Are Pericentric Inversions Reorganizing Wedge Shell Genomes?
Abstract
:1. Introduction
2. Materials and Methods
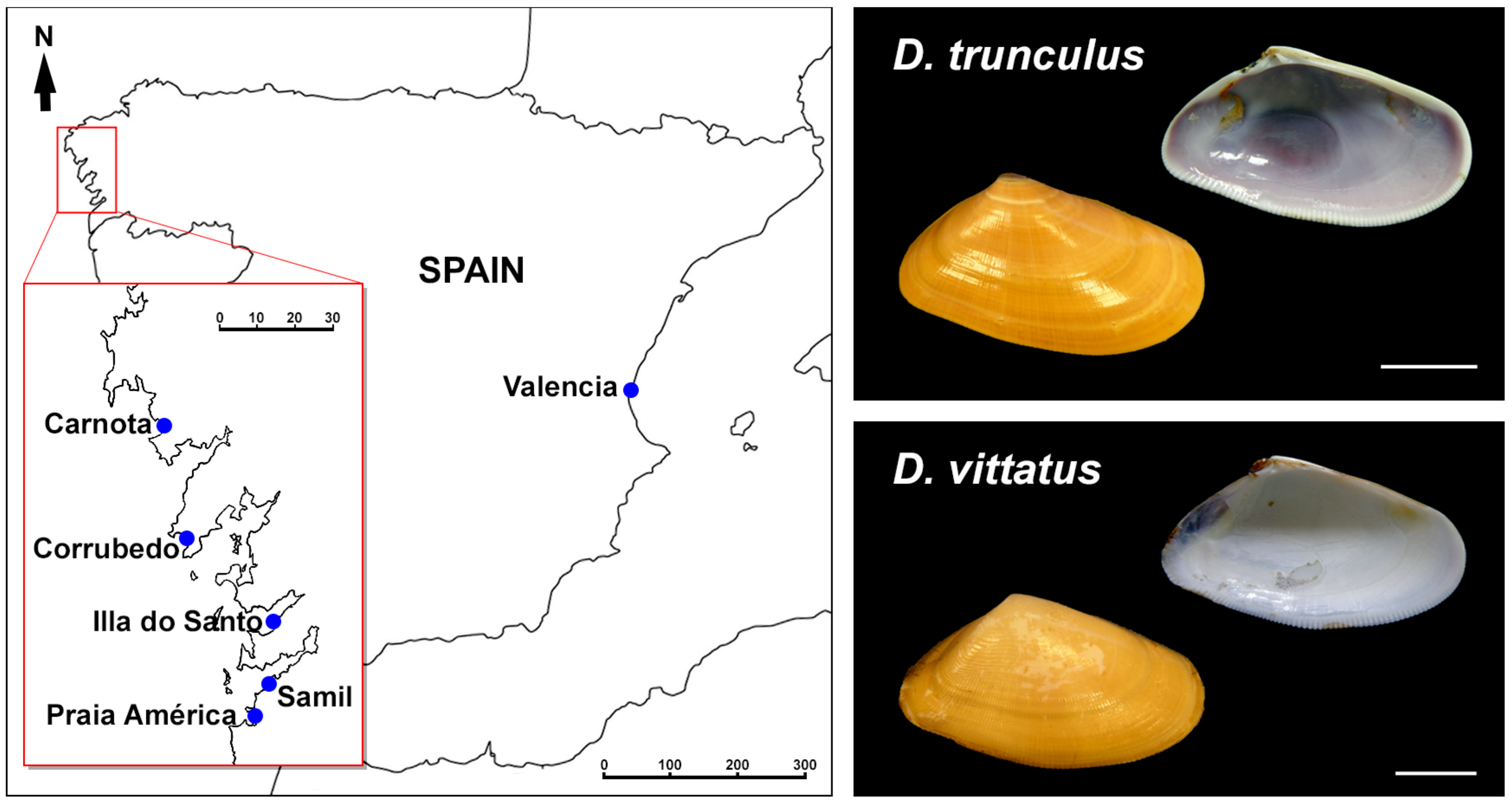
2.1. Biological Material
2.2. Mitotic Chromosome Preparation and Fluorochrome Staining
2.3. DNA Extraction, PCR Amplification and DNA Sequencing
2.4. Fluorescent in Situ Hybridization (FISH)
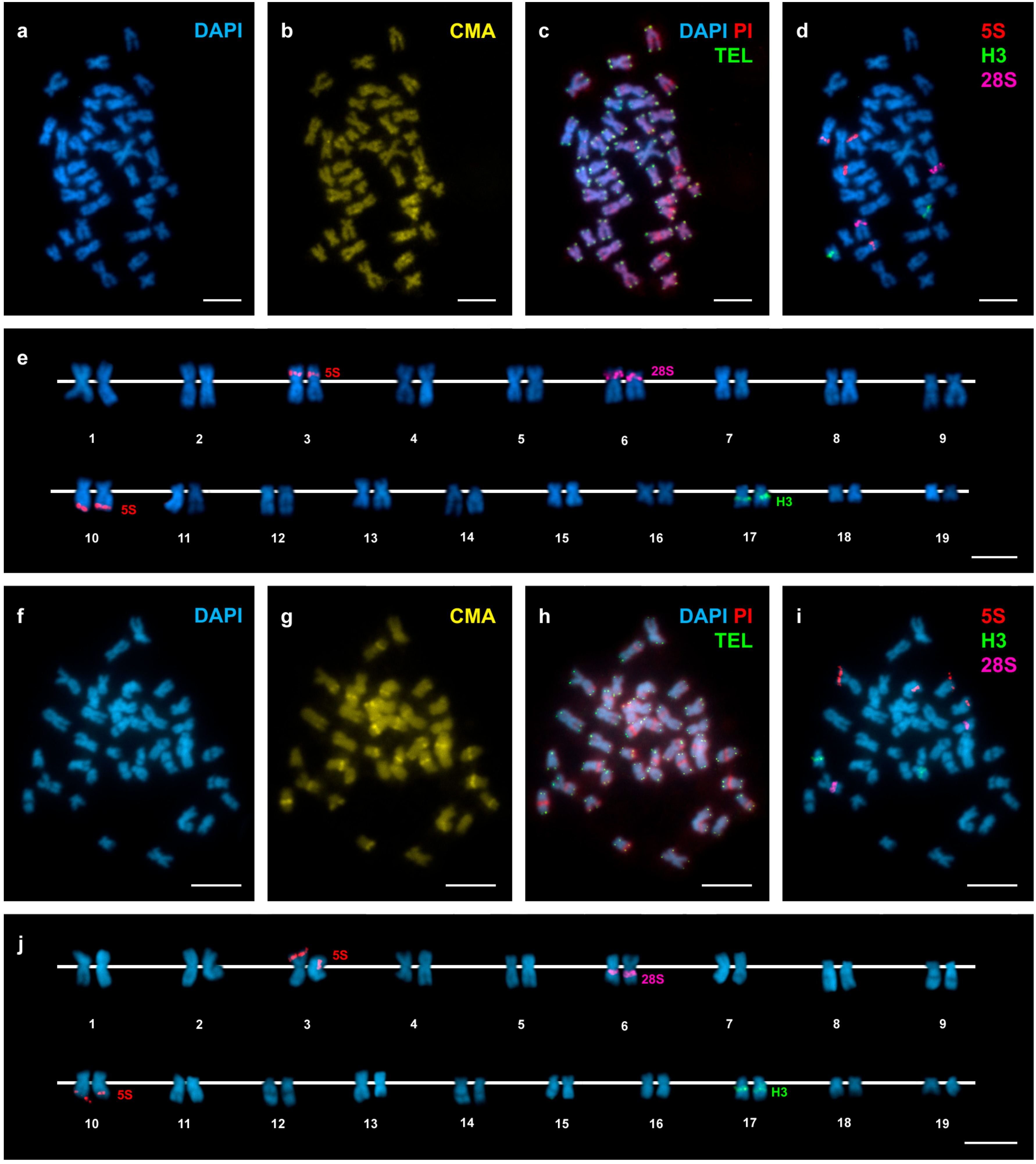
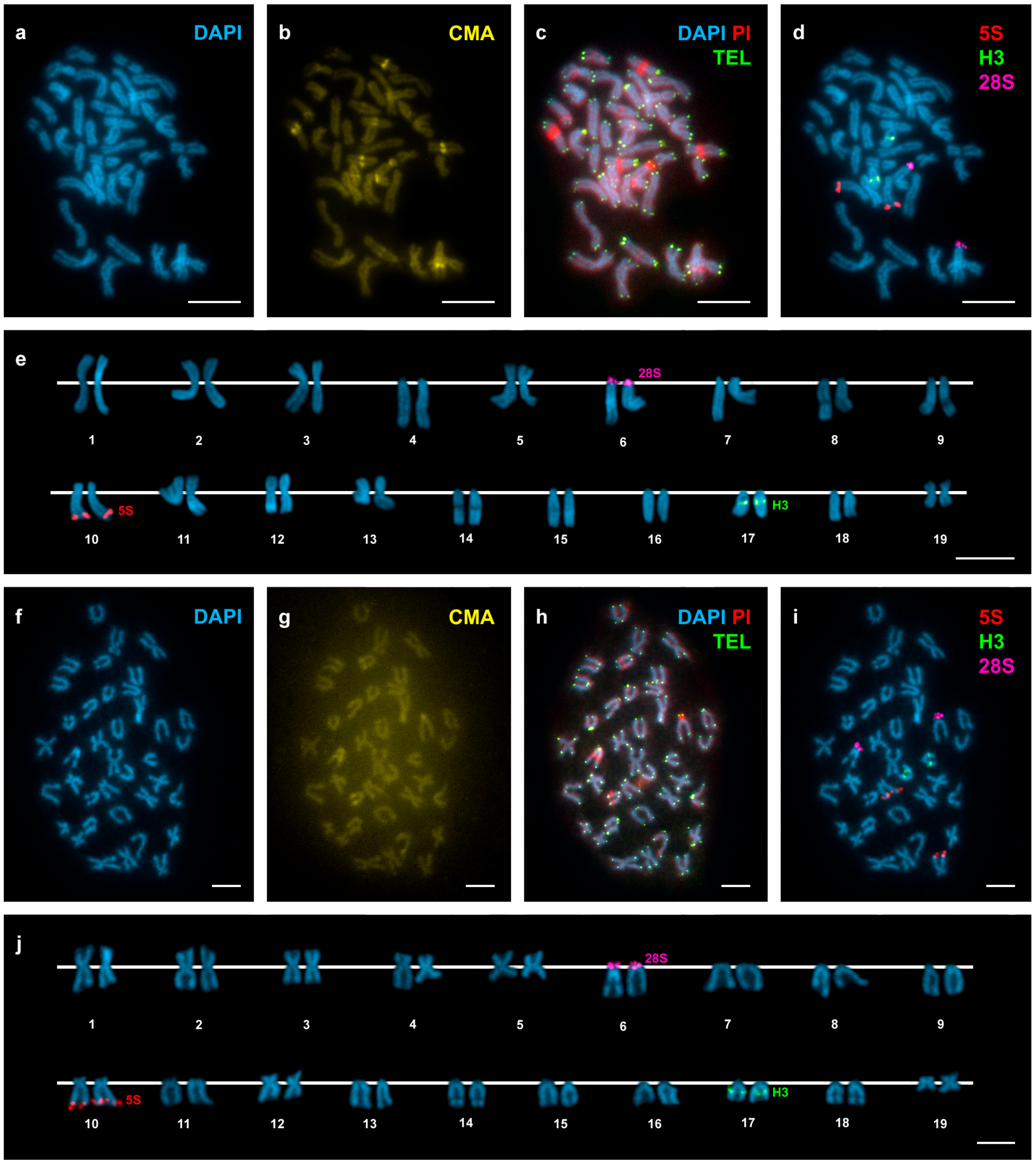
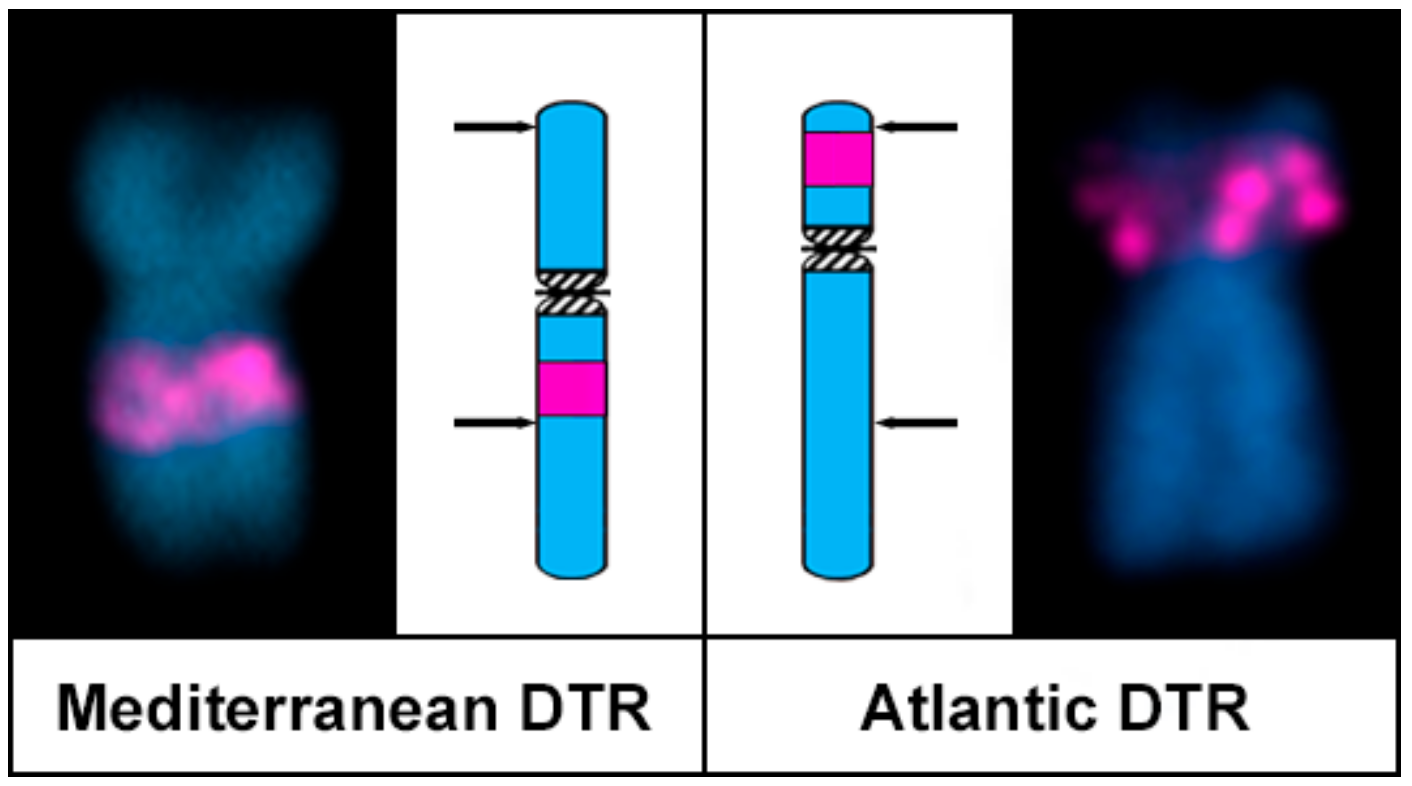
3. Results
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Kirkpatrick, M. How and why chromosome inversions evolve. PLoS Biol. 2010, 8, e1000501. [Google Scholar] [CrossRef] [PubMed]
- Alves, J.M.; Lopes, A.M.; Chikhi, L.; Amorim, A. On the structural plasticity of the human genome: Chromosomal inversions revisited. Curr. Genom. 2012, 13, 623–632. [Google Scholar] [CrossRef] [PubMed]
- Coghlan, A.; Eichler, E.E.; Oliver, S.G.; Paterson, A.H.; Stein, L. Chromosome evolution in eukaryotes: A multi-kingdom perspective. Trends Genet. 2005, 21, 673–682. [Google Scholar] [CrossRef] [PubMed]
- Ayala, F.J.; Coluzzi, M. Chromosome speciation: Humans, Drosophila, and mosquitoes. Proc. Natl. Acad. Sci. USA 2005, 102, 6535–6542. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, A.A.; Rieseberg, L. Revisiting the impact of inversions in evolution: From population genetic markers to drivers of adaptive shifts and speciation? Annu. Rev. Ecol. Evol. Syst. 2008, 39, 21–42. [Google Scholar] [CrossRef] [PubMed]
- Ansell, A.D. The biology of the genus Donax. In Developments in Hydrobiology. Volume 19: Sandy Beaches as Ecosystems; McLanchlan, A., Erasmus, T., Eds.; Junk Publishers: The Hague, The Netherlands, 1983; pp. 607–636. [Google Scholar]
