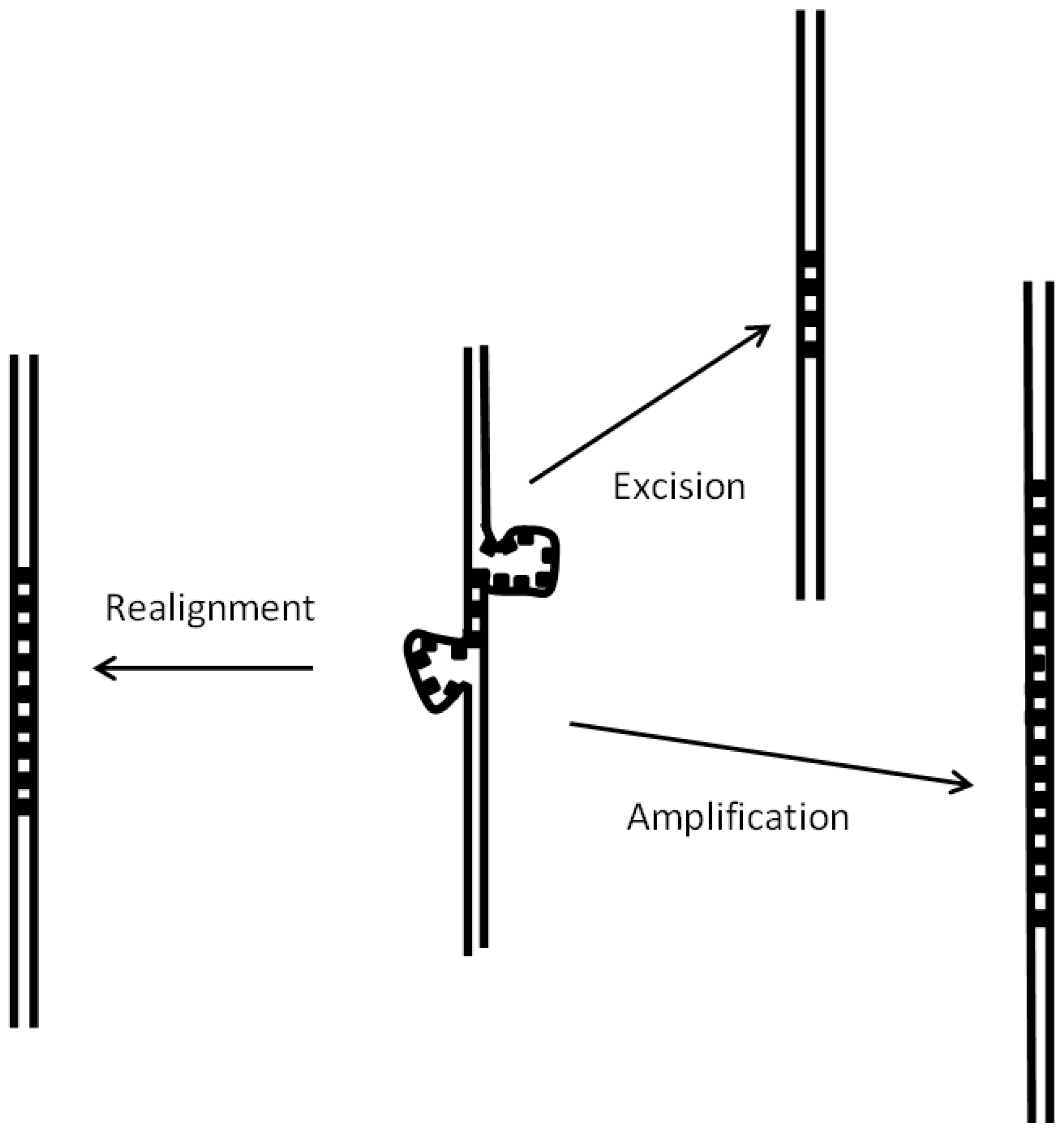
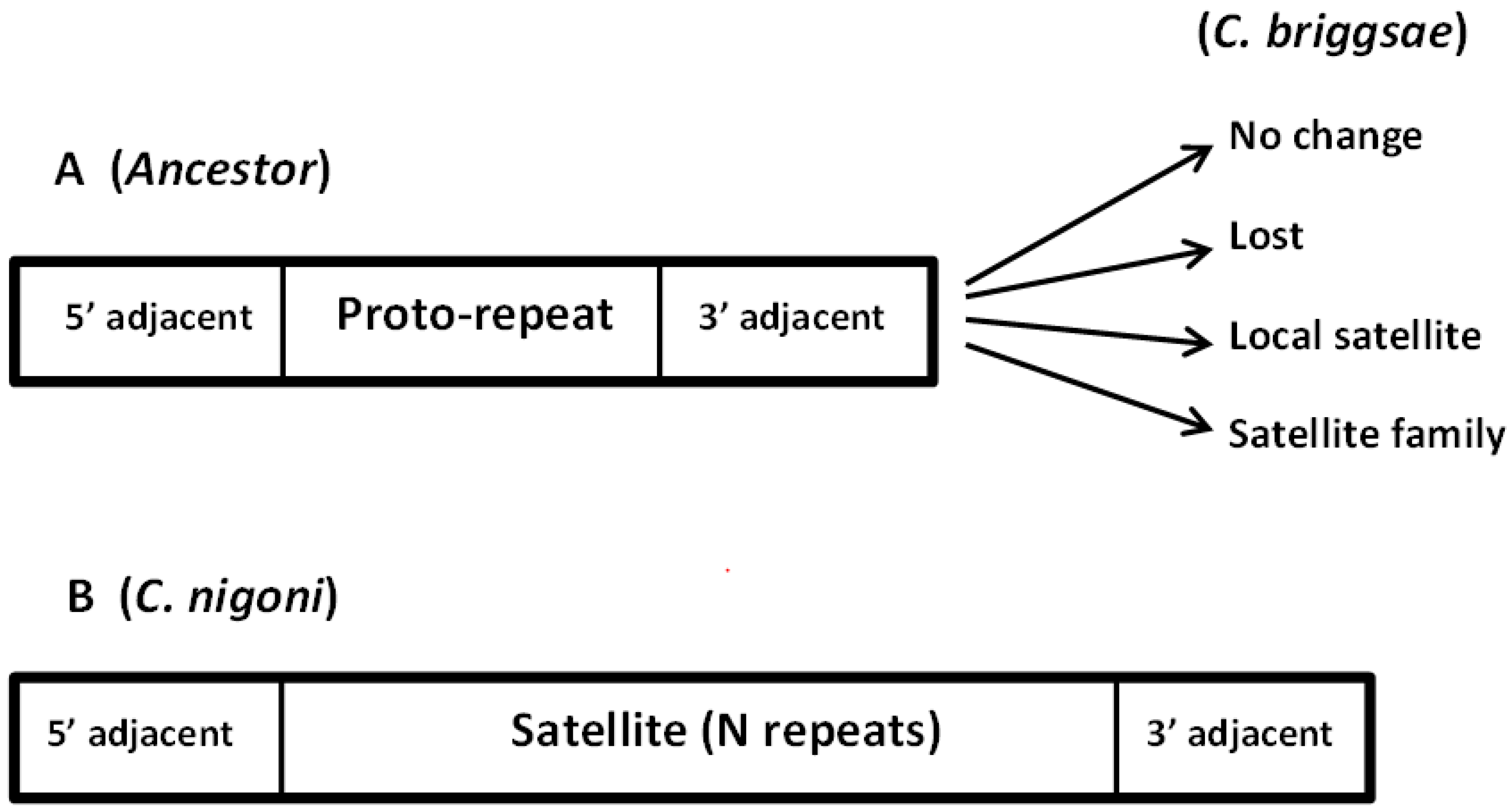
3.4. Birth and Growth of New Satellites
3.5. New Satellite Families: Precise Tandem Repeats and Conserved Adjacent Sequences
3.6. The X Chromosome Contains Characteristic Satellites
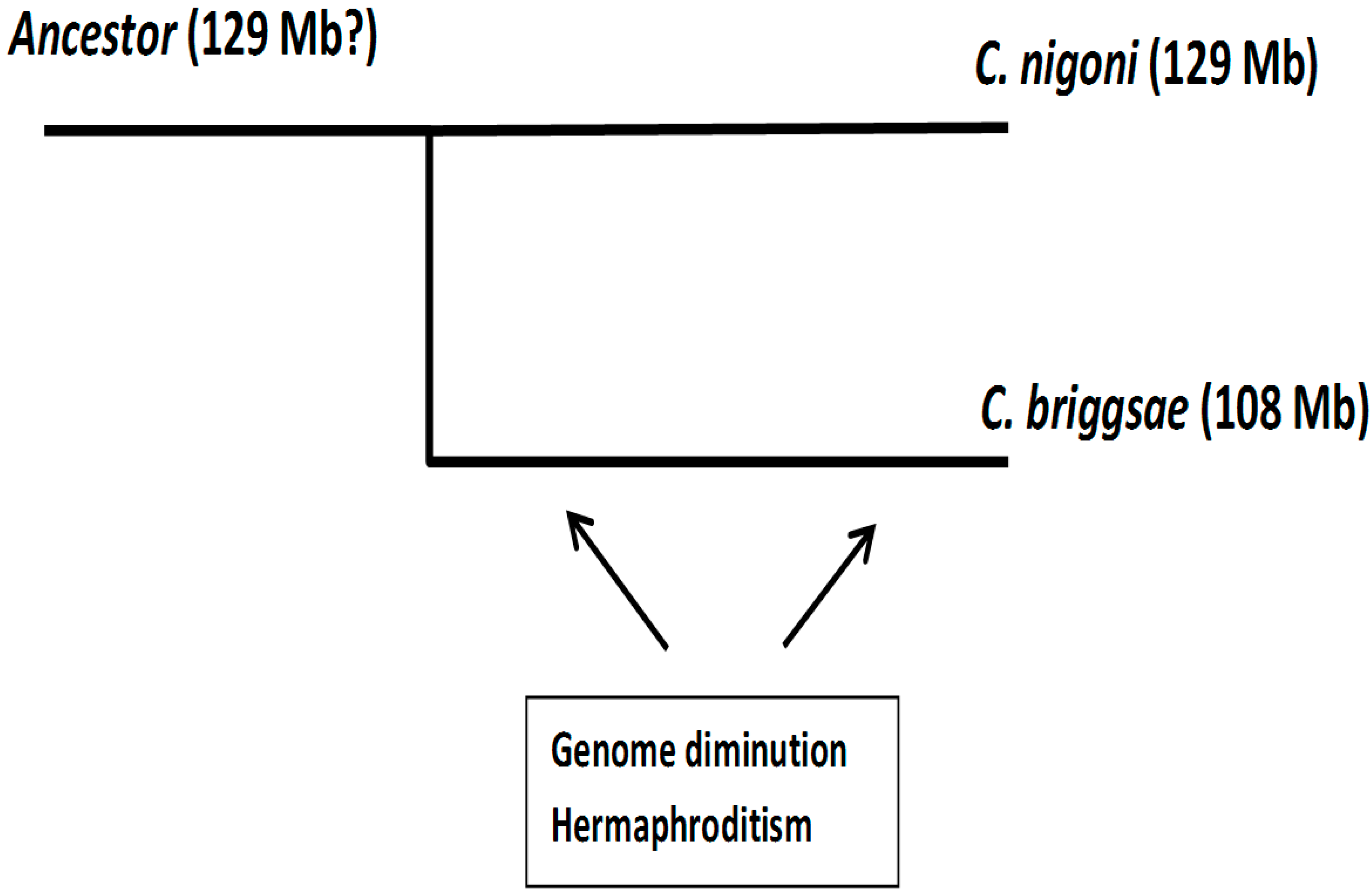
3.7. Satellites and Hermaphroditism
4. Conclusions
- We have detected 4834 satellites in C. briggsae and 7803 in C. nigoni. Satellites with repeats ≥28 nt form 194 families of related satellites from both species. We also find 573 unique satellites, unrelated in sequence with any other satellite (362 in C. nigoni and 211 in C. briggsae).
- Some satellites in C. nigoni have developed from a proto-repeat present in the ancestor species and conserved as an isolated sequence in C. briggsae.
- The rate of neutral mutation in C. briggsae and C. nigoni since they diverged from their ancestor is practically identical, as judged by the changes found in conserved satellites (Supplementary Table S5).
