Solving the Telomere Replication Problem
Abstract
:1. Introduction
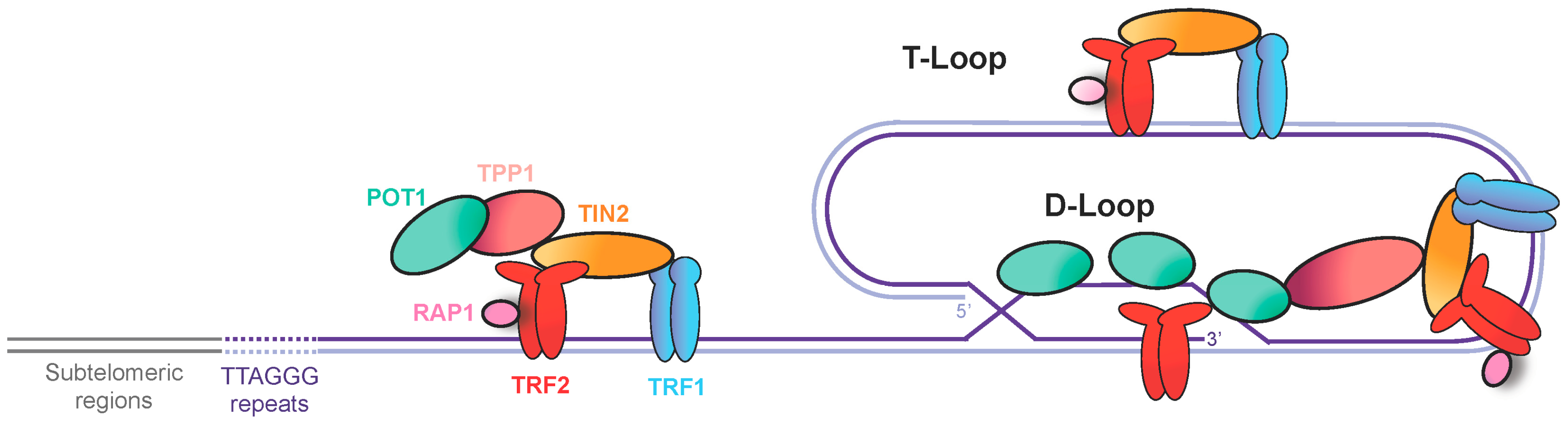
2. Telomeres, a Source of Replication Stress
3. Secondary Structures at Telomeres
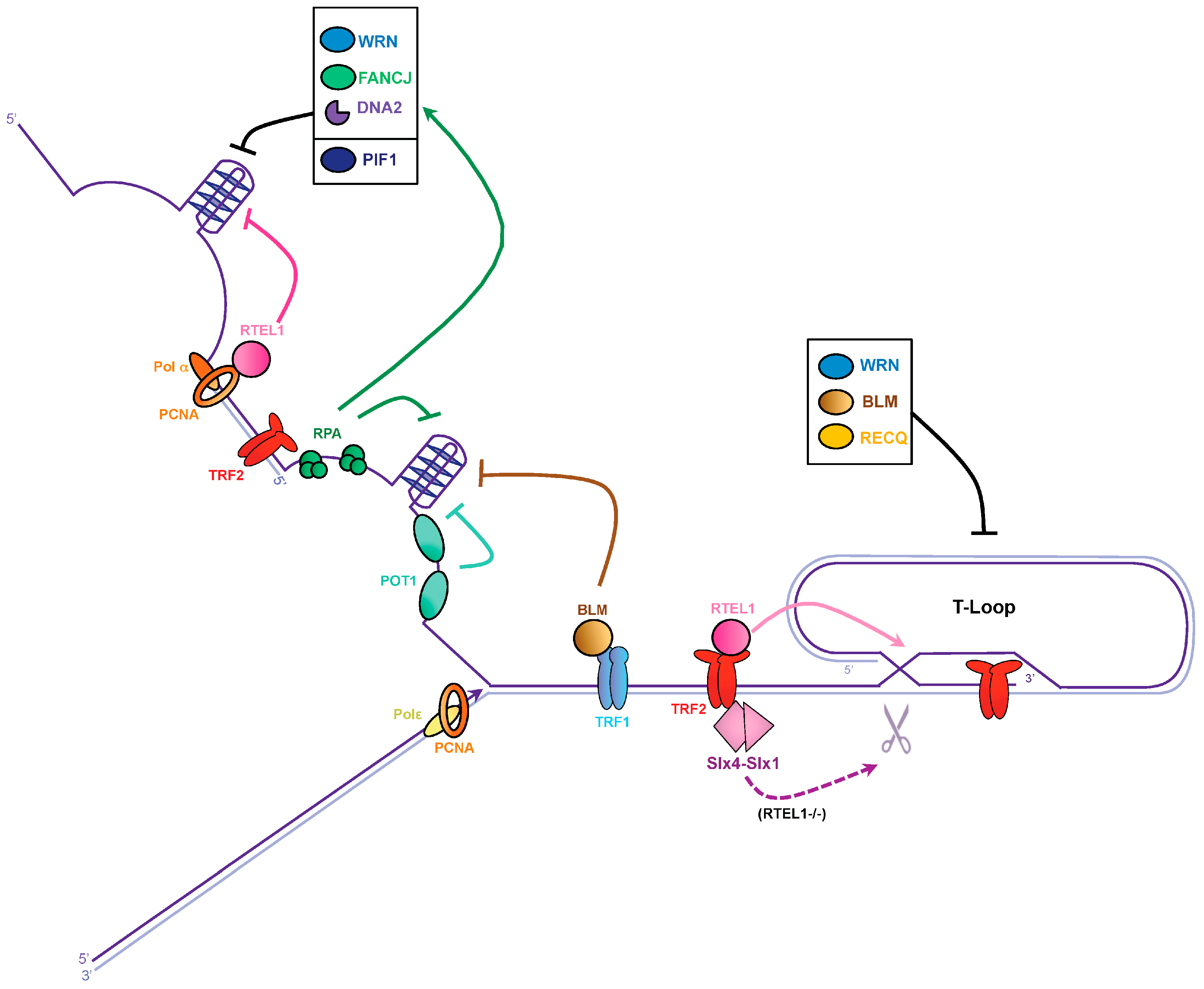
3.1. G-Quadruplex Dissolution by Helicases
3.2. G-Quadruplex Dissolution by Single-Strand Binding Proteins
3.3. T-Loop Dissolution
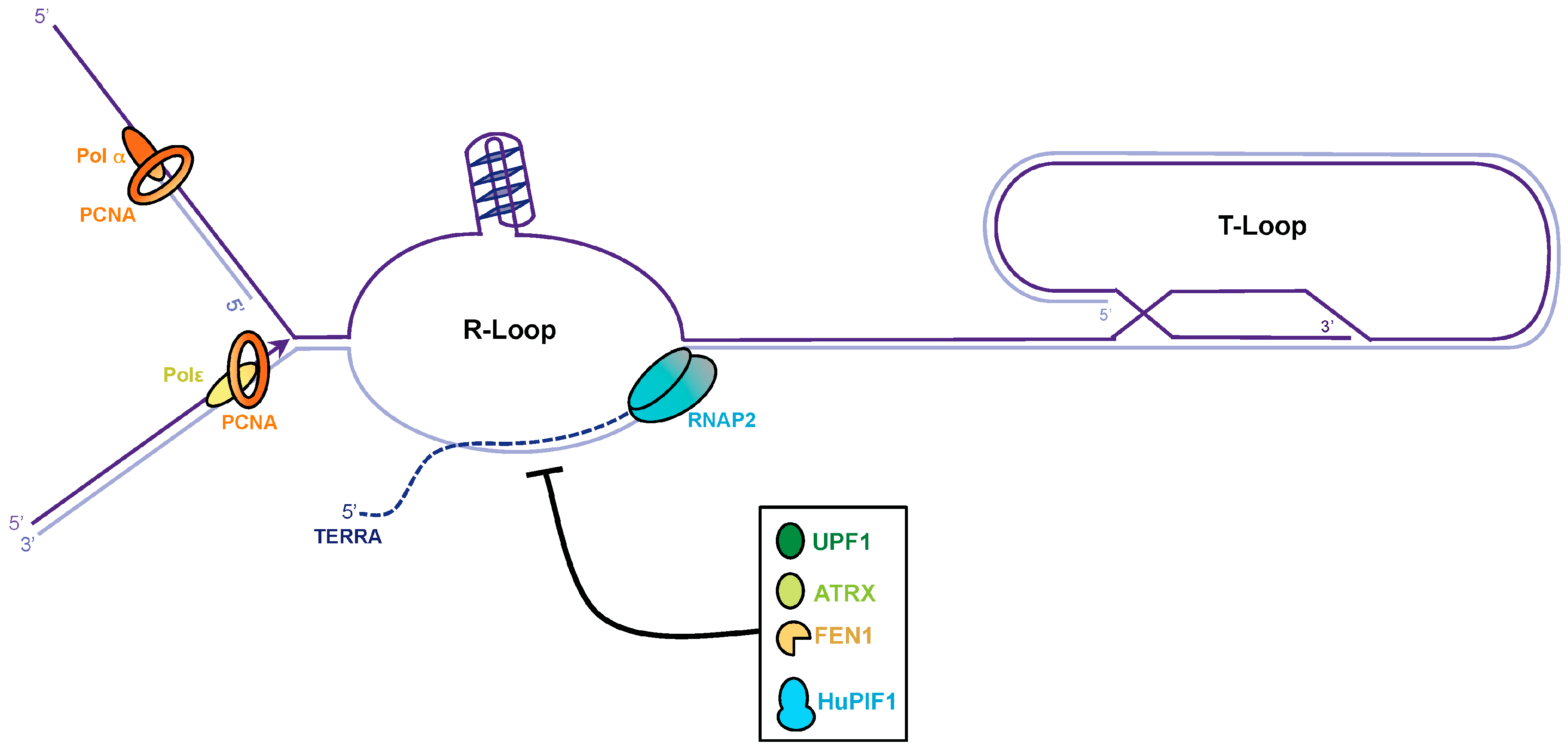
4. TERRA Transcription
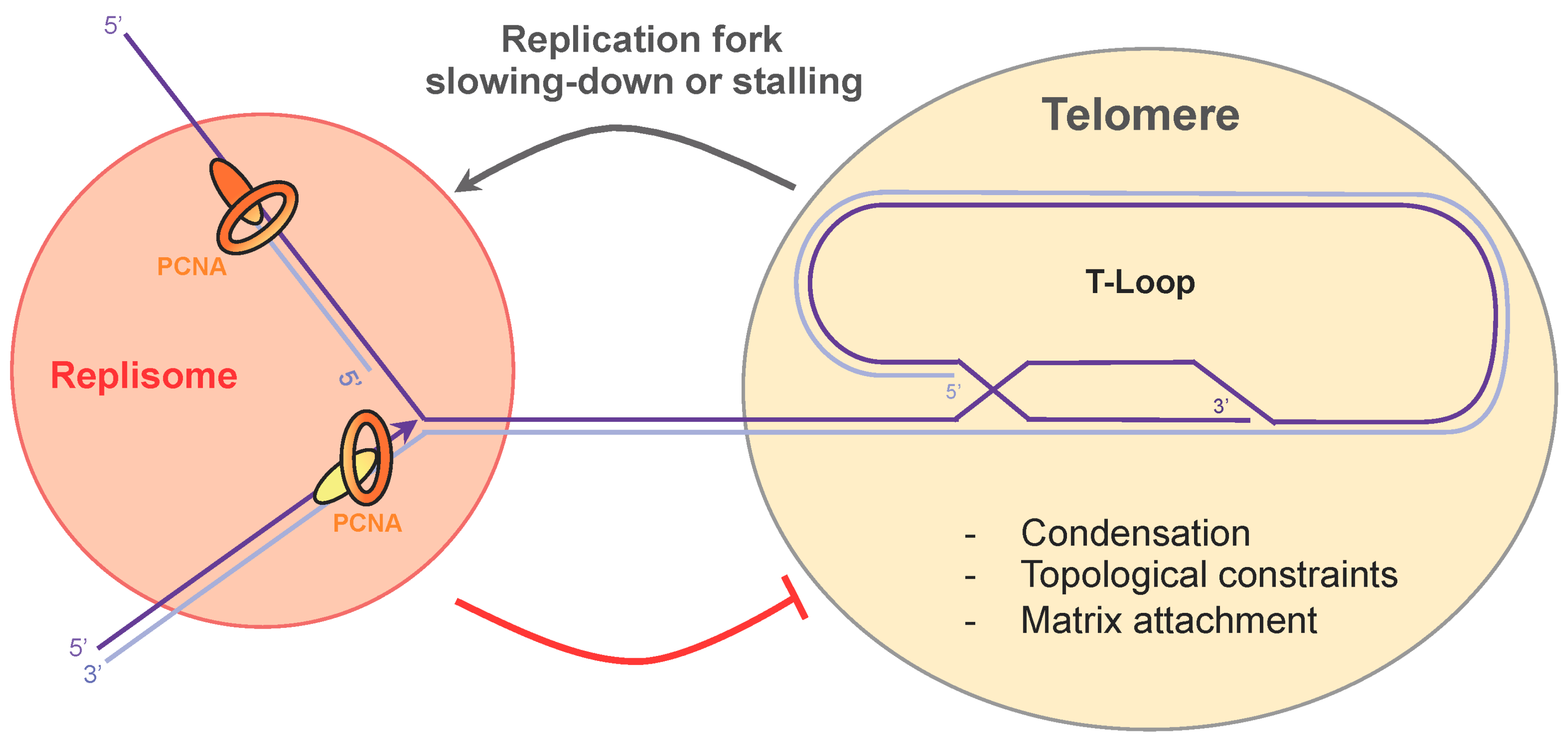
5. Telomere Compaction and Anchoring
6. Concluding Remarks
Acknowledgments
Conflicts of Interest
References
- Ishikawa, F.; Naito, T. Why do we have linear chromosomes? A matter of Adam and Eve. Mutat. Res. 1999, 434, 99–107. [Google Scholar] [CrossRef]
- De Lange, T. Opinion: T-loops and the origin of telomeres. Nat. Rev. Mol. Cell Biol. 2004, 5, 323–329. [Google Scholar] [CrossRef] [PubMed]
- de Lange, T. Shelterin: The protein complex that shapes and safeguards human telomeres. Genes Dev. 2005, 19, 2100–2110. [Google Scholar] [CrossRef] [PubMed]
- Arnoult, N.; Karlseder, J. Complex interactions between the DNA-damage response and mammalian telomeres. Nat. Struct. Mol. Biol. 2015, 22, 859–866. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, M.T.; Cesare, A.J.; Rivera, T.; Karlseder, J. Cell death during crisis is mediated by mitotic telomere deprotection. Nature 2015, 522, 492–496. [Google Scholar] [CrossRef] [PubMed]
- Lingner, J.; Cooper, J.P.; Cech, T.R. Telomerase and DNA end replication: no longer a lagging strand problem? Science 1995, 269, 1533–1534. [Google Scholar] [CrossRef] [PubMed]
- Gilson, E.; Géli, V. How telomeres are replicated. Nat. Rev. Mol. Cell Biol. 2007, 8, 825–838. [Google Scholar] [CrossRef] [PubMed]
- Soudet, J.; Jolivet, P.; Teixeira, M.T. Elucidation of the DNA end-replication problem in Saccharomyces cerevisiae. Mol. Cell 2014, 53, 954–964. [Google Scholar] [CrossRef] [PubMed]
- Zhong, F.L.; Batista, L.F.Z.; Freund, A.; Pech, M.F.; Venteicher, A.S.; Artandi, S.E. TPP1 OB-Fold Domain Controls Telomere Maintenance by Recruiting Telomerase to Chromosome Ends. Cell 2012, 150, 481–494. [Google Scholar] [CrossRef] [PubMed]
- Nandakumar, J.; Bell, C.F.; Weidenfeld, I.; Zaug, A.J.; Leinwand, L.A.; Cech, T.R. The TEL patch of telomere protein TPP1 mediates telomerase recruitment and processivity. Nature 2012, 492, 285–289. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-Y.; Redon, S.; Lingner, J. The human CST complex is a terminator of telomerase activity. Nature 2012, 488, 540–544. [Google Scholar] [CrossRef] [PubMed]
- Casteel, D.E.; Zhuang, S.; Zeng, Y.; Perrino, F.W.; Boss, G.R.; Goulian, M.; Pilz, R.B. A DNA Polymerase-α·Primase Cofactor with Homology to Replication Protein A-32 Regulates DNA Replication in Mammalian Cells. J. Biol. Chem. 2008, 284, 5807–5818. [Google Scholar] [CrossRef] [PubMed]
- Herbig, U.; Jobling, W.A.; Chen, B.P.C.; Chen, D.J.; Sedivy, J.M. Telomere Shortening Triggers Senescence of Human Cells through a Pathway Involving ATM, p53, and p21CIP1, but Not p16INK4a. Mol. Cell 2004, 14, 501–513. [Google Scholar] [CrossRef]
- Verdun, R.E.; Karlseder, J. The DNA damage machinery and homologous recombination pathway act consecutively to protect human telomeres. Cell 2006, 127, 709–720. [Google Scholar] [CrossRef] [PubMed]
- Arnoult, N.; Schluth-Bolard, C.; Letessier, A.; Drascovic, I.; Bouarich-Bourimi, R.; Campisi, J.; Kim, S.-H.; Boussouar, A.; Ottaviani, A.; Magdinier, F.; et al. Replication timing of human telomeres is chromosome arm-specific, influenced by subtelomeric structures and connected to nuclear localization. PLoS Genet. 2010, 6, e1000920. [Google Scholar] [CrossRef] [PubMed]
