Deciphering the Relationship between Obesity and Various Diseases from a Network Perspective
Abstract
:1. Introduction
2. Materials and Methods
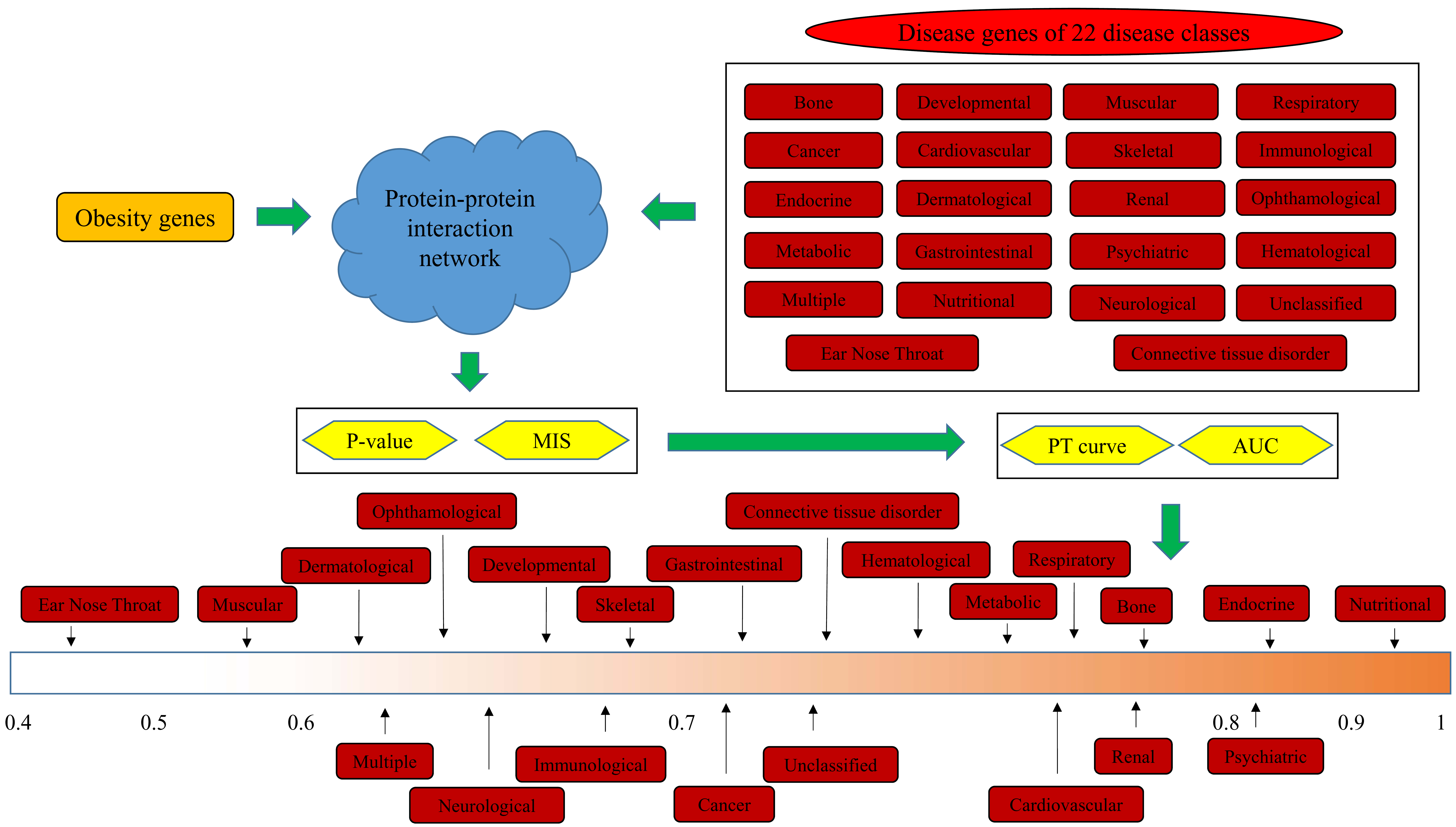
2.1. Disease Genes of 22 Disease Classes
2.2. Obesity Genes
2.3. PPI Network
2.4. Interaction Method
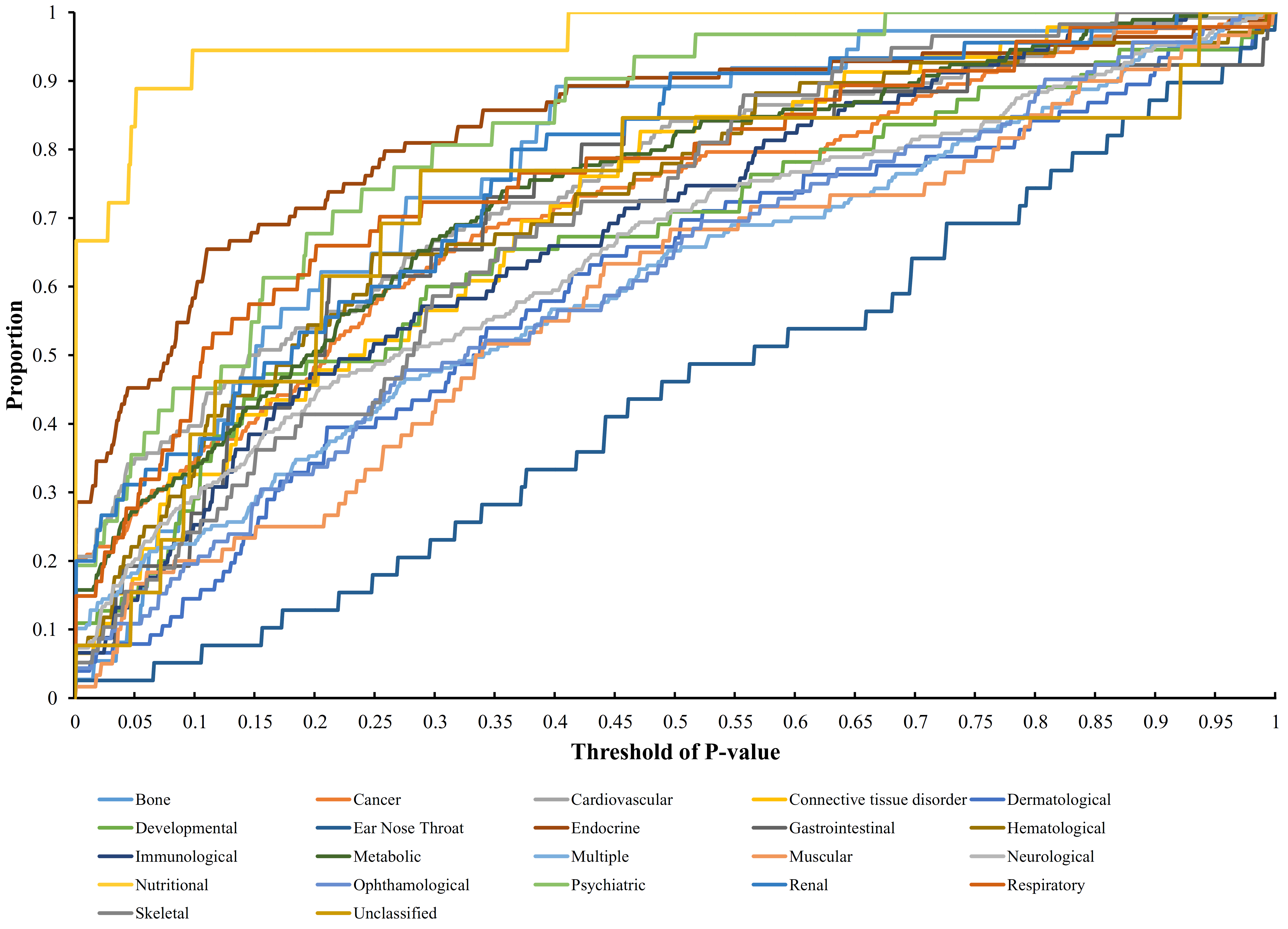
2.5. Quantitative Analysis Method
3. Results
3.1. Results of the Interaction Method
3.2. Quantitative Analysis Results
4. Discussion
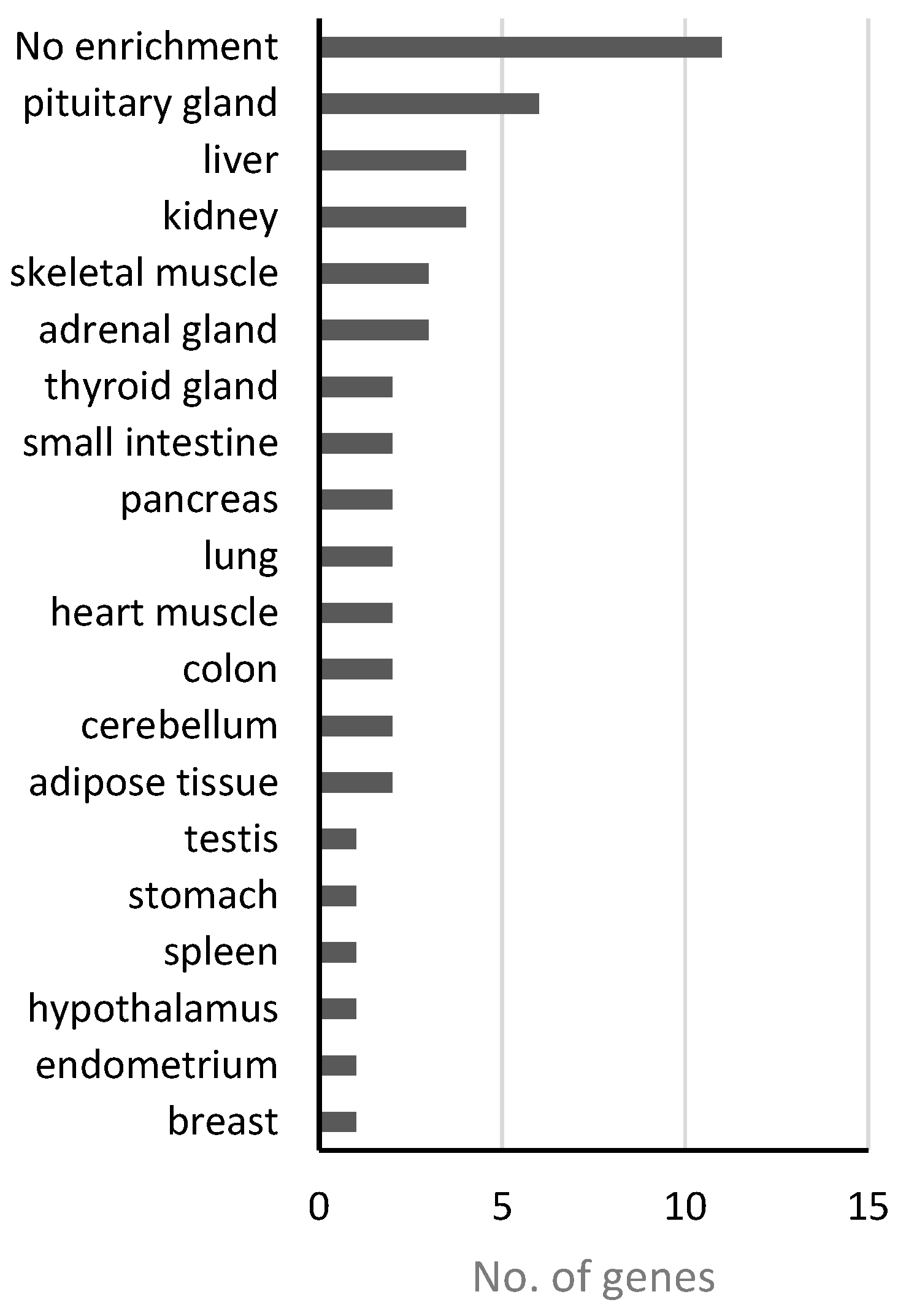
4.1. Analysis of Disease Genes of the “Nutritional” Class
4.2. Analysis of Disease Genes of the “Endocrine” Class
4.3. Analysis of Disease Genes of the “Psychiatric” Class
4.4. Analysis of Disease Genes of the “Bone” Class
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Phillips, C.M. Metabolically healthy obesity: Definitions, determinants and clinical implications. Rev. Endocr. Metab. Disord. 2013, 14, 219–227. [Google Scholar] [CrossRef] [PubMed]
- Lotta, L.A.; Abbasi, A.; Sharp, S.J.; Sahlqvist, A.S.; Waterworth, D.; Brosnan, J.M.; Scott, R.A.; Langenberg, C.; Wareham, N.J. Definitions of metabolic health and risk of future type 2 diabetes in bmi categories: A systematic review and network meta-analysis. Diabetes Care 2015, 38, 2177–2187. [Google Scholar] [CrossRef] [PubMed]
- Porto, L.G.G.; Nogueira, R.M.; Nogueira, E.C.; Molina, G.E.; Farioli, A.; Junqueira, L.F.; Kales, S.N. Agreement between bmi and body fat obesity definitions in a physically active population. Arch. Endocrinol. Metab. 2016, 60, 515–525. [Google Scholar] [CrossRef] [PubMed]
- Keke, L.M.; Samouda, H.; Jacobs, J.; di Pompeo, C.; Lemdani, M.; Hubert, H.; Zitouni, D.; Guinhouya, B.C. Body mass index and childhood obesity classification systems: A comparison of the french, international obesity task force (IOTF) and world health organization (WHO) references. Rev. Epidemiol. Sante 2015, 63, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Buhendwa, R.A.; Roelants, M.; Thomis, M.; Nkiama, C.E. Nutritional status and height, weight and BMI centiles of school-aged children and adolescents of 6-18-years from kinshasa (DRC). Ann. Hum. Biol. 2017, 44, 554–561. [Google Scholar] [CrossRef] [PubMed]
- Wilson, W.M.; Bulkan, J.; Piperata, B.A.; Hicks, K.; Ehlers, P. Nutritional status of makushi amerindian children and adolescents of Guyana. Ann. Hum. Biol. 2011, 38, 615–629. [Google Scholar] [CrossRef] [PubMed]