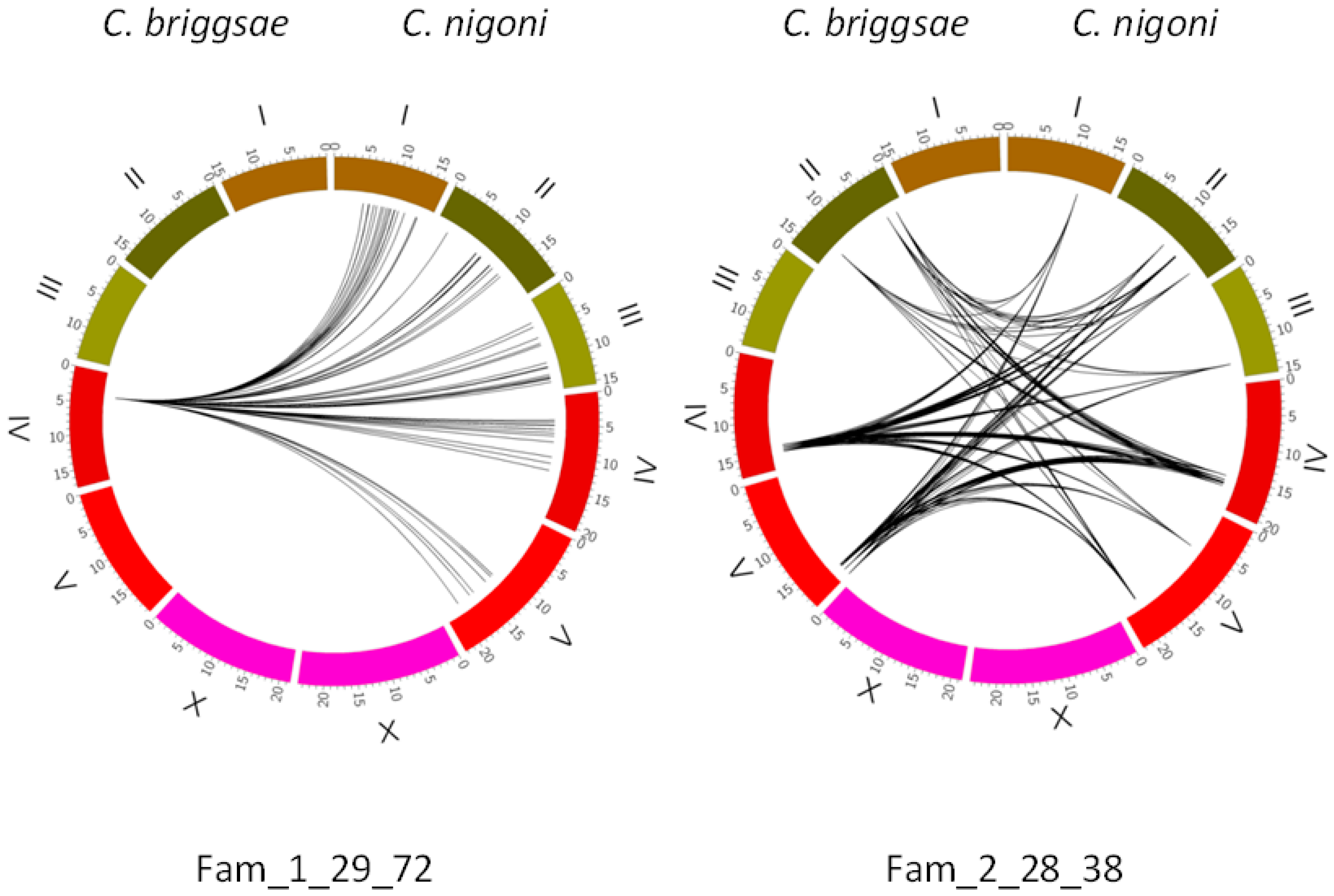
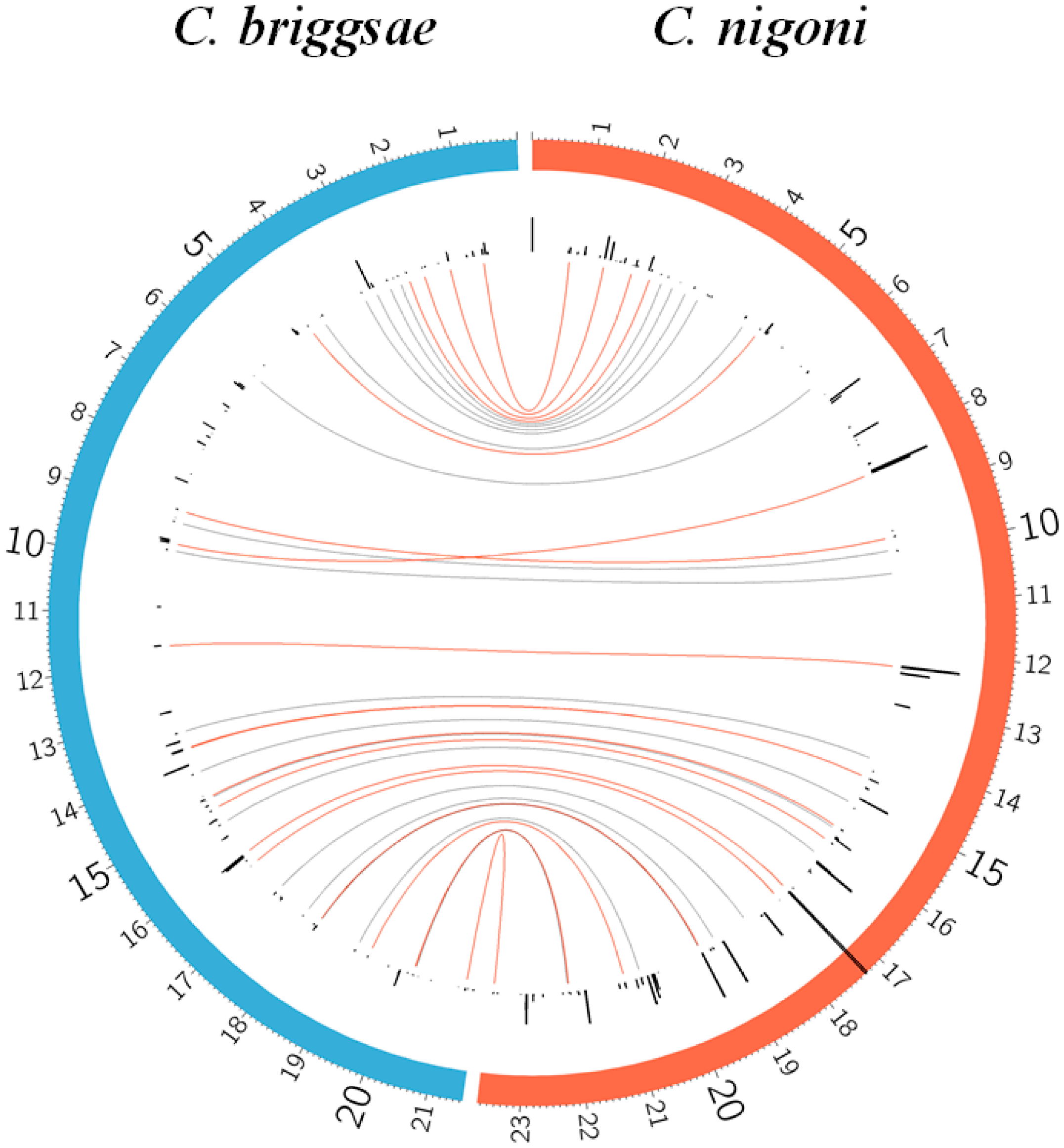
- Transposition of satellites creates satellite families. It follows a different mechanism than transposition of coding sequences, since satellites may be transposed between different chromosomes. An additional mechanism favors transposition of satellites between closely spaced genome regions, which gives rise to clusters of related satellites.
- Transposition of satellites is limited in C. briggsae; very few new families are found when compared with C. nigoni (Figure 3).
- Transposition of satellites to form new satellite families in C. nigoni is associated with conserved 5′ and 3′ adjacent regions at both ends of the satellites (Table 2).
- The identical size of satellites during homologue pairing in meiosis in hermaphrodites may prevent its growth by unequal recombination (Figure 3).
- Birth of new satellites is facilitated in unisexual species, with respect to hermaphrodites, since double-stranded break repair appears to be a major mechanism for the initial duplication of short nucleotide sequences.