- Raghuraman, M.K. Replication Dynamics of the Yeast Genome. Science 2001, 294, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Moser, B.A.; Subramanian, L.; Chang, Y.-T.; Noguchi, C.; Noguchi, E.; Nakamura, T.M. Differential arrival of leading and lagging strand DNA polymerases at fission yeast telomeres. EMBO J. 2009, 28, 810–820. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, A.; Shore, D. Early replication of short telomeres in budding yeast. Cell 2007, 128, 1051–1062. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.-T.; Moser, B.A.; Nakamura, T.M. Fission Yeast Shelterin Regulates DNA Polymerases and Rad3ATR Kinase to Limit Telomere Extension. PLoS Genet. 2013, 9, e1003936. [Google Scholar] [CrossRef] [PubMed]
- Sridhar, A.; Kedziora, S.; Donaldson, A.D. At short telomeres Tel1 directs early replication and phosphorylates Rif1. PLoS Genet. 2014, 10, e1004691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lian, H.-Y.; Robertson, E.D.; Hiraga, S.-I.; Alvino, G.M.; Collingwood, D.; McCune, H.J.; Sridhar, A.; Brewer, B.J.; Raghuraman, M.K.; Donaldson, A.D. The effect of Ku on telomere replication time is mediated by telomere length but is independent of histone tail acetylation. Mol. Biol. Cell 2011, 22, 1753–1765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayano, M.; Kanoh, Y.; Matsumoto, S.; Renard-Guillet, C.; Shirahige, K.; Masai, H. Rif1 is a global regulator of timing of replication origin firing in fission yeast. Genes Dev. 2012, 26, 137–150. [Google Scholar] [CrossRef] [PubMed]
- Miller, K.M.; Rog, O.; Cooper, J.P. Semi-conservative DNA replication through telomeres requires Taz1. Nature 2006, 440, 824–828. [Google Scholar] [CrossRef] [PubMed]
- Ivessa, A.S.; Zhou, J.-Q.; Schulz, V.P.; Monson, E.K.; Zakian, V.A. Saccharomyces Rrm3p, a 5′ to 3′ DNA helicase that promotes replication fork progression through telomeric and subtelomeric DNA. Genes Dev. 2002, 16, 1383–1396. [Google Scholar] [CrossRef] [PubMed]
- Makovets, S.; Herskowitz, I.; Blackburn, E.H. Anatomy and Dynamics of DNA Replication Fork Movement in Yeast Telomeric Regions. Mol. Cell. Biol. 2004, 24, 4019–4031. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sfeir, A.; Kosiyatrakul, S.T.; Hockemeyer, D.; MacRae, S.L.; Karlseder, J.; Schildkraut, C.L.; de Lange, T. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell 2009, 138, 90–103. [Google Scholar] [CrossRef] [PubMed]
- Ohki, R.; Ishikawa, F. Telomere-bound TRF1 and TRF2 stall the replication fork at telomeric repeats. Nucleic Acids Res. 2004, 32, 1627–1637. [Google Scholar] [CrossRef] [PubMed]
- Martínez, P.; Thanasoula, M.; Muñoz, P.; Liao, C.; Tejera, A.; McNees, C.; Flores, J.M.; Fernández-Capetillo, O.; Tarsounas, M.; Blasco, M.A. Increased telomere fragility and fusions resulting from TRF1 deficiency lead to degenerative pathologies and increased cancer in mice. Genes Dev. 2009, 23, 2060–2075. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Lenain, C.; Bauwens, S.; Rizzo, A.; Saint-Léger, A.; Poulet, A.; Benarroch, D.; Magdinier, F.; Morere, J.; Amiard, S.; et al. TRF2 and apollo cooperate with topoisomerase 2α to protect human telomeres from replicative damage. Cell 2010, 142, 230–242. [Google Scholar] [CrossRef] [PubMed]
- Leman, A.R.; Dheekollu, J.; Deng, Z.; Lee, S.W.; Das, M.M.; Lieberman, P.M.; Noguchi, E. Timeless preserves telomere length by promoting efficient DNA replication through human telomeres. Cell Cycle 2012, 11, 2337–2347. [Google Scholar] [CrossRef] [PubMed]