- Mattes, R. Energy intake and obesity: Ingestive frequency outweighs portion size. Physiol. Behav. 2014, 134, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Milic, S.; Lulic, D.; Stimac, D. Non-alcoholic fatty liver disease and obesity: Biochemical, metabolic and clinical presentations. World J. Gastroenterol. 2014, 20, 9330–9337. [Google Scholar] [PubMed]
- Bastien, M.; Poirier, P.; Lemieux, I.; Despres, J.P. Overview of epidemiology and contribution of obesity to cardiovascular disease. Prog. Cardiovasc. Dis. 2014, 56, 369–381. [Google Scholar] [CrossRef] [PubMed]
- Jung, U.J.; Choi, M.S. Obesity and its metabolic complications: The role of adipokines and the relationship between obesity, inflammation, insulin resistance, dyslipidemia and nonalcoholic fatty liver disease. Int. J. Mol. Sci. 2014, 15, 6184–6223. [Google Scholar] [CrossRef] [PubMed]
- Petrarca, L.; Nenna, R. Coeliac disease and obesity in children. J. Pediatr. Gastroenterol. Nutr. 2015, 61, e4. [Google Scholar] [CrossRef] [PubMed]
- Regnell, S.E.; Peterson, P.; Trinh, L.; Broberg, P.; Leander, P.; Lernmark, A.; Mansson, S.; Elding Larsson, H. Magnetic resonance imaging reveals altered distribution of hepatic fat in children with type 1 diabetes compared to controls. Metabolism 2015, 64, 872–878. [Google Scholar] [CrossRef] [PubMed]
- Friedl, C.; Sampl, E.; Prandl, E.C.; Wagner, D.; Piswanger-Soelkner, J.C.; Rosenkranz, A.R.; Fahrleitner-Pammer, A. Osteoporosis, weight gain and atypical fat accumulations—A typical feature not only for cushing’s, but also madelung’s disease: A case report. Wien. Klin. Wochenschr. 2012, 124, 188–192. [Google Scholar] [CrossRef] [PubMed]
- Hamosh, A.; Scott, A.F.; Amberger, J.S.; Bocchini, C.A.; McKusick, V.A. Online mendelian inheritance in man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005, 33, D514–D517. [Google Scholar] [CrossRef] [PubMed]
- Barabasi, A.L.; Gulbahce, N.; Loscalzo, J. Network medicine: A network-based approach to human disease. Nat. Rev. Genet. 2011, 12, 56–68. [Google Scholar] [CrossRef] [PubMed]
- Oliver, S. Guilt-by-association goes global. Nature 2000, 403, 601–603. [Google Scholar] [CrossRef] [PubMed]
- Kohler, S.; Bauer, S.; Horn, D.; Robinson, P.N. Walking the interactome for prioritization of candidate disease genes. Am. J. Hum. Genet. 2008, 82, 949–958. [Google Scholar] [CrossRef] [PubMed]
- Gormen, T.H.; Leiserson, C.E.; Rivest, R.L.; Stein, C. Introduction to Algorithms; MIT Press: Cambridge, MA, USA, 1990. [Google Scholar]