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Schärer, L. The Varied Ways of being Male and Female. Mol. Reprod. Dev. 2017, 84, 94–104. [Google Scholar] [CrossRef] [PubMed]
- Kiontke, K.C.; Felix, M.A.; Ailion, M.; Rockman, M.V.; Braendle, C.; Penigault, J.B.; Fitch, D.H.A. A phylogeny and molecular barcodes for Caenorhabditis, with numerous new species from rotting fruits. BMC Evol. Biol. 2011, 11, 339. [Google Scholar] [CrossRef] [PubMed]
- Ellis, R.E. “The persistence of memory”-Hermaphroditism in nematodes. Mol. Reprod. Dev. 2017, 84, 144–157. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Ramos, M.A. Satellite DNA: An evolving topic. Genes 2017, 8, 230. [Google Scholar] [CrossRef] [PubMed]
- Subirana, J.A.; Messeguer, X. A satellite explosion in the genome of holocentric nematodes. PLoS ONE 2013, 8, e62221. [Google Scholar] [CrossRef] [PubMed]
- Open Science Framework. The C. nigoni genome has also been submitted to DDBJ/ENA/GenBank (accession number PDUG00000000; version PDUG01000000). Available online: https://doi.org/10.17605/osf.io/dkbwt (accessed on 29 September 2017).