- Leman, A.R.; Noguchi, E. Local and global functions of Timeless and Tipin in replication fork protection. Cell Cycle 2012, 11, 3945–3955. [Google Scholar] [CrossRef] [PubMed]
- Xhemalce, B.; Riising, E.M.; Baumann, P.; Dejean, A.; Arcangioli, B.; Seeler, J.-S. Role of SUMO in the dynamics of telomere maintenance in fission yeast. Proc. Natl. Acad. Sci. USA 2007, 104, 893–898. [Google Scholar] [CrossRef] [PubMed]
- Gadaleta, M.C.; Das, M.M.; Tanizawa, H.; Chang, Y.-T.; Noma, K.-I.; Nakamura, T.M.; Noguchi, E. Swi1Timeless Prevents Repeat Instability at Fission Yeast Telomeres. PLoS Genet. 2016, 12, e1005943. [Google Scholar] [CrossRef] [PubMed]
- Tarsounas, M.; Tijsterman, M. Genomes and G-quadruplexes: for better or for worse. J. Mol. Biol. 2013, 425, 4782–4789. [Google Scholar] [CrossRef] [PubMed]
- Nandakumar, J.; Cech, T.R. Finding the end: recruitment of telomerase to telomeres. Nat. Rev. Mol. Cell Biol. 2013, 14, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Karow, J.K.; Hickson, I.D.; Maizels, N. The Bloom’s Syndrome Helicase Unwinds G4 DNA. J. Biol. Chem. 1998, 273, 27587–27592. [Google Scholar] [CrossRef] [PubMed]
- Mohaghegh, P.; Karow, J.K.; Brosh, R.M.; Bohr, V.A.; Hickson, I.D. The Bloom’s and Werner’s syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 2001, 29, 2843–2849. [Google Scholar] [CrossRef] [PubMed]
- Huber, M.D.; Lee, D.C.; Maizels, N. G4 DNA unwinding by BLM and Sgs1p: substrate specificity and substrate-specific inhibition. Nucleic Acids Res. 2002, 30, 3954–3961. [Google Scholar] [CrossRef] [PubMed]
- Crabbe, L.; Verdun, R.E.; Haggblom, C.I.; Karlseder, J. Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science 2004, 306, 1951–1953. [Google Scholar] [CrossRef] [PubMed]
- Arnoult, N.; Saintomé, C.; Ourliac-Garnier, I.; Riou, J.-F.; Londoño-Vallejo, A. Human POT1 is required for efficient telomere C-rich strand replication in the absence of WRN. Genes Dev. 2009, 23, 2915–2924. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Loeb, L.A. Unwinding the molecular basis of the Werner syndrome. Mech. Ageing Dev. 2001, 122, 921–944. [Google Scholar] [CrossRef]
- Opresko, P.L.; von Kobbe, C.; Laine, J.-P.; Harrigan, J.; Hickson, I.D.; Bohr, V.A. Telomere-binding protein TRF2 binds to and stimulates the Werner and Bloom syndrome helicases. J. Biol. Chem. 2002, 277, 41110–41119. [Google Scholar] [CrossRef] [PubMed]
- Lillard-Wetherell, K. Association and regulation of the BLM helicase by the telomere proteins TRF1 and TRF2. Hum. Mol. Genet. 2004, 13, 1919–1932. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, M.; Kibe, T.; Kabir, S.; de Lange, T. TRF1 negotiates TTAGGG repeat-associated replication problems by recruiting the BLM helicase and the TPP1/POT1 repressor of ATR signaling. Genes Dev. 2014, 28, 2477–2491. [Google Scholar] [CrossRef] [PubMed]
- Vannier, J.-B.; Pavicic-Kaltenbrunner, V.; Petalcorin, M.I.R.; Ding, H.; Boulton, S.J. RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell 2012, 149, 795–806. [Google Scholar] [CrossRef] [PubMed]
- Vannier, J.-B.; Sarek, G.; Boulton, S.J. RTEL1: functions of a disease-associated helicase. Trends Cell Biol. 2014, 24, 416–425. [Google Scholar] [CrossRef] [PubMed]
- Vannier, J.B.; Sandhu, S.; Petalcorin, M.I.; Wu, X.; Nabi, Z.; Ding, H.; Boulton, S.J. RTEL1 Is a Replisome-Associated Helicase That Promotes Telomere and Genome-Wide Replication. Science 2013, 342, 239–242. [Google Scholar] [CrossRef] [PubMed]
