Alternative Splicing Profile and Sex-Preferential Gene Expression in the Female and Male Pacific Abalone Haliotis discus hannai
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Preparation
2.2. RNA Extraction and Library Construction
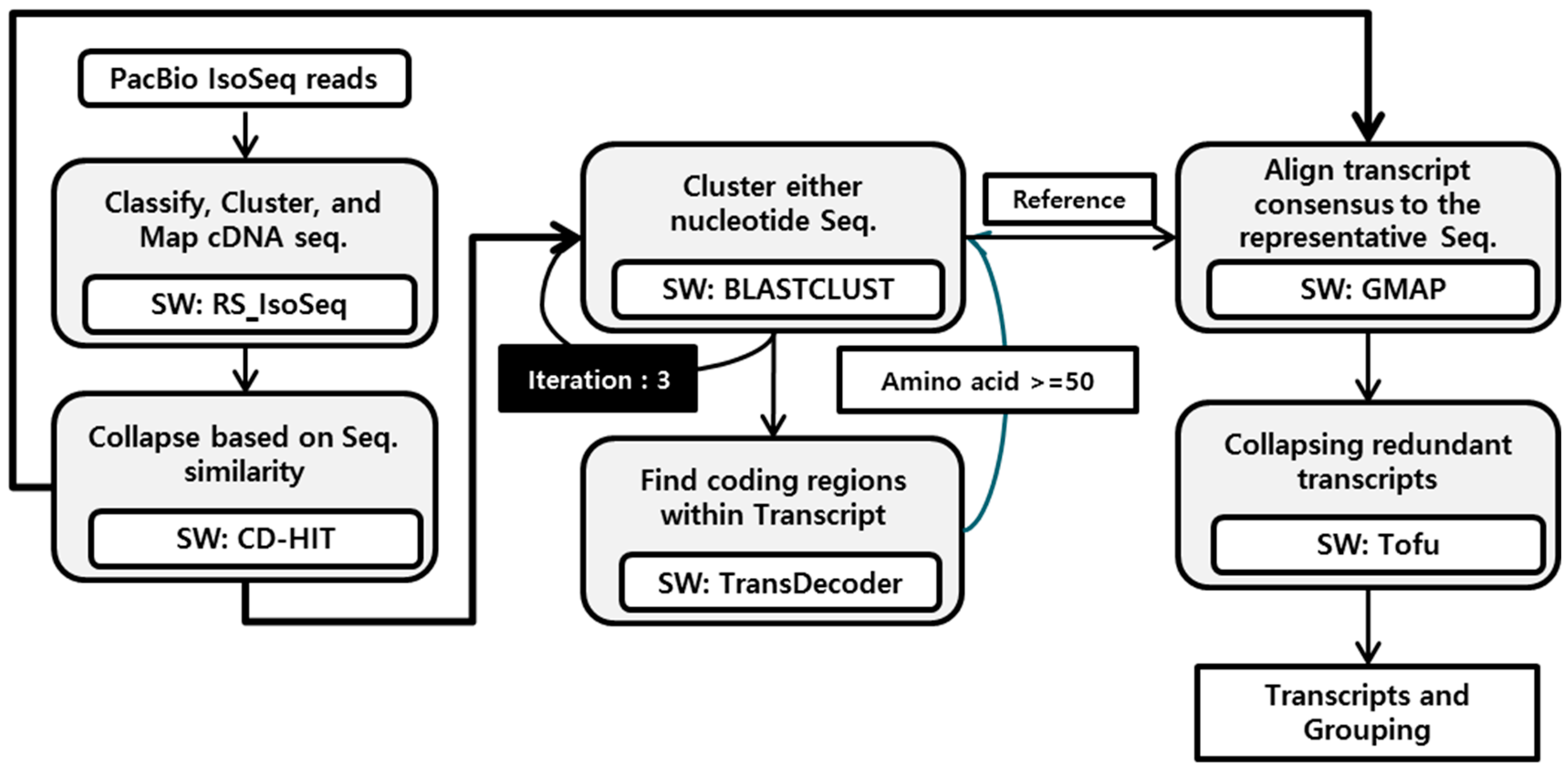
2.3. Gene Annotation and Isoform Analysis
2.4. Gene Ontology and Kyoto Encyclopedia of Genes and Genomes Pathway Analysis
2.5. Isoform Grouping
2.6. Quantitative Real-Time RT-PCR
2.7. Statistical Analysis
3. Results and Discussion
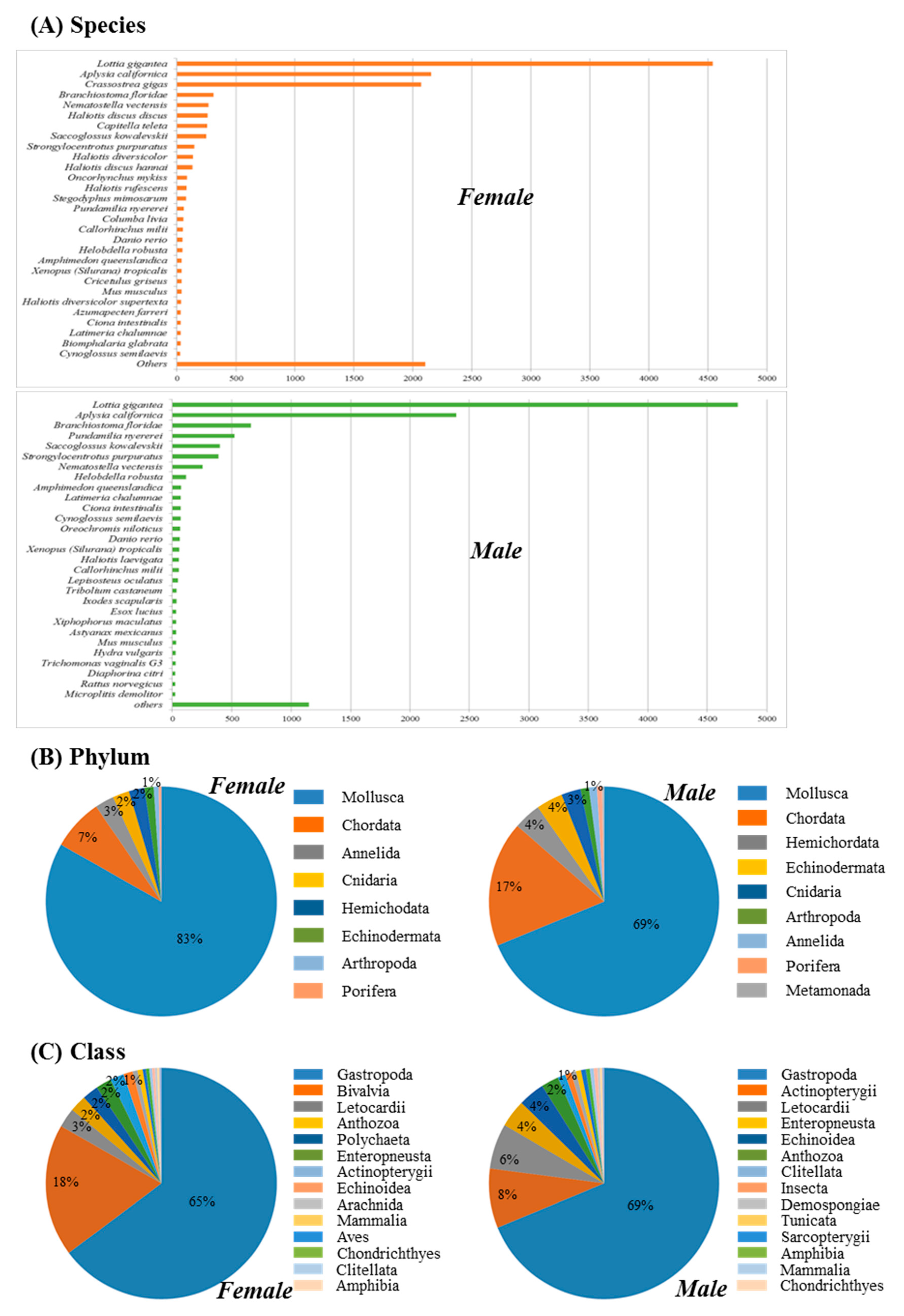
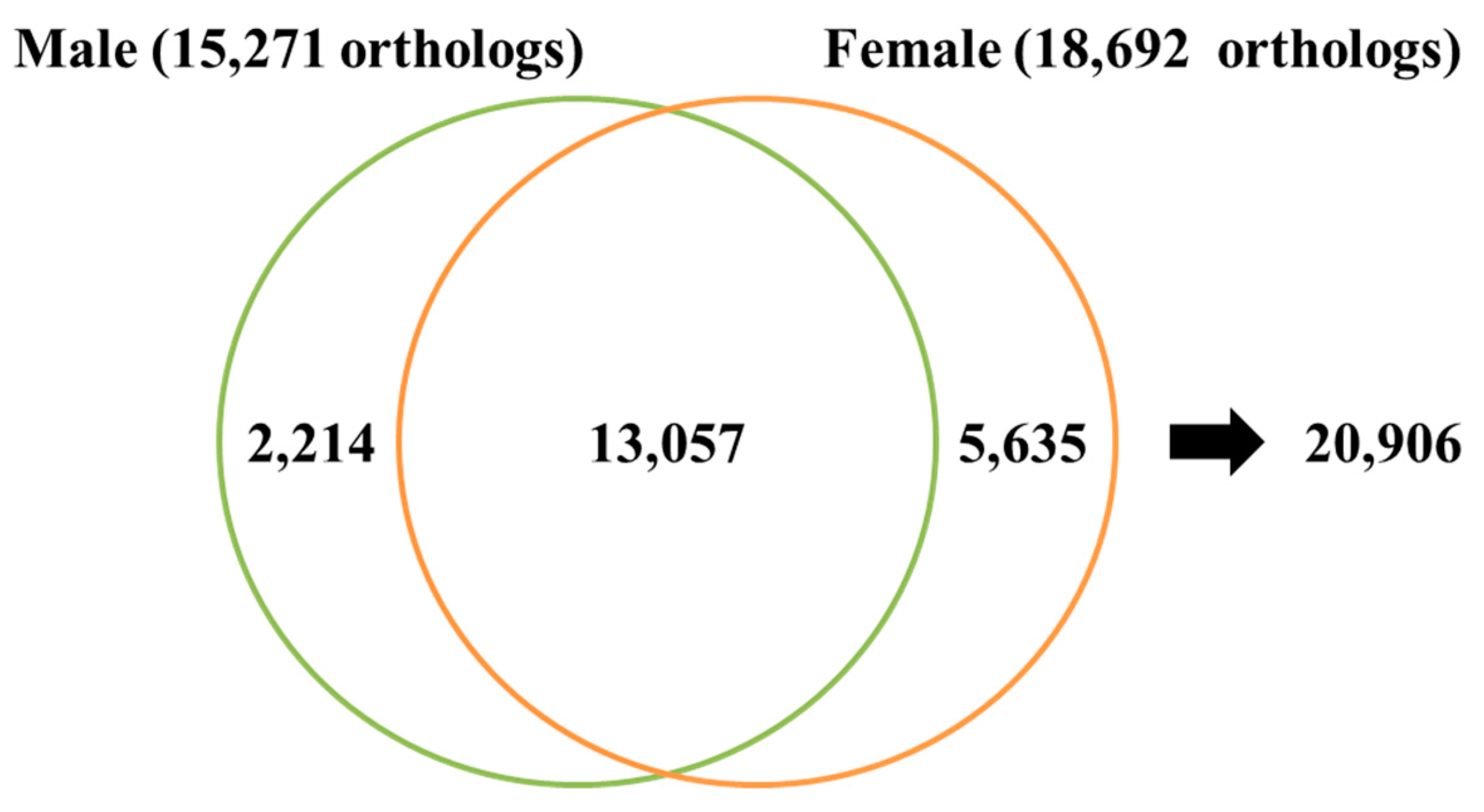
3.1. Transcriptome Assembly and Gene Annotation
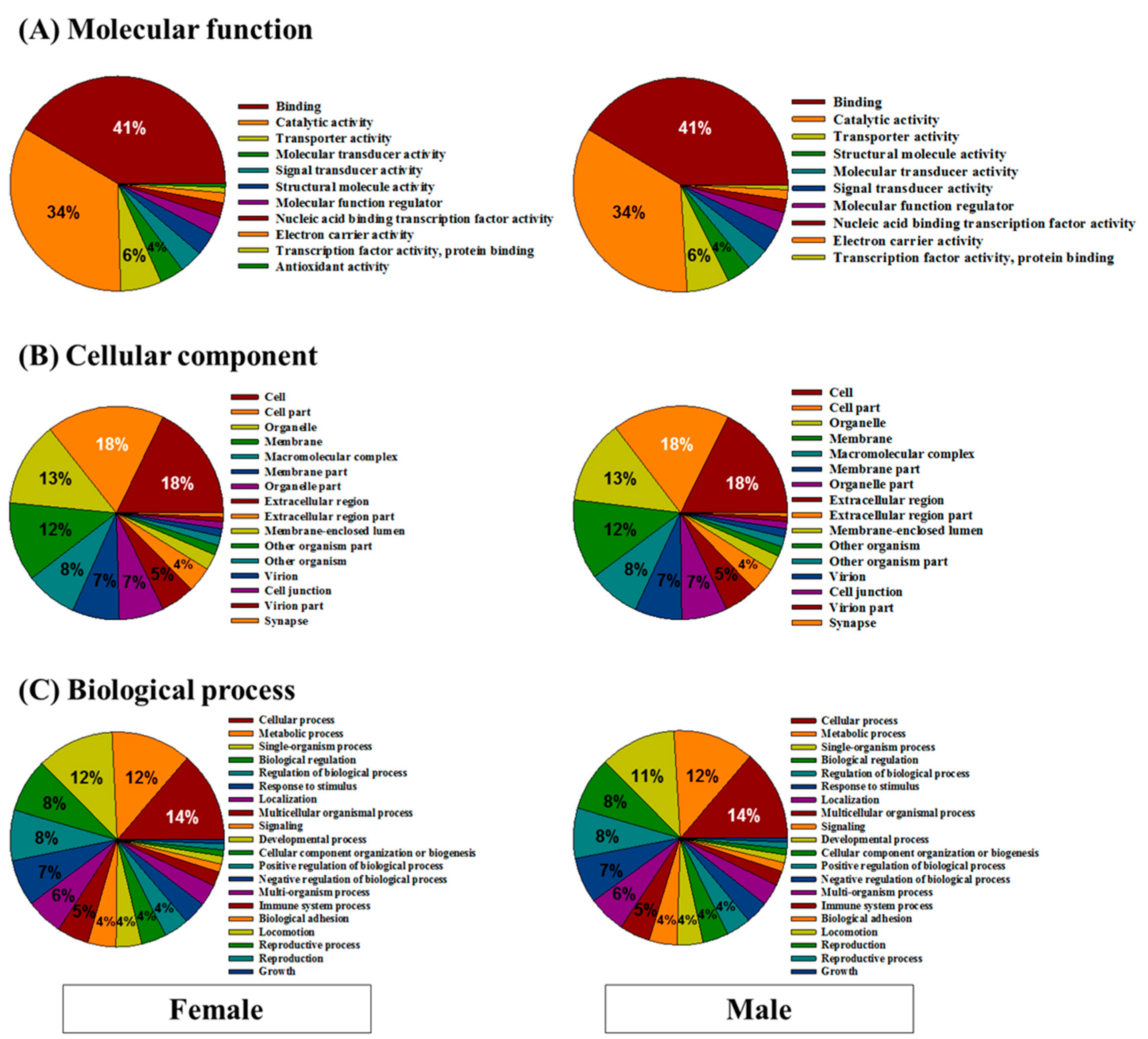
3.2. Functional Annotation
3.3. Isoform Analysis
3.4. Sex-Preferential Genes Expression
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Estes, J.A.; Lindberg, D.R.; Wray, C. Evolution of large body size in abalones (Haliotis): Patterns and implications. Paleobiology 2005, 31, 591–606. [Google Scholar] [CrossRef]
- Suleria, H.A.; Masci, P.P.; Gobe, G.C.; Osborne, S.A. Therapeutic potential of abalone and status of bioactive molecules: A comprehensive review. Crit. Rev. Food Sci. Nutr. 2015, 57, 1742–1748. [Google Scholar] [CrossRef] [PubMed]
- Cook, P.A.; Gordon, H.R. World abalone supply, markets, and pricing. J. Shellfish Res. 2010, 29, 569–571. [Google Scholar] [CrossRef]
- Cook, P.A. The worldwide abalone industry. Mod. Econ. 2014, 5, 1181–1186. [Google Scholar] [CrossRef]