- Subirana, J.A.; Albà, M.M.; Messeguer, X. High evolutionary turnover of satellite families in Caenorhabditis. BMC Evol. Biol. 2015, 15, 218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, C.G.; Wang, W.; Jovelin, R.; Ghosh, R.; Lomasko, T.; Trinh, Q.; Kruglyak, L.; Stein, L.D.; Cutter, A.D. Full-genome evolutionary histories of selfing, splitting and selection in Caenorhabditis. Genome Res. 2015, 25, 667–678. [Google Scholar] [CrossRef] [PubMed]
- Kozlowska, J.L.; Ahmad, A.R.; Jahesh, E.; Cutter, A.D. Genetic variation for postzygotic reproductive isolation between Caenorhabditis briggsae and Caenorhabditis sp. 9. Evolution 2011, 66, 1180–1195. [Google Scholar] [CrossRef] [PubMed]
- Woodruff, G.C.; Eke, O.; Baird, S.E.; Félix, M.; Haag, E.S. Insights into species divergence and the evolution of hermaphroditism from fertile interspecies hybrids of Caenorhabditis nematodes. Genetics 2010, 186, 997–1012. [Google Scholar] [CrossRef] [PubMed]
- Chang, W.; Tilmann, C.; Thoemke, K.; Markussen, F.; Mathies, L.D.; Kimble, J.; Zarkower, D. A forkhead protein controls sexual identity of the C. elegans male somatic gonad. Development 2003, 131, 1425–1436. [Google Scholar] [CrossRef] [PubMed]
- Graunstein, A.; Gaspar, J.M.; Walters, J.R.; Palopoli, M.F. Levels of DNA polymorphism vary with mating system in the nematode genus Caenorhabditis. Genetics 2002, 161, 99–107. [Google Scholar]
- Yin, D.; Schwarz, E.M.; Thomas, C.G.; Felde, R.L.; Korf, I.F.; Cutter, A.D.; Schartner, C.M.; Ralston, E.J.; Meyer, B.J.; Haag, E.S. Rapid genome shrinkage in a self-fertile nematode reveals sperm competition proteins. Science 2017, in press. [Google Scholar]
- Thomas, C.G.; Li, R.; Smith, H.E.; Woodruff, G.C.; Oliver, B.; Haag, E.S. Simplification and desexualization of gene expression in self-fertile nematodes. Curr. Biol. 2012, 22, 2167–2172. [Google Scholar] [CrossRef] [PubMed]
- Fierst, J.L.; Willis, J.H.; Thomas, C.G.; Wang, W.; Reynolds, R.M.; Ahearne, T.E.; Cutter, A.D.; Phillips, P.C. Reproductive mode and the evolution of genome size and structure in Caenorhabditis nematodes. PLoS Genet. 2015, 11, e1005323. [Google Scholar]
- Stein, L.D.; Bao, Z.; Blasiar, D.; Blumenthal, T.; Brent, M.R.; Chen, N.; Chinwalla, A.; Clarke, L.; Clee, C.; Coghlan, A.; et al. The genome sequence of Caenorhabditis briggsae: A platform for comparative genomics. PLoS Biol. 2003, 1, e45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wormbase. Available online: http://www.wormbase.org (accessed on 15 September 2017).
- Ross, J.A.; Koboldt, D.C.; Staisch, J.E.; Chamberlin, H.M.; Gupta, B.P.; Miller, R.D.; Baird, S.E.; Haag, E.S. Caenorhabditis briggsae recombinant inbred line genotypes reveal inter-strain incompatibility and the evolution of recombination. PLoS Genet. 2011, 7, e1002174. [Google Scholar] [CrossRef] [PubMed]
- Treangen, T.J.; Messeguer, X. M-GCAT: interactively and efficiently constructing large-scale multiple genome comparison frameworks in closely related species. BMC Bioinform. 2006, 7, 433. [Google Scholar] [CrossRef] [PubMed]
- M-GCAT. Available online: http://alggen.lsi.upc.edu/recerca/align/mgcat/mgcat_win.zip (accessed on 15 June 2017).
- Sonnhammer, E.L.; Durbin, R. A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene 1995, 167, GC1–GC10. [Google Scholar] [CrossRef]
- Dotter. Available online: http://www.sanger.ac.uk/science/tools/seqtools (accessed on 15 June 2017).
- Algorithmics and Genetics Group. Available online: http://alggen.lsi.upc.edu (accessed on 26 September 2017).
- SATFIND and MALIG. Available online: http://dx.doi.org/10.5061/dryad.h5s2q (accessed on 26 September 2017).