- Bochman, M.L.; Sabouri, N.; Zakian, V.A. Unwinding the functions of the Pif1 family helicases. DNA Repair (Amst.) 2010, 9, 237–249. [Google Scholar] [CrossRef] [PubMed]
- Ribeyre, C.; Lopes, J.; Boulé, J.-B.; Piazza, A.; Guédin, A.; Zakian, V.A.; Mergny, J.-L.; Nicolas, A. The yeast Pif1 helicase prevents genomic instability caused by G-quadruplex-forming CEB1 Sequences In Vivo. PLoS Genet. 2009, 5, e1000475. [Google Scholar] [CrossRef] [PubMed]
- Paeschke, K.; Capra, J.A.; Zakian, V.A. DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell 2011, 145, 678–691. [Google Scholar] [CrossRef] [PubMed]
- Boulé, J.-B.; Vega, L.R.; Zakian, V.A. The yeast Pif1p helicase removes telomerase from telomeric DNA. Nature 2005, 438, 57–61. [Google Scholar] [CrossRef] [PubMed]
- Boulé, J.-B.; Zakian, V.A. The yeast Pif1p DNA helicase preferentially unwinds RNA DNA substrates. Nucleic Acids Res. 2007, 35, 5809–5818. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Zhang, J.; Bochman, M.L.; Zakian, V.A.; Ha, T. Periodic DNA patrolling underlies diverse functions of Pif1 on R-loops and G-rich DNA. eLife 2014, 3, e02190. [Google Scholar] [CrossRef] [PubMed]
- Azvolinsky, A.; Dunaway, S.; Torres, J.Z.; Bessler, J.B.; Zakian, V.A. The S. cerevisiae Rrm3p DNA helicase moves with the replication fork and affects replication of all yeast chromosomes. Genes Dev. 2006, 20, 3104–3116. [Google Scholar] [CrossRef] [PubMed]
- Paeschke, K.; Bochman, M.L.; Garcia, P.D.; Cejka, P.; Friedman, K.L.; Kowalczykowski, S.C.; Zakian, V.A. Pif1 family helicases suppress genome instability at G-quadruplex motifs. Nature 2013, 497, 458–462. [Google Scholar] [CrossRef] [PubMed]
- Sanders, C.M. Human Pif1 helicase is a G-quadruplex DNA-binding protein with G-quadruplex DNA-unwinding activity. Biochem. J. 2010, 430, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, R.; Miller, K.M.; Forment, J.V.; Bradshaw, C.R.; Nikan, M.; Britton, S.; Oelschlaegel, T.; Xhemalce, B.; Balasubramanian, S.; Jackson, S.P. Small-molecule–induced DNA damage identifies alternative DNA structures in human genes. Nat. Chem. Biol. 2012, 8, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Mateyak, M.K.; Zakian, V.A. Human PIF helicase is cell cycle regulated and associates with telomerase. Cell Cycle 2006, 5, 2796–2804. [Google Scholar] [CrossRef] [PubMed]
- Snow, B.E.; Mateyak, M.; Paderova, J.; Wakeham, A.; Iorio, C.; Zakian, V.; Squire, J.; Harrington, L. Murine Pif1 interacts with telomerase and is dispensable for telomere function in vivo. Mol. Cell Biol. 2007, 27, 1017–1026. [Google Scholar] [CrossRef] [PubMed]
- Pinter, S.F.; Aubert, S.D.; Zakian, V.A. The Schizosaccharomyces pombe Pfh1p DNA helicase is essential for the maintenance of nuclear and mitochondrial DNA. Mol. Cell. Biol. 2008, 28, 6594–6608. [Google Scholar] [CrossRef] [PubMed]
- Sabouri, N.; McDonald, K.R.; Webb, C.J.; Cristea, I.M.; Zakian, V.A. DNA replication through hard-to-replicate sites, including both highly transcribed RNA Pol II and Pol III genes, requires the S. pombe Pfh1 helicase. Genes Dev. 2012, 26, 581–593. [Google Scholar] [CrossRef] [PubMed]
- Sabouri, N.; Capra, J.A.; Zakian, V.A. The essential Schizosaccharomyces pombe Pfh1 DNA helicase promotes fork movement past G-quadruplex motifs to prevent DNA damage. BMC Biol. 2014, 12, 101. [Google Scholar] [CrossRef] [PubMed]
- McDonald, K.R.; Sabouri, N.; Webb, C.J.; Zakian, V.A. The Pif1 family helicase Pfh1 facilitates telomere replication and has an RPA-dependent role during telomere lengthening. DNA Repair (Amst.) 2014, 24, 80–86. [Google Scholar] [CrossRef] [PubMed]