- Huber, M. Compendium of Bivalves. A Full-Color Guide to 3300 of the World’s Marine Bivalves. A Status on Bivalvia after 250 Years of Research; ConchBooks: Hackenheim, Germany, 2010. [Google Scholar]
- Carstensen, D.; Laudien, J.; Leese, F.; Arntz, W.; Held, C. Genetic variability, shell and sperm morphology suggest that surf clams Donax marincovichi and D. obeselus are one species. J. Molluscan. Stud. 2009, 75, 381–390. [Google Scholar] [CrossRef]
- Salas-Casanova, C. The Donacidae of the bay of Malaga (Spain). Taxonomy. Basteria 1987, 51, 33–50. [Google Scholar]
- Tirado, C.; Salas, C. Reproduction of Donax. venustus Poli, 1795, Donax semistriatus Poli, 1795 and intermediate morphotypes (Bivalvia: Donacidae) in the littoral of Málaga (Southern Spain). Mar. Ecol. 1999, 20, 111–130. [Google Scholar] [CrossRef]
- Adamkewicz, S.L.; Harasewych, M.G. Use of random amplified polymorphic DNA (RAPD) markers to assess relationships among beach clams of the genus Donax. Nautilus 1994, 108 (Suppl. 2), 51–60. [Google Scholar]
- Pereira, A.M.; Fernández-Tajes, J.; Gaspar, M.B.; Méndez, J. Identification of the wedge clam Donax trunculus by a simple PCR technique. Food Control 2012, 23, 268–270. [Google Scholar] [CrossRef]
- Nantón, A.; Arias-Pérez, A.; Méndez, J.; Freire, R. Characterization of nineteen microsatellite markers and development of multiplex PCRs for the wedge clam Donax trunculus (Mollusca: Bivalvia). Mol. Biol. Rep. 2014, 41, 5351–5357. [Google Scholar] [CrossRef] [PubMed]
- Nantón, A.; Freire, R.; Arias-Pérez, A.; Gaspar, M.B.; Méndez, J. Identification of four Donax species by PCR–RFLP analysis of cytochrome c oxidase subunit I (COI). Eur. Food Res. Technol. 2015, 240, 1129–1133. [Google Scholar] [CrossRef]