- Khost, D.E.; Eickbush, D.E.; Larracuente, A.M. Single-molecule sequencing resolves the detailed structure of complex satellite DNA loci in Drosophila melanogaster. Genome Res. 2017, 27, 709–721. [Google Scholar] [CrossRef] [PubMed]
- Wicky, C.; Villeneuve, A.M.; Lauper, N.; Codourey, L.; Tobler, H.; Müller, F. Telomeric repeats (TTAGGC)n are sufficient for chromosome capping function in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 1996, 93, 8983–8988. [Google Scholar] [CrossRef] [PubMed]
- Hillier, L.W.; Miller, R.D.; Baird, S.E.; Chinwalla, A.; Fulton, L.A.; Koboldt, D.C.; Waterston, R.H. Comparison of C. elegans and C. briggsae genome sequences reveals extensive conservation of chromosome organization and synteny. PLoS Biol. 2007, 5, e167. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Farré, M.; Auvil, L.; Capitanu, B.; Larkin, D.M.; Ma, J.; Lewin, H.A. Reconstruction and evolutionary history of eutherian chromosomes. Proc. Natl. Acad. Sci. USA 2017, 114, E5379–E5388. [Google Scholar] [CrossRef] [PubMed]
- D’Andrea, L.D.; Regan, L. TPR proteins: The versatile helix. Trends Biochem. Sci. 2003, 28, 655–662. [Google Scholar] [CrossRef] [PubMed]
- Northam, M.R.; Moore, E.A.; Mertz, T.M.; Binz, S.K.; Stith, C.M.; Stepchenkova, E.I.; Wendt, K.L.; Burgers, P.M.J.; Shcherbakova, P.V. DNA polymerases zeta and Rev1 mediate error-prone bypass of non-B DNA structures. Nucleic Acids Res. 2014, 42, 290–306. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Brazier, J.A.; Sugimoto, N. Topological impact of noncanonical DNA structures on Klenow fragment of DNA polymerase. Proc. Natl. Acad. Sci. USA 2017, 114, 9605–9610. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Chang, C.; Kao, Y.; Chin, C.; Ni, C.; Hsu, H.; Hu, N.; Hsieh, L.; Chou, S.; Lee, I.; et al. Parity-dependent hairpin configurations of repetitive DNA sequence promote slippage associated with DNA expansion. Proc. Natl. Acad. Sci. USA 2017, 114, 9535–9540. [Google Scholar] [CrossRef] [PubMed]
- Gadgil, R.; Barthelemy, J.; Lewis, T.; Leffak, M. Replication stalling and DNA microsatellite instability. Biophys. Chem. 2017, 225, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Ellegren, H. Microsatellites: Simple sequences with complex evolution. Nat. Rev. Genet. 2004, 5, 435–445. [Google Scholar] [CrossRef] [PubMed]
- Onozawa, M.; Zhang, Z.; Kim, Y.J.; Goldberg, L.; Varga, T.; Bergsagel, P.L.; Kuehl, W.M.; Aplan, P.D. Repair of DNA double-strand breaks by templated nucleotide sequence insertions derived from distant regions of the genome. Proc. Natl. Acad. Sci. USA 2014, 111, 7729–7734. [Google Scholar] [CrossRef] [PubMed]
- Korolev, S. Advances in structural studies of recombination mediator proteins. Biophys. Chem. 2017, 225, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Azeroglu, B.; Leach, D.R.F. RegG controls DNA amplification at double-strand breaks and arrested replication forks. FEBS Lett. 2017, 591, 1101–1113. [Google Scholar] [CrossRef] [PubMed]
- Seeber, A.; Gasser, S.M. Chromatin organization and dynamics in double-strand break repair. Curr. Opin. Genet. Dev. 2017, 43, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Smolikov, S.; Eizinger, A.; Hurlburt, A.; Rogers, E.; Villeneuve, A.M.; Colaiácovo, M.P. Synapsis-defective mutants reveal a correlation between chromosome conformation and the mode of double-strand break repair during Caenorhabditis elegans meiosis. Genetics 2007, 176, 2027–2033. [Google Scholar] [CrossRef] [PubMed]
- Rubnitz, J.; Subramani, S. The minimum amount of homology required for homologous recombination in mammalian cells. Mol. Cell. Biol. 1984, 4, 2253–2258. [Google Scholar] [CrossRef] [PubMed]
- Opperman, R.; Emmanuel, E.; Levy, A.A. The effect of sequence divergence on recombination between direct repeats in Arabidopsis. Genetics 2004, 168, 2207–2215. [Google Scholar] [CrossRef] [PubMed]