- Sekino, M.; Hara, M. Linkage maps for the Pacific abalone (genus Haliotis) based on microsatellite DNA markers. Genetics 2007, 175, 945–958. [Google Scholar] [CrossRef] [PubMed]
- Franchini, P.; van der Merwe, M.; Roodt-Wilding, R. Transcriptome characterization of the South African abalone Haliotis midae using sequencing-by-synthesis. BMC Res. Notes 2011, 4, 59. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.X.; Chen, Z.S.; Ke, C.H.; Zhao, J.; You, W.W.; Zhang, J.; Dong, W.T.; Chen, J. Pyrosequencing of Haliotis diversicolor transcriptomes: Insights into early developmental molluscan gene expression. PLoS ONE 2012, 7, e51279. [Google Scholar] [CrossRef] [PubMed]
- Palmer, M.R.; McDowall, M.H.; Stewart, L.; Ouaddi, A.; MacCoss, M.J.; Swanson, W.J. Mass spectrometry and next-generation sequencing reveal an abundant and rapidly evolving abalone sperm protein. Mol. Reprod. Dev. 2013, 80, 460–465. [Google Scholar] [CrossRef] [PubMed]
- Mendoza-Porras, O.; Botwright, N.A.; McWilliam, S.M.; Cook, M.T.; Harris, J.O.; Wijffels, G.; Colgrave, M.L. Exploiting genomic data to identify proteins involved in abalone reproduction. J. Proteom. 2014, 108, 337–353. [Google Scholar] [CrossRef] [PubMed]