- Fernández-Pérez, J.; Froufe, E.; Nantón, A.; Gaspar, M.B.; Méndez, J. Genetic diversity and population genetic analysis of Donax vittatus (Mollusca: Bivalvia) and phylogeny of the genus with mitochondrial and nuclear markers. Estuar. Coast. Shelf Sci. 2017, 197, 126–135. [Google Scholar] [CrossRef]
- Fernández-Pérez, J.; Nantón, A.; Ruiz-Ruano, F.J.; Camacho, J.P.M.; Méndez, J. First complete female mitochondrial genome in four bivalve species genus Donax and their phylogenetic relationships within the Veneroida order. PLoS ONE 2017, 12, e0184464. [Google Scholar] [CrossRef] [PubMed]
- Plohl, M.; Cornudella, L. Characterization of a complex satellite DNA in the mollusc Donax trunculus: Analysis of sequence variations and divergence. Gene 1996, 169, 157–164. [Google Scholar] [CrossRef]
- Plohl, M.; Cornudella, L. Characterization of interrelated sequence motifs in four satellite DNAs and their distribution in the genome of the mollusc Donax trunculus. J. Mol. Evol. 1997, 44, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Plohl, M.; Prats, E.; Martínez-Lage, A.; González-Tizón, A.; Méndez, J.; Cornudella, L. Telomeric localization of the vertebrate-type hexamer repeat (TTAGGG)n in the wedgeshell clam Donax trunculus and other marine invertebrate genomes. J. Biol. Chem. 2002, 277, 19839–19846. [Google Scholar] [CrossRef] [PubMed]
- Petrović, V.; Plohl, M. Sequence divergence and conservation in organizationally distinct subfamilies of Donax trunculus satellite DNA. Gene 2005, 362, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Petrović, V.; Pérez-García, C.; Pasantes, J.J.; Šatović, E.; Prats, E.; Plohl, M. A GC-rich satellite DNA and karyology of the bivalve mollusk Donax. trunculus: A dominance of GC-rich heterochromatin. Cytogenet. Genome Res. 2009, 124, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Šatović, E.; Plohl, M. Tandem repeat-containing MITEs in the clam Donax trunculus. Genome Biol. Evol. 2013, 5, 2549–2559. [Google Scholar] [CrossRef] [PubMed]