- Chen, L.; Yang, J.; Xing, Z.; Yuan, F.; Shu, Y.; Zhang, Y.; Kong, X.; Huang, T.; Li, H.; Cai, Y.D. An integrated method for the identification of novel genes related to oral cancer. PLoS ONE 2017, 12, e0175185. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Yang, J.; Huang, T.; Kong, X.; Lu, L.; Cai, Y.-D. Mining for novel tumor suppressor genes using a shortest path approach. J. Biomol. Struct. Dyn. 2016, 34, 664–675. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Xing, Z.; Huang, T.; Shu, Y.; Huang, G.; Li, H.-P. Application of the shortest path algorithm for the discovery of breast cancer related genes. Curr. Bioinform. 2016, 11, 51–58. [Google Scholar] [CrossRef]
- Zhang, J.; Yang, J.; Huang, T.; Shu, Y.; Chen, L. Identification of novel proliferative diabetic retinopathy related genes on protein–protein interaction network. Neurocomputing 2016, 217, 63–72. [Google Scholar] [CrossRef]
- Shi, H.B.; Xu, J.; Zhang, G.D.; Xu, L.D.; Li, C.Q.; Wang, L.; Zhao, Z.; Jiang, W.; Guo, Z.; Li, X. Walking the interactome to identify human miRNA-disease associations through the functional link between miRNA targets and disease genes. BMC Syst. Biol. 2013, 7, 101. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Liu, M.X.; Yan, G.Y. Drug-target interaction prediction by random walk on the heterogeneous network. Mol. BioSyst. 2012, 8, 1970–1978. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Pan, H.; Zhang, Y.-H.; Feng, K.; Kong, X.; Huang, T.; Cai, Y.-D. Network-based method for identifying co-regeneration genes in bone, dentin, nerve and vessel tissues. Genes 2017, 8, 252. [Google Scholar] [CrossRef] [PubMed]
- Gui, T.; Dong, X.; Li, R.; Li, Y.; Wang, Z. Identification of hepatocellular carcinoma–related genes with a machine learning and network analysis. J. Comput. Biol. 2015, 22, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Liu, C.-L.; Li, L.-L.; Cai, M.-H.; Chen, W.-Z.; Xu, Y.-F.; O’Reilly, P.F.; Cai, L.; He, L. A new method for identifying causal genes of schizophrenia and anti-tuberculosis drug-induced hepatotoxicity. Sci. Rep. 2016, 6, 32571. [Google Scholar] [CrossRef] [PubMed]
- Goh, K.I.; Cusick, M.E.; Valle, D.; Childs, B.; Vidal, M.; Barabasi, A.L. The human disease network. Proc. Natl. Acad. Sci. USA 2007, 104, 8685–8690. [Google Scholar] [CrossRef] [PubMed]
- Amberger, J.S.; Bocchini, C.A.; Schiettecatte, F.; Scott, A.F.; Hamosh, A. Omim.Org: Online mendelian inheritance in man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015, 43, D789–D798. [Google Scholar] [CrossRef] [PubMed]
- Online Mendelian Inheritance in Man. Available online: http://www.omim.org/ (accessed on 15 January 2014).