- Shiel, B.P.; Hall, N.E.; Cooke, I.R.; Robinson, N.A.; Strugnell, J.M. De novo characterisation of the greenlip abalone transcriptome (Haliotis laevigata) with a focus on the heat shock protein 70 (HSP70) family. Mar. Biotechnol. 2015, 17, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Harney, E.; Dubief, B.; Boudry, P.; Basuyaux, O.; Schilhabel, M.B.; Huchette, S.; Paillard, C.; Nunes, F.L. De novo assembly and annotation of the European abalone Haliotis tuberculata transcriptome. Mar. Genom. 2016, 28, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; You, W.; Zhang, X.; Xu, J.; Jiang, Y.; Wang, K.; Zhao, Z.; Chen, B.; Zhao, Y.; Mahboob, S.; et al. Construction of the BAC library of small abalone (Haliotis diversicolor) for gene screening and genome characterization. Mar. Biotechnol. 2016, 18, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Bathige, S.D.; Umasuthan, N.; Jayasinghe, J.D.; Godahewa, G.I.; Park, H.C.; Lee, J. Three novel C1q domain containing proteins from the disk abalone Haliotis discus discus: Genomic organization and analysis of the transcriptional changes in response to bacterial pathogens. Fish Shellfish Immunol. 2016, 56, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Nuurai, P.; McDougall, C.; York, P.S.; Bose, U.; Degnan, B.M.; Cummins, S.F. Identification of a female spawn-associated Kazal-type inhibitor from the tropical abalone Haliotis asinina. J. Pept. Sci. 2016, 22, 461–470. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Liu, X.; Guo, X.; Gao, Q.; Zhao, H.; Zhang, G. A preliminary genetic linkage map of the Pacific abalone Haliotis discus hannai Ino. Mar. Biotechnol. 2006, 8, 386–397. [Google Scholar] [CrossRef] [PubMed]