- Menzel, R.W. Chromosome number in nine families of marine pelecypod mollusks. Nautilus 1968, 82, 45–50. [Google Scholar]
- Cornet, M.; Soulard, C. Chromosome number and karyotype of Donax trunculus L. (Mollusca, Bivalvia, Tellinacea). Genetica 1990, 82, 93–97. [Google Scholar] [CrossRef]
- Martínez, A.; Marinas, L.; González-Tizón, A.; Méndez, J. Cytogenetic characterization of Donax trunculus (Bivalvia: Donacidae) by means of karyotyping, fluorochrome banding and fluorescent in situ hybridization. J. Molluscan Stud. 2002, 68, 393–396. [Google Scholar] [CrossRef]
- Pasantes, J.; Martínez-Expósito, M.J.; Martínez-Lage, A.; Méndez, J. Chromosomes of Galician mussels. J. Molluscan Stud. 1990, 56, 123–126. [Google Scholar] [CrossRef]
- García-Souto, D. Cytogenetic Characterization of Veneroid Bivalves and Some of Their Parasites. Ph.D. Thesis, University of Vigo, Vigo, Spain, 14 July 2017. [Google Scholar]
- Martínez-Expósito, M.J.; Pasantes, J.J.; Méndez, J. NOR activity in larval and juvenile mussels (Mytilus galloprovincialis Lmk.). J. Exp. Mar. Biol. Ecol. 1994, 175, 155–165. [Google Scholar] [CrossRef]
- Martínez-Expósito, M.J.; Pasantes, J.J.; Méndez, J. Proliferation kinetics of mussel (Mytilus galloprovincialis) gill cells. Mar. Biol. 1994, 120, 41–45. [Google Scholar] [CrossRef]
- Pérez-García, C.; Guerra-Varela, J.; Morán, P.; Pasantes, J.J. Chromosomal mapping of rRNA genes, core histone genes and telomeric sequences in Brachidontes puniceus and Brachidontes rodriguezi (Bivalvia: Mytilidae). BMC Genet. 2010, 11, 109. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Pérez-García, C.; Morán, P.; Pasantes, J.J. Cytogenetic characterization of the invasive mussel species Xenostrobus. securis Lmk. (Bivalvia: Mytilidae). Genome 2011, 54, 771–778. [Google Scholar] [CrossRef] [PubMed]