- Rankinen, T.; Zuberi, A.; Chagnon, Y.C.; Weisnagel, S.J.; Argyropoulos, G.; Walts, B.; Pérusse, L.; Bouchard, C. The human obesity gene map: The 2005 update. Obesity 2006, 14, 529–644. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.-D.; Zhang, Q.; Zhang, Y.-H.; Chen, L.; Huang, T. Identification of genes associated with breast cancer metastasis to bone on a protein–protein interaction network with a shortest path algorithm. J. Proteome Res. 2017, 16, 1027–1038. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Zhang, Y.H.; Su, F.; Chen, L.; Huang, T.; Cai, Y.D. A shortest-path-based method for the analysis and prediction of fruit-related genes in arabidopsis thaliana. PLoS ONE 2016, 11, e0159519. [Google Scholar] [CrossRef] [PubMed]
- Ng, K.L.; Ciou, J.S.; Huang, C.H. Prediction of protein functions based on function-function correlation relations. Comput. Biol. Med. 2010, 40, 300–305. [Google Scholar] [CrossRef] [PubMed]
- Yuan, F.; Zhang, Y.H.; Wan, S.; Wang, S.; Kong, X.Y. Mining for candidate genes related to pancreatic cancer using protein–protein interactions and a shortest path approach. Biomed. Res. Int. 2015, 2015, 623121. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.; Chu, C.; Huang, T.; Kong, X.; Zhang, Y.; Zhang, N.; Cai, Y.D. Exploring mouse protein function via multiple approaches. PLoS ONE 2016, 11, e0166580. [Google Scholar] [CrossRef] [PubMed]
- Xenarios, I.; Rice, D.W.; Salwinski, L.; Baron, M.K.; Marcotte, E.M.; Eisenberg, D. Dip: The database of interacting proteins. Nucleic Acids Res. 2000, 28, 289–291. [Google Scholar] [CrossRef] [PubMed]
- Stark, C.; Breitkreutz, B.J.; Reguly, T.; Boucher, L.; Breitkreutz, A.; Tyers, M. Biogrid: A general repository for interaction datasets. Nucleic Acids Res. 2006, 34, D535–D539. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. String v10: Protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef] [PubMed]
- STRING. Available online: http://string-db.Org/ (accessed on 3 June 2016).
- Swets, J.A. Indices of discrimination or diagnostic accuracy: Their ROCs and implied models. Psychol. Bull. 1986, 99, 100–117. [Google Scholar] [CrossRef] [PubMed]
- Diethelm, K.; Huybrechts, I.; Moreno, L.; De Henauw, S.; Manios, Y.; Beghin, L.; Gonzalez-Gross, M.; Le Donne, C.; Cuenca-Garcia, M.; Castillo, M.J.; et al. Nutrient intake of European adolescents: Results of the HELENA (healthy lifestyle in Europe by nutrition in adolescence) study. Public Health Nutr. 2014, 17, 486–497. [Google Scholar] [CrossRef] [PubMed]
- Steyn, N.P.; McHiza, Z.J. Obesity and the nutrition transition in Sub-Saharan Africa. Ann. N. Y. Acad. Sci. 2014, 1311, 88–101. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Chanda, D.; Sim, J.; Park, Y.Y.; Choi, H.S. Structure and function of the atypical orphan nuclear receptor small heterodimer partner. Int. Rev. Cytol. 2007, 261, 117–158. [Google Scholar] [PubMed]
- Zhi, X.Y.; Zhou, X.E.; He, Y.Z.; Zechner, C.; Suino-Powell, K.M.; Kliewer, S.A.; Melcher, K.; Mangelsdorf, D.J.; Xu, H.E. Structural insights into gene repression by the orphan nuclear receptor SHP. Proc. Natl. Acad. Sci. USA 2014, 111, 839–844. [Google Scholar] [CrossRef] [PubMed]
- Myronovych, A.; Salazar-Gonzalez, R.M.; Ryan, K.K.; Miles, L.; Zhang, W.J.; Jha, P.; Wang, L.; Setchell, K.D.R.; Seeley, R.J.; Kohli, R. The role of small heterodimer partner in nonalcoholic fatty liver disease improvement after sleeve gastrectomy in mice. Obesity 2014, 22, 2301–2311. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.X.; Yang, Q.; Xu, C.L.; Shou, P.S.; Cao, J.C.; Jiang, M.H.; Chen, Q.; Cao, G.; Han, Y.Y.; Li, F.Y.; et al. CD11b regulates obesity-induced insulin resistance via limiting alternative activation and proliferation of adipose tissue macrophages. Proc. Natl. Acad. Sci. USA 2015, 112, E7239–E7248. [Google Scholar] [CrossRef] [PubMed]
- Tseng, H.T.; Park, Y.J.; Lee, Y.K.; Moore, D.D. The orphan nuclear receptor small heterodimer partner is required for thiazolidinedione effects in leptin-deficient mice. J. Biomed. Sci. 2015, 22. [Google Scholar] [CrossRef] [PubMed]
- Tolson, K.P.; Gemelli, T.; Meyer, D.; Yazdani, U.; Kozlitina, J.; Zinn, A.R. Inducible neuronal inactivation of sim1 in adult mice causes hyperphagic obesity. Endocrinology 2014, 155, 2436–2444. [Google Scholar] [CrossRef] [PubMed]
- Montagne, L.; Raimondo, A.; Delobel, B.; Duban-Bedu, B.; Noblet, F.S.; Dechaume, A.; Bersten, D.C.; Meyre, D.; Whitelaw, M.L.; Froguel, P.; et al. Identification of two novel loss-of-function SIM1 mutations in two overweight children with developmental delay. Obesity 2014, 22, 2621–2624. [Google Scholar] [PubMed]