- Wallgren, M.; Mohammad, J.B.; Yan, K.-P.; Pourbozorgi-Langroudi, P.; Ebrahimi, M.; Sabouri, N. G-rich telomeric and ribosomal DNA sequences from the fission yeast genome form stable G-quadruplex DNA structures in vitro and are unwound by the Pfh1 DNA helicase. Nucleic Acids Res. 2016, 44, 6213–6231. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.; Sampathi, S.; Dai, H.; Liu, C.; Zhou, M.; Hu, J.; Huang, Q.; Campbell, J.; Shin-Ya, K.; Zheng, L.; et al. Mammalian DNA2 helicase/nuclease cleaves G-quadruplex DNA and is required for telomere integrity. EMBO J. 2013, 32, 1425–1439. [Google Scholar] [CrossRef] [PubMed]
- Masuda-Sasa, T.; Polaczek, P.; Peng, X.P.; Chen, L.; Campbell, J.L. Processing of G4 DNA by Dna2 Helicase/Nuclease and Replication Protein A (RPA) Provides Insights into the Mechanism of Dna2/RPA Substrate Recognition. J. Biol. Chem. 2008, 283, 24359–24373. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Pourmal, S.; Pavletich, N.P. Dna2 nuclease-helicase structure, mechanism and regulation by Rpa. eLife 2015, 4, e09832. [Google Scholar] [CrossRef] [PubMed]
- Zaug, A.J.; Podell, E.R.; Cech, T.R. Human POT1 disrupts telomeric G-quadruplexes allowing telomerase extension in vitro. Proc. Natl. Acad. Sci. USA 2005, 102, 10864–10869. [Google Scholar] [CrossRef] [PubMed]
- Torigoe, H.; Furukawa, A. Tetraplex Structure of Fission Yeast Telomeric DNA and Unfolding of the Tetraplex on the Interaction with Telomeric DNA Binding Protein Pot1. J. Biochem. 2006, 141, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Nora, G.J.; Ghodke, H.; Opresko, P.L. Single Molecule Studies of Physiologically Relevant Telomeric Tails Reveal POT1 Mechanism for Promoting G-quadruplex Unfolding. J. Biol. Chem. 2011, 286, 7479–7489. [Google Scholar] [CrossRef] [PubMed]
- Salas, T.R.; Petruseva, I.; Lavrik, O.; Bourdoncle, A.; Mergny, J.-L.; Favre, A.; Saintomé, C. Human replication protein A unfolds telomeric G-quadruplexes. Nucleic Acids Res. 2006, 34, 4857–4865. [Google Scholar] [CrossRef] [PubMed]
- Safa, L.; Gueddouda, N.M.; Thiebaut, F.; Delagoutte, E.; Petruseva, I.; Lavrik, O.; Mendoza, O.; Bourdoncle, A.; Alberti, P.; Riou, J.-F.; et al. 5′ to 3′ unfolding directionality of DNA secondary structures by replication protein A: G-quadruplexes and duplexes. J. Biol. Chem. 2016, 291, 21246–21256. [Google Scholar] [CrossRef] [PubMed]
- Audry, J.; Maestroni, L.; Delagoutte, E.; Gauthier, T.; Nakamura, T.M.; Gachet, Y.; Saintomé, C.; Géli, V.; Coulon, S. RPA prevents G-rich structure formation at lagging-strand telomeres to allow maintenance of chromosome ends. EMBO J. 2015, 34, 1942–1958. [Google Scholar] [CrossRef] [PubMed]
- Ono, Y.; Tomita, K.; Matsuura, A.; Nakagawa, T.; Masukata, H.; Uritani, M.; Ushimaru, T.; Ueno, M. A novel allele of fission yeast rad11 that causes defects in DNA repair and telomere length regulation. Nucleic Acids Res. 2003, 31, 7141–7149. [Google Scholar] [CrossRef] [PubMed]
- Brosh, R.M.; Orren, D.K.; Nehlin, J.O.; Ravn, P.H.; Kenny, M.K.; Machwe, A.; Bohr, V.A. Functional and Physical Interaction between WRN Helicase and Human Replication Protein A. J. Biol. Chem. 1999, 274, 18341–18350. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Sharma, S.; Sommers, J.A.; Kenny, M.K.; Cantor, S.B.; Brosh, R.M. FANCJ (BACH1) helicase forms DNA damage inducible foci with replication protein A and interacts physically and functionally with the single-stranded DNA-binding protein. Blood 2007, 110, 2390–2398. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Shin-ya, K.; Brosh, R.M. FANCJ helicase defective in fanconia anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability. Mol. Cell. Biol. 2008, 28, 4116–4128. [Google Scholar] [CrossRef] [PubMed]