- Adachi, K.; Okumura, S.I. Determination of genome size of Haliotis discus hannai and H. diversicolor aquatilis (Haliotidae) and phylogenetic examination of this family. Fish. Sci. 2012, 78, 849–852. [Google Scholar] [CrossRef]
- Nam, B.H.; Jung, M.; Subramaniyam, S.; Yoo, S.I.; Markkandan, K.; Moon, J.Y.; Kim, Y.O.; Kim, D.G.; An, C.M.; Shin, Y.; et al. Transcriptome analysis revealed changes of multiple genes involved in Haliotis discus hannai innate immunity during Vibrio parahemolyticus infection. PLoS ONE 2016, 11, e0153474. [Google Scholar] [CrossRef] [PubMed]
- Ino, T. Biological studies on the propagation of Japanese abalone (Genus Haliotis). Bulletin Tokai Reg. Fish. Res. Lab. 1952, 5, 1–102. [Google Scholar]
- Won, S.-H.; Kim, S.-K.; Kim, S.-C.; Yang, B.-K.; Lim, B.-S.; Lee, J.-H.; Lim, H.K.; Lee, J.-S.; Lee, J.-S. The morphological characteristics of four Korean abalone species in Nordotis. Korean J. Malacol. 2014, 30, 87–93. [Google Scholar] [CrossRef]
- Wu, T.D.; Watanabe, C.K. GMAP: A genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 2005, 21, 1859–1875. [Google Scholar] [CrossRef] [PubMed]
- De Sousa, J.T.; Milan, M.; Bargelloni, L.; Pauletto, M.; Matias, D.; Joaquim, S.; Matias, A.M.; Quillien, V.; Leitão, A.; Huvet, A. A microarray-based analysis of gametogenesis in two Portuguese populations of the European clam Ruditapes decussatus. PLoS ONE 2014, 9, e92202. [Google Scholar] [CrossRef] [PubMed]