- Luscombe-Marsh, N.D.; Seimon, R.V.; Bollmeyer, E.; Wishart, J.M.; Wittert, G.A.; Horowitz, M.; Bellon, M.; Feinle-Bisset, C. Acute effects of oral preloads with increasing energy density on gastric emptying, gut hormone release, thermogenesis and energy intake, in overweight and obese men. Asia Pac. J. Clin. Nutr. 2013, 22, 380–390. [Google Scholar] [PubMed]
- Muller, M.J.; Bosy-Westphal, A. Adaptive thermogenesis with weight loss in humans. Obesity 2013, 21, 218–228. [Google Scholar] [CrossRef] [PubMed]
- Chouchani, E.T.; Kazak, L.; Jedrychowski, M.P.; Lu, G.N.Z.; Erickson, B.K.; Szpyt, J.; Pierce, K.A.; Laznik-Bogoslavski, D.; Vetrivelan, R.; Clish, C.B.; et al. Mitochondrial ROS regulate thermogenic energy expenditure and sulfenylation of UCP1. Nature 2016, 532, 112–116. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.; Chung, J.Y.; Cho, S.K.; Shin, H.W.; Jang, I.J.; Park, J.W.; Yu, K.S.; Cho, J.Y. Antihyperglycemic mechanism of metformin occurs via the AMPK/LXR ALPHA/POMC pathway. Sci. Rep. 2015, 5, 8145. [Google Scholar] [CrossRef] [PubMed]
- Martins, L.M.; de Oliveira, A.R.S.; Cruz, K.J.C.; de Aratujo, C.G.B.; de Oliveira, F.E.; de Sousa, G.S.; Nogueira, N.D.; Marreiro, D.D. Influence of cortisol on zinc metabolism in morbidly obese women. Nutr. Hosp. 2014, 29, 57–63. [Google Scholar]
- Rodriguez, A.C.I.; Epel, E.S.; White, M.L.; Standen, E.C.; Seckl, J.R.; Tomiyama, A.J. Hypothalamic-pituitary-adrenal axis dysregulation and cortisol activity in obesity: A systematic review. Psychoneuroendocrino 2015, 62, 301–318. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, T.J.; Lin, E. The Pro12Ala polymorphism in the peroxisome proliferator-activated receptor gamma (PPARG) gene in relation to obesity and metabolic phenotypes in a Taiwanese population. Endocrine 2015, 48, 786–793. [Google Scholar] [CrossRef] [PubMed]
- Carlos, F.F.; Silva-Nunes, J.; Flores, O.; Brito, M.; Doria, G.; Veiga, L.; Baptista, P.V. Association of FTO and PPARG polymorphisms with obesity in portuguese women. Diabetes Metab. Syndr. Obes. 2013, 6, 241–245. [Google Scholar] [PubMed]
- Queiroz, E.M.; Candido, A.P.; Castro, I.M.; Bastos, A.Q.; Machado-Coelho, G.L.; Freitas, R.N. IGF2, LEPR, POMC, PPARG, and PPARGC1 gene variants are associated with obesity-related risk phenotypes in brazilian children and adolescents. Braz. J. Med. Biol. Res. 2015, 48, 595–602. [Google Scholar] [CrossRef] [PubMed]
- Pepino, M.Y.; Klein, S. Response to comment on Pepino et al. Sucralose affects glycemic and hormonal responses to an oral glucose load. Diabetes care 2013;36:2530–2535. Diabetes Care 2014, 37, e149. [Google Scholar]
- Uhlen, M.; Oksvold, P.; Fagerberg, L.; Lundberg, E.; Jonasson, K.; Forsberg, M.; Zwahlen, M.; Kampf, C.; Wester, K.; Hober, S.; et al. Towards a knowledge-based human protein atlas. Nat. Biotechnol. 2010, 28, 1248–1250. [Google Scholar] [CrossRef] [PubMed]
- The GTEx Consortium. The genotype-tissue expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science 2015, 348, 648–660. [Google Scholar]
- Lizio, M.; Harshbarger, J.; Shimoji, H.; Severin, J.; Kasukawa, T.; Sahin, S.; Abugessaisa, I.; Fukuda, S.; Hori, F.; Ishikawa-Kato, S.; et al. Gateways to the FANTOM5 promoter level mammalian expression atlas. Gen. Biol. 2015, 16, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zayani, N.; Hamdouni, H.; Boumaiza, I.; Achour, O.; Neffati, F.; Omezzine, A.; Najjar, M.F.; Bouslama, A. Resistin polymorphims, plasma resistin levels and obesity in Tunisian volunteers. J. Clin. Lab. Anal. 2017. [Google Scholar] [CrossRef] [PubMed]
- D’Incao, R.B.; Tovo, C.V.; Mattevi, V.S.; Borges, D.O.; Ulbrich, J.M.; Coral, G.P.; Ramos, M.J.; Meinhardt, N.G. Adipokine levels versus hepatic histopathology in bariatric surgery patients. Obes. Surg. 2017, 27, 2151–2158. [Google Scholar] [CrossRef] [PubMed]
- Lackey, D.E.; Lazaro, R.G.; Li, P.; Johnson, A.; Hernandez-Carretero, A.; Weber, N.; Vorobyova, I.; Tsukomoto, H.; Osborn, O. The role of dietary fat in obesity-induced insulin resistance. Am. J. Physiol. Endocrinol. Metab. 2016, 311, E989–E997. [Google Scholar] [CrossRef] [PubMed]
- Otero, Y.F.; Stafford, J.M.; McGuinness, O.P. Pathway-selective insulin resistance and metabolic disease: The importance of nutrient flux. J. Biol. Chem. 2014, 289, 20462–20469. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, C.; Doss, C.G.; Bandyopadhyay, S.; Agoramoorthy, G. Influence of miRNA in insulin signaling pathway and insulin resistance: Micro-molecules with a major role in type-2 diabetes. Wiley Interdiscip. Rev. RNA 2014, 5, 697–712. [Google Scholar] [CrossRef] [PubMed]