- London, T.B.C.; Barber, L.J.; Mosedale, G.; Kelly, G.P.; Balasubramanian, S.; Hickson, I.D.; Boulton, S.J.; Hiom, K. FANCJ Is a Structure-specific DNA Helicase Associated with the Maintenance of Genomic G/C Tracts. J. Biol. Chem. 2008, 283, 36132–36139. [Google Scholar] [CrossRef] [PubMed]
- Castillo Bosch, P.; Segura-Bayona, S.; Koole, W.; van Heteren, J.T.; Dewar, J.M.; Tijsterman, M.; Knipscheer, P. FANCJ promotes DNA synthesis through G-quadruplex structures. EMBO J. 2014, 33, 2521–2533. [Google Scholar] [CrossRef] [PubMed]
- Déjardin, J.; Kingston, R.E. Purification of proteins associated with specific genomic Loci. Cell 2009, 136, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Uringa, E.-J.; Lisaingo, K.; Pickett, H.A.; Brind'Amour, J.; Rohde, J.-H.; Zelensky, A.; Essers, J.; Lansdorp, P.M. RTEL1 contributes to DNA replication and repair and telomere maintenance. Mol. Biol. Cell 2012, 23, 2782–2792. [Google Scholar] [CrossRef] [PubMed]
- Sarek, G.; Vannier, J.-B.; Panier, S.; Petrini, J.H.J.; Boulton, S.J. TRF2 Recruits RTEL1 to Telomeres in S Phase to Promote T-Loop Unwinding. Mol. Cell 2015, 57, 622–635. [Google Scholar] [CrossRef] [PubMed]
- Walker, J.R.; Zhu, X.-D. Post-translational modifications of TRF1 and TRF2 and their roles in telomere maintenance. Mech. Ageing Dev. 2012, 133, 421–434. [Google Scholar] [CrossRef] [PubMed]
- Opresko, P.L.; Otterlei, M.; Graakjaer, J.; Bruheim, P.; Dawut, L.; Kølvraa, S.; May, A.; Seidman, M.M.; Bohr, V.A. The Werner syndrome helicase and exonuclease cooperate to resolve telomeric D loops in a manner regulated by TRF1 and TRF2. Mol. Cell 2004, 14, 763–774. [Google Scholar] [CrossRef] [PubMed]
- Opresko, P.L.; Mason, P.A.; Podell, E.R.; Lei, M.; Hickson, I.D.; Cech, T.R.; Bohr, V.A. POT1 Stimulates RecQ Helicases WRN and BLM to Unwind Telomeric DNA Substrates. J. Biol. Chem. 2005, 280, 32069–32080. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.K.; Rossi, M.L.; Singh, D.K.; Dunn, C.; Ramamoorthy, M.; Croteau, D.L.; Liu, Y.; Bohr, V.A. RECQL4, the Protein Mutated in Rothmund-Thomson Syndrome, Functions in Telomere Maintenance. J. Biol. Chem. 2011, 287, 196–209. [Google Scholar] [CrossRef] [PubMed]
- Lenain, C.; Bauwens, S.; Amiard, S.; Brunori, M.; Giraud-Panis, M.-J.; Gilson, E. The Apollo 5' exonuclease functions together with TRF2 to protect telomeres from DNA repair. Curr. Biol. 2006, 16, 1303–1310. [Google Scholar] [CrossRef] [PubMed]
- van Overbeek, M.; de Lange, T. Apollo, an Artemis-Related Nuclease, Interacts with TRF2 and Protects Human Telomeres in S Phase. Curr. Biol. 2006, 16, 1295–1302. [Google Scholar] [CrossRef] [PubMed]
- Azzalin, C.M.; Reichenbach, P.; Khoriauli, L.; Giulotto, E.; Lingner, J. Telomeric Repeat Containing RNA and RNA Surveillance Factors at Mammalian Chromosome Ends. Science 2007, 318, 798–801. [Google Scholar] [CrossRef] [PubMed]
- Schoeftner, S.; Blasco, M.A. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nature 2007, 10, 228–236. [Google Scholar] [CrossRef] [PubMed]
- Costantino, L.; Koshland, D. The Yin and Yang of R-loop biology. Curr. Opin. Cell Biol. 2015, 34, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Rippe, K.; Luke, B. TERRA and the state of the telomere. Nat. Struct. Mol. Biol. 2015, 22, 853–858. [Google Scholar] [CrossRef] [PubMed]