- García-Souto, D.; Pérez-García, C.; Morán, P.; Pasantes, J.J. Divergent evolutionary behavior of H3 histone gene and rDNA clusters in venerid clams. Mol. Cytogenet. 2015, 8, 40. [Google Scholar] [CrossRef] [PubMed]
- García-Souto, D.; Pérez-García, C.; Kendall, J.; Pasantes, J.J. Molecular cytogenetics in trough shells (Mactridae, Bivalvia): Divergent GC-rich heterochromatin content. Genes 2016, 7, 47. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, H.K. A review of molluscan cytogenetic information based on the CISMOCH-computerized system for molluscan chromosomes. Bivalvia, Polyplacophora and Cephalopoda. Venus 1985, 44, 193–225. [Google Scholar]
- Thiriot-Quiévreux, C. Advances in cytogenetics of aquatic organisms. In Genetics and Evolution of Aquatic Organisms; Beaumont, A.R., Ed.; Chapman and Hall: London, UK, 1994; pp. 369–388. ISBN 0 412 49370 5. [Google Scholar]
- Thiriot-Quiévreux, C. Review of the literature on bivalve cytogenetics in the last ten years. Cah. Biol. Mar. 2002, 43, 17–26. [Google Scholar]
- Leitão, A.; Chaves, R. Banding for chromosomal identification in bivalves: A 20-year history. In Aquaculture 1. Dynamic Biochemistry, Process Biotechnology and Molecular Biology 2 (Special Issue 1); Russo, R., Ed.; Global Science Books: Ikenobe, Japan, 2008; pp. 44–49. [Google Scholar]
- Carrilho, J.; Pérez-García, C.; Leitão, A.; Malheiro, I.; Pasantes, J.J. Cytogenetic characterization and mapping of rDNAs, core histone genes and telomeric sequences in Venerupis aurea and Tapes rhomboides (Bivalvia: Veneridae). Genetica 2011, 139, 823–830. [Google Scholar] [CrossRef] [PubMed]