- Oliva-Olivera, W.; Lhamyani, S.; Coin-Araguez, L.; Castellano-Castillo, D.; Alcaide-Torres, J.; Yubero-Serrano, E.M.; El Bekay, R.; Tinahones, F.J. Neovascular deterioration, impaired NADPH oxidase and inflammatory cytokine expression in adipose-derived multipotent cells from subjects with metabolic syndrome. Metabolism 2017, 71, 132–143. [Google Scholar] [CrossRef] [PubMed]
- Muratsu, J.; Iwabayashi, M.; Sanada, F.; Taniyama, Y.; Otsu, R.; Rakugi, H.; Morishita, R. Hepatocyte growth factor prevented high-fat diet-induced obesity and improved insulin resistance in mice. Sci. Rep. 2017, 7, 130. [Google Scholar] [CrossRef] [PubMed]
- Costantini, S.; Malerba, G.; Contreas, G.; Corradi, M.; Marin Vargas, S.P.; Giorgetti, A.; Maffeis, C. Genetic and bioinformatics analysis of four novel GCK missense variants detected in Caucasian families with GCK-MODY phenotype. Clin. Genet. 2015, 87, 440–447. [Google Scholar] [CrossRef] [PubMed]
- Stanik, J.; Kusekova, M.; Huckova, M.; Valentinova, L.; Masindova, I.; Stanikova, D.; Ferenczova, J.; Gasperikova, D.; Klimes, I. Impact of type 2 diabetes on Glucokinase diabetes (GCK-MODY) phenotype in a Roma (GYPSY) family—Case report. Endocr. Regul. 2012, 46, 99–105. [Google Scholar] [CrossRef] [PubMed]
- Carmody, D.; Naylor, R.N.; Bell, C.D.; Berry, S.; Montgomery, J.T.; Tadie, E.C.; Hwang, J.L.; Greeley, S.A.; Philipson, L.H. GCK-MODY in the us national monogenic diabetes registry: Frequently misdiagnosed and unnecessarily treated. Acta Diabetol. 2016, 53, 703–708. [Google Scholar] [CrossRef] [PubMed]
- Szopa, M.; Osmenda, G.; Wilk, G.; Matejko, B.; Skupien, J.; Zapala, B.; Mlynarski, W.; Guzik, T.; Malecki, M.T. Intima-media thickness and endothelial dysfunction in GCK and hnf1a-mody patients. Eur. J. Endocrinol. 2015, 172, 277–283. [Google Scholar] [CrossRef] [PubMed]
- Stanley, C.A. Perspective on the genetics and diagnosis of congenital hyperinsulinism disorders. J. Clin. Endocrinol. Metab. 2016, 101, 815–826. [Google Scholar] [CrossRef] [PubMed]
- Fisher, M.M.; Watkins, R.A.; Blum, J.; Evans-Molina, C.; Chalasani, N.; DiMeglio, L.A.; Mather, K.J.; Tersey, S.A.; Mirmira, R.G. Elevations in circulating methylated and unmethylated preproinsulin DNA in new-onset type 1 diabetes. Diabetes 2015, 64, 3867–3872. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Babcook, M.A.; Shukla, S.; Shankar, E.; Wang, Z.; Liu, G.; Erokwu, B.O.; Flask, C.A.; Lu, L.; Daneshgari, F.; et al. Obesity-initiated metabolic syndrome promotes urinary voiding dysfunction in a mouse model. Prostate 2016, 76, 964–976. [Google Scholar] [CrossRef] [PubMed]
- Weaver, J.U.; Kopelman, P.G.; McLoughlin, L.; Forsling, M.L.; Grossman, A. Hyperactivity of the hypothalamo-pituitary-adrenal axis in obesity: A study of ACTH, AVP, beta-lipotrophin and cortisol responses to insulin-induced hypoglycaemia. Clin. Endocrinol. 1993, 39, 345–350. [Google Scholar] [CrossRef]
- Gassanov, N.; Semmo, N.; Semmo, M.; Nia, A.M.; Fuhr, U.; Er, F. Arginine vasopressin (AVP) and treatment with arginine vasopressin receptor antagonists (VAPTANS) in congestive heart failure, liver cirrhosis and syndrome of inappropriate antidiuretic hormone secretion (SIADH). Eur. J. Clin. Pharmacol. 2011, 67, 333–346. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, M.; Hidaka, K.; Tada, S.; Tasaki, Y.; Yamaguchi, T. Full-length CDNA cloning and distribution of human dopamine D4 receptor. Mol. Brain Res. 1995, 29, 157–162. [Google Scholar] [CrossRef]
- Broderick, P.A.; Piercey, M.F. Clozapine, haloperidol, and the D4 antagonist PNU-101387G: In vivo effects on mesocortical, mesolimbic, and nigrostriatal dopamine and serotonin release. J. Neural Transm. 1998, 105, 749–767. [Google Scholar] [CrossRef] [PubMed]
- Levitan, R.D.; Masellis, M.; Lam, R.W.; Kaplan, A.S.; Davis, C.; Tharmalingam, S.; Mackenzie, B.; Basile, V.S.; Kennedy, J.L. A birth-season/drd4 gene interaction predicts weight gain and obesity in women with seasonal affective disorder: A seasonal thrifty phenotype hypothesis. Neuropsychopharmacol 2006, 31, 2498–2503. [Google Scholar] [CrossRef] [PubMed]
- Frodl, T.; Szyf, M.; Carballedo, A.; Ly, V.; Dymov, S.; Vaisheva, F.; Morris, D.; Fahey, C.; Meaney, J.; Gill, M.; et al. DNA methylation of the serotonin transporter gene (SLC6A4) is associated with brain function involved in processing emotional stimuli. J. Psychiatry Neurosci. 2015, 40, 296–305. [Google Scholar] [CrossRef] [PubMed]