- Zetka, M. Homologue Pairing, Recombination and Segregation in Caenorhabditis elegans. In Meiosis; Benavente, R., Volff, J., Eds.; Karger: Basel, Switzerland, 2009; Volume 5, pp. 43–55. [Google Scholar]
- Shah, K.A.; Shishkin, A.A.; Voineagu, I.; Pavlov, Y.I.; Shcherbakova, P.V.; Mirkin, S.M. Role of DNA polymerases in repeat-mediated genome instability. Cell Rep. 2012, 2, 1088–1095. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, K.J.; Farslow, J.C.; Fitzpatrick, K.A.; Lynch, M.; Katju, V.; Bergthorsson, U. High spontaneous rate of gene duplication in Caenorhabditis elegans. Curr. Biol. 2011, 21, 306–310. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.C.; Mirkin, S.M. The balancing act of DNA repeat expansions. Curr. Opin. Genet. Dev. 2013, 23, 280–288. [Google Scholar] [CrossRef] [PubMed]
- Hahn, M.W. Distinguishing among evolutionary models for the maintenance of gene duplicates. J. Hered. 2009, 100, 605–617. [Google Scholar] [CrossRef] [PubMed]
- Van, M.V.; Larson, B.J.; Engebrecht, J. To break or not to break: sex chromosome hemizygosity during meiosis in Caenorhabditis. Genetics 2016, 204, 999–1013. [Google Scholar] [CrossRef] [PubMed]
| Chromosome | Satellite Distribution | |||||
|---|---|---|---|---|---|---|
| Name | Species | Size (Mb) | Repeat 10–27 | Repeat ≥28 | Total | Satellites/Mb |
| I | C. briggsae | 15.46 | 634 | 66 | 700 | 45.3 |
| C. nigoni | 16.74 | 923 | 145 | 1068 | 63.8 | |
| II | C. briggsae | 16.63 | 1172 | 63 | 1235 | 74.3 |
| C. nigoni | 19.23 | 1904 | 145 | 2049 | 106.6 | |
| III | C. briggsae | 14.58 | 500 | 67 | 567 | 38.9 |
| C. nigoni | 15.54 | 740 | 129 | 869 | 55.9 | |
| IV | C. briggsae | 17.49 | 497 | 71 | 568 | 32.5 |
| C. nigoni | 20.39 | 816 | 134 | 950 | 46.6 | |
| V | C. briggsae | 19.50 | 866 | 85 | 951 | 48.8 |
| C. nigoni | 22.29 | 1360 | 141 | 1501 | 67.3 | |
| X | C. briggsae | 21.54 | 279 | 101 | 380 | 17.6 |
| C. nigoni | 23.65 | 482 | 125 | 607 | 25.7 | |
| Unplaced | C. briggsae | 3.12 | 373 | 60 | 433 | 138.8 |
| C. nigoni | 11.66 | 604 | 181 | 785 | 67.3 | |
| Total | C. briggsae | 108.42 | 4321 | 513 | 4834 | 44.6 |
| C. nigoni | 129.49 | 6829 | 974 | 7803 | 60.3 | |
| Family | 5′ Adjacent Region | Start-Satellite-End | 3′ Adjacent Region |
|---|---|---|---|
| 4_137_15 | AATCGGTAGGACACAGTTCGAT | GATGTTTGTG---GACACCGTTC | ATGGGTTTTTGAAGGTGATCCC |
| 33_107_4 | CCCCAtTGTtCTCagTCTGATC | AGTATTCGCA---GCCTACTTTC | ACAAACGTCGAACAACTGAAaT |
| 10_176_10 | ATCAnCATCTTGAAAAAATCAA | AATTTTGACG---ATTTTTAGCG | variable |
| 11_155_10 | ATCTcCCctTTCCTCCTncCnC | TAGCATTCGA---CTCCTAACTC | variable |
| 12_154_9 | Variable | GTCCGTCCGA---ACTCTCTTTT | (TC)nCATCCACAATAGTCWGC |
| 5_85_13 | ACCTGACAAAAAACCCCTATCA | GGATATATGG---TCTRGCATCC | naGGGCGTCTnGCGACACCCTG |
| C. briggsae | C. nigoni | ||
|---|---|---|---|
| Chromosome | 468 Sequence | Satellite Position | |
| Start | Size | ||
| I | 1610666 | 144 | I_2234586 |
| III | 14143016 | 342 | III_15035724 |
| IV | 241446 | 224 | IV_317549 |
| IV | 17352776 | 169 | IV_20134955 |
| V | 19137314 | 332 | V_21406428 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Subirana, J.A.; Messeguer, X. Evolution of Tandem Repeat Satellite Sequences in Two Closely Related Caenorhabditis Species. Diminution of Satellites in Hermaphrodites. Genes 2017, 8, 351. https://doi.org/10.3390/genes8120351
Subirana JA, Messeguer X. Evolution of Tandem Repeat Satellite Sequences in Two Closely Related Caenorhabditis Species. Diminution of Satellites in Hermaphrodites. Genes. 2017; 8(12):351. https://doi.org/10.3390/genes8120351
Chicago/Turabian StyleSubirana, Juan A., and Xavier Messeguer. 2017. "Evolution of Tandem Repeat Satellite Sequences in Two Closely Related Caenorhabditis Species. Diminution of Satellites in Hermaphrodites" Genes 8, no. 12: 351. https://doi.org/10.3390/genes8120351