- Teaniniuraitemoana, V.; Huvet, A.; Levy, P.; Klopp, C.; Lhuillier, E.; Gaertner-Mazouni, N.; Gueguen, Y.; Le Moullac, G. Gonad transcriptome analysis of pearl oyster Pinctada margaritifera: Identification of potential sex differentiation and sex determining genes. BMC Genom. 2014, 15, 491. [Google Scholar] [CrossRef] [PubMed]
- Teaniniuraitemoana, V.; Huvet, A.; Levy, P.; Gaertner-Mazouni, N.; Gueguen, Y.; Le Moullac, G. Molecular signatures discriminating the male and the female sexual pathways in the pearl oyster Pinctada margaritifera. PLoS ONE 2015, 10, e0122819. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Hui, M.; Cui, Z.; Luo, D.; Song, C.; Li, Y.; Liu, L. Comparative transcriptome analysis reveals sex-biased gene expression in juvenile Chinese mitten crab Eriocheir sinensis. PLoS ONE 2015, 10, e0133068. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Picone, B.; Rhode, C.; Roodt-Wilding, R. Transcriptome profiles of wild and cultured South African abalone, Haliotis midae. Mar. Genom. 2015, 20, 3–6. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Mai, K.; Wu, G. Thiamin requirement of juvenile abalone, Haliotis discus hannai Ino. Aquaculture 2002, 207, 331–343. [Google Scholar] [CrossRef]