- Dias, H.; Muc, M.; Padez, C.; Manco, L. Association of polymorphisms in 5-HTT (SLC6A4) and maoa genes with measures of obesity in young adults of Portuguese origin. Arch. Physiol. Biochem. 2016, 122, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Galindo-Moreno, J.; Iurlaro, R.; El Mjiyad, N.; Diez-Perez, J.; Gabaldon, T.; Munoz-Pinedo, C. Apolipoprotein L2 contains a BH3-like domain but it does not behave as a BH3-only protein. Cell Death Dis. 2014, 5, e1275. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.H.; Kim, J.H.; Song, G.G. Pathway analysis of a genome-wide association study in schizophrenia. Gene 2013, 525, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Cui, Y.H.; Han, Y.H.; Fagerness, J.A.; Galloway, B.; Shen, Y.C.; Kojima, T.; Uchiyama, M.; Faraone, S.V.; Tsuang, M.T. Association of SNPS and haplotypes in APOL1, 2 and 4 with schizophrenia. Schizophr. Res. 2008, 104, 153–164. [Google Scholar] [CrossRef] [PubMed]
- Nagaraj, S.; Raghavan, A.V.; Rao, S.N.; Manjappara, U.V. Obestatin and nt8u influence glycerolipid metabolism and PPAR gamma signaling in mice. Int. J. Biochem. Cell Biol. 2014, 53, 414–422. [Google Scholar] [CrossRef] [PubMed]
- Ridker, P.M.; Chasman, D.I.; Rose, L.; Loscalzo, J.; Elias, J.A. Plasma levels of the proinflammatory chitin-binding glycoprotein YKL-40, variation in the chitinase 3-like 1 gene (CHI3L1), and incident cardiovascular events. J. Am. Heart Assoc. 2014, 3, e000897. [Google Scholar] [CrossRef] [PubMed]
- Alcolea, D.; Vilaplana, E.; Pegueroles, J.; Montal, V.; Sanchez-Juan, P.; Gonzalez-Suarez, A.; Pozueta, A.; Rodriguez-Rodriguez, E.; Bartres-Faz, D.; Vidal-Pineiro, D.; et al. Relationship between cortical thickness and cerebrospinal fluid YKL-40 in predementia stages of Alzheimer’s disease. Neurobiol. Aging 2015, 36, 2018–2023. [Google Scholar] [CrossRef] [PubMed]
- Melah, K.E.; Lu, S.Y.; Hoscheidt, S.M.; Alexander, A.L.; Adluru, N.; Destiche, D.J.; Carlsson, C.M.; Zetterberg, H.; Blennow, K.; Okonkwo, O.C.; et al. Cerebrospinal fluid markers of alzheimer’s disease pathology and microglial activation are associated with altered white matter microstructure in asymptomatic adults at risk for alzheimer’s disease. J. Alzheimers Dis. 2016, 50, 873–886. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.J.; Beasley, K.N.; Acevedo, E.O.; Franco, R.L.; Jones, T.L.; Mari, D.C.; Shibata, Y. Chitin enhances obese inflammation ex vivo. Hum. Immunol. 2014, 75, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Mansego, M.L.; De Marco, G.; Ivorra, C.; Lopez-Izquierdo, R.; Morcillo, S.; Rojo-Martinez, G.; Gonzalez-Albert, V.; Martinez, F.; Soriguer, F.; Martin-Escudero, J.C.; et al. The nutrigenetic influence of the interaction between dietary vitamin E and TXN and COMT gene polymorphisms on waist circumference: A case control study. J. Transl. Med. 2015, 13, 286. [Google Scholar] [CrossRef] [PubMed]
- Williams, C.J. The role of ankh in pathologic mineralization of cartilage. Curr. Opin. Rheumatol. 2016, 28, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.A. Update on ankylosing spondylitis: Current concepts in pathogenesis. Curr. Allergy Asthma Rep. 2015, 15, 489. [Google Scholar] [CrossRef] [PubMed]
- Pimentel-Santos, F.M.; Ligeiro, D.; Matos, M.; Mourao, A.F.; Vieira de Sousa, E.; Pinto, P.; Ribeiro, A.; Santos, H.; Barcelos, A.; Godinho, F.; et al. Ankh and susceptibility to and severity of ankylosing spondylitis. J. Rheumatol. 2012, 39, 131–134. [Google Scholar] [CrossRef] [PubMed]
- Korostishevsky, M.; Cohen, Z.; Malkin, I.; Ermakov, S.; Yarenchuk, O.; Livshits, G. Morphological and biochemical features of obesity are associated with mineralization genes’ polymorphisms. Int. J. Obes. 2010, 34, 1308–1318. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.H.; Ko, H.M.; Moon, J.S.; Yoo, H.I.; Jung, J.Y.; Kim, M.S.; Koh, J.T.; Kim, W.J.; Kim, S.H. Osteoprotegerin expressed by osteoclasts: An autoregulator of osteoclastogenesis. J. Dent. Res. 2014, 93, 1116–1123. [Google Scholar] [CrossRef] [PubMed]
- Brunetti, G.; Marzano, F.; Colucci, S.; Ventura, A.; Cavallo, L.; Grano, M.; Faienza, M.F. Genotype-phenotype correlation in juvenile Paget disease: Role of molecular alterations of the TNFRSF11B gene. Endocrine 2012, 42, 266–271. [Google Scholar] [CrossRef] [PubMed]
- Balsa, J.A.; Lafuente, C.; Gomez-Martin, J.M.; Galindo, J.; Peromingo, R.; Garcia-Moreno, F.; Rodriguez-Velasco, G.; Martinez-Botas, J.; Gomez-Coronado, D.; Escobar-Morreale, H.F.; et al. The role of serum osteoprotegerin and receptor-activator of nuclear factor-kappaB ligand in metabolic bone disease of women after obesity surgery. J. Bone Miner. Metab. 2016, 34, 655–661. [Google Scholar] [CrossRef] [PubMed]