- Flynn, R.L.; Cox, K.E.; Jeitany, M.; Wakimoto, H.; Bryll, A.R.; Ganem, N.J.; Bersani, F.; Pineda, J.R.; Suva, M.L.; Benes, C.H.; et al. Alternative lengthening of telomeres renders cancer cells hypersensitive to ATR inhibitors. Science 2015, 347, 273–277. [Google Scholar] [CrossRef] [PubMed]
- Luke, B. Institute of Molecular Biology (IMB), Mainz Germany. Personal communication, 2016. [Google Scholar]
- Arudchandran, A.; Cerritelli, S.; Narimatsu, S.; Itaya, M.; Shin, D.Y.; Shimada, Y.; Crouch, R.J. The absence of ribonuclease H1 or H2 alters the sensitivity of Saccharomyces cerevisiae to hydroxyurea, caffeine and ethyl methanesulphonate: implications for roles of RNases H in DNA replication and repair. Genes Cells 2000, 5, 789–802. [Google Scholar] [CrossRef] [PubMed]
- Balk, B.; Maicher, A.; Dees, M.; Klermund, J.; Luke-Glaser, S.; Bender, K.; Luke, B. Telomeric RNA-DNA hybrids affect telomere-length dynamics and senescence. Nat. Struct. Mol. Biol. 2013, 20, 1199–1205. [Google Scholar] [CrossRef] [PubMed]
- Arora, R.; Lee, Y.; Wischnewski, H.; Brun, C.M.; Schwarz, T.; Azzalin, C.M. RNaseH1 regulates TERRA-telomeric DNA hybrids and telomere maintenance in ALT tumour cells. Nat. Commun. 2014, 5, 5220. [Google Scholar] [CrossRef] [PubMed]
- Lovejoy, C.A.; Li, W.; Reisenweber, S.; Thongthip, S.; Bruno, J.; de Lange, T.; De, S.; Petrini, J.H.J.; Sung, P.A.; Jasin, M.; et al. ALT Starr Cancer Consortium Loss of ATRX, genome instability, and an altered DNA damage response are hallmarks of the alternative lengthening of telomeres pathway. PLoS Genet. 2012, 8, e1002772. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, A.D.; Banaszynski, L.A.; Noh, K.-M.; Lewis, P.W.; Elsaesser, S.J.; Stadler, S.; Dewell, S.; Law, M.; Guo, X.; Li, X.; et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 2010, 140, 678–691. [Google Scholar] [CrossRef] [PubMed]
- Law, M.J.; Lower, K.M.; Voon, H.P.J.; Hughes, J.R.; Garrick, D.; Viprakasit, V.; Mitson, M.; De Gobbi, M.; Marra, M.; Morris, A.; et al. ATR-X syndrome protein targets tandem repeats and influences allele-specific expression in a size-dependent manner. Cell 2010, 143, 367–378. [Google Scholar] [CrossRef] [PubMed]
- Clynes, D.; Jelinska, C.; Xella, B.; Ayyub, H.; Scott, C.; Mitson, M.; Taylor, S.; Higgs, D.R.; Gibbons, R.J. Suppression of the alternative lengthening of telomere pathway by the chromatin remodelling factor ATRX. Nat. Commun. 2015, 6, 7538. [Google Scholar] [CrossRef] [PubMed]
- Azzalin, C.M.; Lingner, J. The human RNA surveillance factor UPF1 is required for S phase progression and genome stability. Curr. Biol. 2006, 16, 433–439. [Google Scholar] [CrossRef] [PubMed]
- Chawla, R.; Redon, S.; Raftopoulou, C.; Wischnewski, H.; Gagos, S.; Azzalin, C.M. Human UPF1 interacts with TPP1 and telomerase and sustains telomere leading-strand replication. EMBO J. 2011, 30, 4047–4058. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.-H.; Zhou, B.; Huang, Y.; Xu, L.-X.; Zhou, J.-Q. The human Pif1 helicase, a potential Escherichia coli RecD homologue, inhibits telomerase activity. Nucleic Acids Res. 2006, 34, 1393–1404. [Google Scholar] [CrossRef] [PubMed]
- Chib, S.; Byrd, A.K.; Raney, K.D. Yeast Helicase Pif1 Unwinds RNA:DNA Hybrids with Higher Processivity than DNA:DNA Duplexes. J. Biol. Chem. 2016, 291, 5889–5901. [Google Scholar] [CrossRef] [PubMed]
- Teasley, D.C.; Parajuli, S.; Nguyen, M.; Moore, H.R.; Alspach, E.; Lock, Y.J.; Honaker, Y.; Saharia, A.; Piwnica-Worms, H.; Stewart, S.A. Flap Endonuclease 1 Limits Telomere Fragility on the Leading Strand. J. Biol. Chem. 2015, 290, 15133–15145. [Google Scholar] [CrossRef] [PubMed]