- Fyfe, J.C.; Madsen, M.; Højrup, P.; Christensen, E.I.; Tanner, S.M.; de la Chapelle, A.; He, Q.; Moestrup, S.K. The functional cobalamin (vitamin B12)-intrinsic factor receptor is a novel complex of cubilin and amnionless. Blood 2004, 103, 1573–1579. [Google Scholar] [CrossRef] [PubMed]
- Moestrup, S.K.; Kozyraki, R.; Kristiansen, M.; Kaysen, J.H.; Rasmussen, H.H.; Brault, D.; Pontillon, F.; Goda, F.O.; Christensen, E.I.; Hammond, T.G.; et al. The intrinsic factor-vitamin B12 receptor and target of teratogenic antibodies is a megalin-binding peripheral membrane protein with homology to developmental proteins. J. Biol. Chem. 1998, 273, 5235–5242. [Google Scholar] [CrossRef] [PubMed]
- Verroust, P.J.; Birn, H.; Nielsen, R.; Kozyraki, R.; Christensen, E.I. The tandem endocytic receptors megalin and cubilin are important proteins in renal pathology. Kidney Int. 2002, 62, 745–756. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Mai, K.; Xu, W.; Ai, Q.; Tan, B.; Liufu, Z.; Ma, H. Effects of vitamin A and D on shell biomineralization of abalone Haliotis discus hannai, Ino. J. Shellfish Res. 2004, 23, 1065–1071. [Google Scholar]
- Zhang, W.; Mai, K.; Xu, W.; Tan, B.; Ai, Q.; Liufu, Z.; Ma, H.; Wang, X. Interaction between vitamins A and D on growth and metabolic responses of abalone Haliotis discus hannai, Ino. J. Shellfish Res. 2007, 26, 51–58. [Google Scholar] [CrossRef]
- Gesto, M.; Castro, L.F.; Reis-Henriques, M.A.; Santos, M.M. Retinol metabolism in the mollusk Osilinus lineatus indicates an ancient origin for retinyl ester storage capacity. PLoS ONE 2012, 7, e35138. [Google Scholar] [CrossRef] [PubMed]