- Halade, G.V.; El Jamali, A.; Williams, P.J.; Fajardo, R.J.; Fernandes, G. Obesity-mediated inflammatory microenvironment stimulates osteoclastogenesis and bone loss in mice. Exp. Gerontol. 2011, 46, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Millan, J.L. The role of phosphatases in the initiation of skeletal mineralization. Calcif. Tissue Int. 2013, 93, 299–306. [Google Scholar] [CrossRef] [PubMed]
- Yadav, M.C.; Huesa, C.; Narisawa, S.; Hoylaerts, M.F.; Moreau, A.; Farquharson, C.; Millan, J.L. Ablation of osteopontin improves the skeletal phenotype of phospho1(-/-) mice. J. Bone Miner. Res. 2014, 29, 2369–2381. [Google Scholar] [CrossRef] [PubMed]
- Nemr, R.; Al-Busaidi, A.S.; Sater, M.S.; Echtay, A.; Saldanha, F.L.; Racoubian, E.; Keleshian, S.H.; Almawi, W.Y. Lack of replication of common EXT2 gene variants with susceptibility to type 2 diabetes in lebanese arabs. Diabetes Metab. 2013, 39, 532–536. [Google Scholar] [CrossRef] [PubMed]
- Kang, Q.L.; Xu, J.; Zhang, Z.; He, J.W.; Fu, W.Z.; Zhang, Z.L. Mutation screening for the EXT1 and EXT2 genes in Chinese patients with multiple osteochondromas. Arch. Med. Res. 2013, 44, 542–548. [Google Scholar] [CrossRef] [PubMed]

| OMIM Disease Class | Number of Disease Genes |
|---|---|
| Bone | 38 |
| Cancer | 172 |
| Cardiovascular | 126 |
| Connective tissue disorder | 46 |
| Dermatological | 76 |
| Developmental | 55 |
| Ear Nose Throat | 39 |
| Endocrine | 84 |
| Gastrointestinal | 26 |
| Hematological | 68 |
| Immunological | 91 |
| Metabolic | 184 |
| Multiple | 187 |
| Muscular | 60 |
| Neurological | 232 |
| Nutritional | 18 |
| Ophthamological | 92 |
| Psychiatric | 31 |
| Renal | 45 |
| Respiratory | 47 |
| Skeletal | 58 |
| Unclassified | 14 |
| Gene Symbol | MIS | p-Value |
|---|---|---|
| UCP1 | 969 | <0.001 |
| POMC | 998 | <0.001 |
| PPARG | 999 | <0.001 |
| AGRP | 999 | <0.001 |
| MC4R | 999 | <0.001 |
| PCSK1 | 970 | <0.001 |
| PPARGC1B | 978 | <0.001 |
| UCP3 | 960 | <0.001 |
| GHRL | 994 | <0.001 |
| ADRB3 | 964 | <0.001 |
| PYY | 993 | <0.001 |
| ENPP1 | 979 | <0.001 |
| NR0B2 | 993 | 0.027 |
| SIM1 | 879 | 0.044 |
| HTR2A | 949 | 0.046 |
| Gene Symbol | MIS | p-Value | Gene Symbol | MIS | p-Value |
|---|---|---|---|---|---|
| RETN | 981 | <0.001 | FSHB | 999 | <0.001 |
| PPP1R3A | 954 | <0.001 | CYP11B1 | 973 | <0.001 |
| NEUROD1 | 994 | <0.001 | IRS1 | 999 | <0.001 |
| MC2R | 998 | <0.001 | KCNJ11 | 999 | <0.001 |
| CYP17A1 | 994 | <0.001 | AR | 999 | <0.001 |
| AVP | 997 | <0.001 | ABCC8 | 999 | <0.001 |
| INS | 999 | <0.001 | IL6 | 999 | <0.001 |
| PPARG | 999 | <0.001 | STAT5B | 999 | <0.001 |
| LIPC | 984 | <0.001 | HNF4A | 999 | <0.001 |
| AVPR2 | 804 | <0.001 | ENPP1 | 979 | <0.001 |
| PTPN1 | 999 | <0.001 | IRS2 | 999 | <0.001 |
| IGF2BP2 | 989 | <0.001 | CAPN10 | 923 | <0.001 |
| SSTR5 | 968 | 0.013 | PAX8 | 954 | 0.014 |
| GCK | 989 | 0.017 | TBX19 | 937 | 0.017 |
| TSHR | 980 | 0.017 | SLC2A4 | 994 | 0.026 |
| PAX4 | 925 | 0.03 | STAR | 942 | 0.032 |
| HGF | 992 | 0.033 | BMP15 | 699 | 0.034 |
| HNF1A | 993 | 0.036 | GPD2 | 924 | 0.038 |
| HMGA1 | 984 | 0.041 | GCGR | 960 | 0.043 |
| Gene Symbol | MIS | p-Value |
|---|---|---|
| DRD4 | 972 | <0.001 |
| SLC6A4 | 961 | <0.001 |
| AKT1 | 999 | <0.001 |
| APOL4 | 742 | <0.001 |
| COMT | 981 | <0.001 |
| BDNF | 999 | <0.001 |
| SLC6A3 | 984 | 0.017 |
| CHI3L1 | 842 | 0.024 |
| APOL2 | 662 | 0.035 |
| HCRT | 967 | 0.043 |
| HTR2A | 949 | 0.046 |
| Gene Symbol | MIS | p-Value |
|---|---|---|
| ALPL | 969 | <0.001 |
| ANKH | 610 | 0.015 |
| TNFRSF11B | 895 | 0.034 |
| EXT1 | 968 | 0.043 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, L.; Zhang, Y.-H.; Li, J.; Wang, S.; Zhang, Y.; Huang, T.; Cai, Y.-D. Deciphering the Relationship between Obesity and Various Diseases from a Network Perspective. Genes 2017, 8, 392. https://doi.org/10.3390/genes8120392
Chen L, Zhang Y-H, Li J, Wang S, Zhang Y, Huang T, Cai Y-D. Deciphering the Relationship between Obesity and Various Diseases from a Network Perspective. Genes. 2017; 8(12):392. https://doi.org/10.3390/genes8120392
Chicago/Turabian StyleChen, Lei, Yu-Hang Zhang, JiaRui Li, ShaoPeng Wang, YunHua Zhang, Tao Huang, and Yu-Dong Cai. 2017. "Deciphering the Relationship between Obesity and Various Diseases from a Network Perspective" Genes 8, no. 12: 392. https://doi.org/10.3390/genes8120392