- Azzalin, C.M.; Lingner, J. Telomere functions grounding on TERRA firma. Trends Cell Biol. 2015, 25, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Maicher, A.; Lockhart, A.; Luke, B. Breaking new ground: Digging into TERRA function. Biochim. Biophys. Acta 2014, 1839, 387–394. [Google Scholar] [CrossRef] [PubMed]
- Feuerhahn, S.; Iglesias, N.; Panza, A.; Porro, A.; Lingner, J. TERRA biogenesis, turnover and implications for function. FEBS Lett. 2010, 584, 3812–3818. [Google Scholar] [CrossRef] [PubMed]
- Bandaria, J.N.; Qin, P.; Berk, V.; Chu, S.; Yildiz, A. Shelterin Protects Chromosome Ends by Compacting Telomeric Chromatin. Cell 2016, 164, 735–746. [Google Scholar] [CrossRef] [PubMed]
- Benarroch-Popivker, D.; Pisano, S.; Mendez-Bermudez, A.; Lototska, L.; Kaur, P.; Bauwens, S.; Djerbi, N.; Latrick, C.M.; Fraisier, V.; Pei, B.; et al. TRF2-Mediated Control of Telomere DNA Topology as a Mechanism for Chromosome-End Protection. Mol. Cell 2016, 61, 274–286. [Google Scholar] [CrossRef] [PubMed]
- Nagai, S.; Heun, P.; Gasser, S.M. Roles for nuclear organization in the maintenance of genome stability. Epigenomics 2010, 2, 289–305. [Google Scholar] [CrossRef] [PubMed]
- Funabiki, H.; Hagan, I.; Uzawa, S.; Yanagida, M. Cell cycle-dependent specific positioning and clustering of centromeres and telomeres in fission yeast. J. Cell Biol. 1993, 121, 961–976. [Google Scholar] [CrossRef] [PubMed]
- Chikashige, Y.; Yamane, M.; Okamasa, K.; Tsutsumi, C.; Kojidani, T.; Sato, M.; Haraguchi, T.; Hiraoka, Y. Membrane proteins Bqt3 and -4 anchor telomeres to the nuclear envelope to ensure chromosomal bouquet formation. J. Cell Biol. 2009, 187, 413–427. [Google Scholar] [CrossRef] [PubMed]
- Steglich, B.; Strålfors, A.; Khorosjutina, O.; Persson, J.; Smialowska, A.; Javerzat, J.-P.; Ekwall, K. The Fun30 chromatin remodeler Fft3 controls nuclear organization and chromatin structure of insulators and subtelomeres in fission yeast. PLoS Genet. 2015, 11, e1005101. [Google Scholar] [CrossRef] [PubMed]
- Ludérus, M.E.; van Steensel, B.; Chong, L.; Sibon, O.C.; Cremers, F.F.; de Lange, T. Structure, subnuclear distribution, and nuclear matrix association of the mammalian telomeric complex. J. Cell Biol. 1996, 135, 867–881. [Google Scholar] [CrossRef] [PubMed]
- Kaminker, P.G.; Kim, S.-H.; Desprez, P.-Y.; Campisi, J. A novel form of the telomere-associated protein TIN2 localizes to the nuclear matrix. Cell Cycle 2009, 8, 931–939. [Google Scholar] [CrossRef] [PubMed]
- Marcand, S.; Gilson, E.; Shore, D. A protein-counting mechanism for telomere length regulation in yeast. Science 1997, 275, 986–990. [Google Scholar] [CrossRef] [PubMed]
- Greider, C.W. Regulating telomere length from the inside out: The replication fork model. Genes Dev. 2016, 30, 1483–1491. [Google Scholar] [CrossRef] [PubMed]
- Diede, S.J.; Gottschling, D.E. Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerases α and δ. Cell 1999, 99, 723–733. [Google Scholar] [CrossRef]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maestroni, L.; Matmati, S.; Coulon, S. Solving the Telomere Replication Problem. Genes 2017, 8, 55. https://doi.org/10.3390/genes8020055
Maestroni L, Matmati S, Coulon S. Solving the Telomere Replication Problem. Genes. 2017; 8(2):55. https://doi.org/10.3390/genes8020055
Chicago/Turabian StyleMaestroni, Laetitia, Samah Matmati, and Stéphane Coulon. 2017. "Solving the Telomere Replication Problem" Genes 8, no. 2: 55. https://doi.org/10.3390/genes8020055