- González-Tizón, A.; Rojo, V.; Vierna, J.; Jensen, K.T.; Egea, E.; Martínez-Lage, A. Cytogenetic characterisation of the razor shells Ensis directus (Conrad, 1843) and E. minor (Chenu, 1843) (Mollusca: Bivalvia). Helgol. Mar. Res. 2013, 67, 73–82. [Google Scholar] [CrossRef]
- Pérez-García, C.; Hurtado, N.S.; Morán, P.; Pasantes, J.J. Evolutionary dynamics of rDNA clusters in chromosomes of five clam species belonging to the family Veneridae (Mollusca, Bivalvia). BioMed Res. Int. 2014, 754012. [Google Scholar] [CrossRef] [PubMed]
- García-Souto, D.; Qarkaxhija, V.; Pasantes, J.J. Resolving the taxonomic status of Chamelea gallina and C. striatula (Veneridae, Bivalvia): A combined molecular cytogenetic and phylogenetic approach. BioMed Res. Int. 2017, 7638790. [Google Scholar] [CrossRef]
- García-Souto, D.; Rios, G.; Pasantes, J.J. Karyotype differentiation in tellin shells (Bivalvia: Tellinidae). BMC Genet. 2017, 18, 66. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Guo, X. Chromosomal mapping of the major ribosomal RNA genes in the dwarf surfclam (Mulinia lateralis Say). J. Shellfish Res. 2008, 27, 307–311. [Google Scholar] [CrossRef]
- Fernández-Tajes, J.; Martínez-Lage, A.; Freire, R.; Guerra, A.; Méndez, J.; González-Tizón, A.M. Genome sizes and karyotypes in the razor clams Ensis arcuatus (Jeffreys, 1985) and E. siliqua (Linnaeus, 1758). Cah. Biol. Mar. 2008, 49, 79–85. [Google Scholar] [CrossRef]
- Ventura, M.; Antonacci, F.; Cardone, M.F.; Stanyon, R.; Addabbo, P.D.; Cellamare, A.; Sprague, L.J.; Eichler, E.E.; Archidiacono, N.; Rocchi, M. Evolutionary formation of new centromeres in macaque. Science 2007, 316, 243–246. [Google Scholar] [CrossRef] [PubMed]
- Schubert, I.; Lysak, M.A. Interpretation of karyotype evolution should consider chromosome structural constraints. Trends Genet. 2010, 27, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Woznicki, P.; Boron, A. Banding chromosome patterns of zebra mussel Dreissena polymorpha (Pallas) from the heated Konin lakes system (Poland). Caryologia 2003, 56, 427–430. [Google Scholar] [CrossRef]
- Boron, A.; Woznicki, P.; Skuza, L.; Zielinski, R. Cytogenetic characterization of the zebra mussel Dreissena polymorpha (Pallas) from Miedwie Lake, Poland. Folia Biol. 2004, 52, 33–38. [Google Scholar]
- García-Souto, D.; Mravinac, B.; Šatović, E.; Plohl, M.; Morán, P.; Pasantes, J.J. Methylation profile of a satellite DNA constituting the intercalary G+C-rich heterochromatin of the cut trough shell Spisula subtruncata (Bivalvia, Mactridae). Sci. Rep. 2017, 7, 6930. [Google Scholar] [CrossRef] [PubMed]
- Novák, P.; Neumann, P.; Pech, J.; Steinhaisl, J.; Macas, J. RepeatExplorer: A Galaxy-based web server for genome-wide characterization of eukaryotic repetitive elements from next-generation sequence reads. Bioinformatics 2013, 29, 792–793. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Ruano, F.J.; López-León, M.D.; Cabrero, J.; Camacho, J.P.M. High-throughput analysis of the satellitome illuminates satellite DNA evolution. Sci. Rep. 2016, 6, 28333. [Google Scholar] [CrossRef] [PubMed]
- Utsunomia, R.; Ruiz-Ruano, F.; Silva, D.M.; Serrano, E.A.; Rosa, I.F.; Scudeler, P.E.; Hashimoto, D.T.; Oliveira, C.; Camacho, J.P.M.; Foresti, F. A glimpse into the satellite DNA library in Characidae fish (Teleostei, Characiformes). Front. Genet. 2017, 8, 103. [Google Scholar] [CrossRef] [PubMed]
- Lowry, D.B.; Willis, J.H. A widespread chromosomal inversion polymorphism contributes to a major life-history transition, local adaptation, and reproductive isolation. PLoS Biol. 2010, 8, e1000500. [Google Scholar] [CrossRef] [PubMed]
- Noor, M.A.; Grams, K.L.; Bertucci, L.A.; Reiland, J. Chromosomal inversions and the reproductive isolation of species. Proc. Natl. Acad. Sci. USA 2001, 98, 12084–12088. [Google Scholar] [CrossRef] [PubMed]
- Knebel, S.; Pasantes, J.J.; Thi, D.A.; Schaller, F.; Schempp, W. Heterogeneity of pericentric inversions of the human Y chromosome. Cytogenet. Genome Res. 2011, 132, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Lange, J.; Noordam, M.J.; van Daalen, S.K.; Skaletsky, H.; Clark, B.A.; Macville, M.V.; Page, D.C.; Repping, S. Intrachromosomal homologous recombination between inverted amplicons on opposing Y-chromosome arms. Genomics 2013, 102, 257–264. [Google Scholar] [CrossRef] [PubMed]

| Sequence | Denaturation | Annealing | Elongation |
|---|---|---|---|
| COI gene | 95 °C, 30 s | 55 °C, 20 s | 72 °C, 20 s |
| 28S rDNA | 95 °C, 20 s | 48 °C, 20 s | 72 °C, 15 s |
| 5S rDNA | 95 °C, 30 s | 48 °C, 30 s | 72 °C, 30 s |
| h3 | 95 °C, 15 s | 48 °C, 15 s | 72 °C, 15 s |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
García-Souto, D.; Pérez-García, C.; Pasantes, J.J. Are Pericentric Inversions Reorganizing Wedge Shell Genomes? Genes 2017, 8, 370. https://doi.org/10.3390/genes8120370
García-Souto D, Pérez-García C, Pasantes JJ. Are Pericentric Inversions Reorganizing Wedge Shell Genomes? Genes. 2017; 8(12):370. https://doi.org/10.3390/genes8120370
Chicago/Turabian StyleGarcía-Souto, Daniel, Concepción Pérez-García, and Juan J. Pasantes. 2017. "Are Pericentric Inversions Reorganizing Wedge Shell Genomes?" Genes 8, no. 12: 370. https://doi.org/10.3390/genes8120370