- Gesto, M.; Ruivo, R.; Páscoa, I.; André, A.; Castro, L.F.; Santos, M.M. Retinoid level dynamics during gonad recycling in the limpet Patella vulgata. Gen. Comp. Endocrinol. 2016, 225, 142–148. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, F.; Katsura, H.; Takenaka, S.; Enomoto, T.; Miyamoto, E.; Nakatsuka, T.; Nakano, Y. Characterization of vitamin B12 compounds from edible shellfish, clam, oyster, and mussel. Int. J. Food Sci. Nutr. 2001, 52, 263–268. [Google Scholar] [CrossRef] [PubMed]
- Tanioka, Y.; Takenaka, S.; Furusho, T.; Yabuta, Y.; Nakano, Y.; Watanabe, F. Identification of vitamin B12 and pseudovitamin B12 from various edible shellfish using liquid chromatography-electrospray ionization/tandem mass spectrometry. Fish. Sci. 2014, 80, 1065–1071. [Google Scholar] [CrossRef]
- Lambert, L.A.; Perri, H.; Halbrooks, P.J.; Mason, A.B. Evolution of the transferrin family: Conservation of residues associated with iron and anion binding. Comp. Biochem. Physiol. B: Biochem. Mol. Biol. 2005, 142, 129–141. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhang, S.; Li, L. A transferrin-like homolog in amphioxus Branchiostoma belcheri: Identification, expression and functional characterization. Mol. Immunol. 2009, 46, 3117–3124. [Google Scholar] [CrossRef] [PubMed]
- Herath, H.M.; Elvitigala, D.A.; Godahewa, G.I.; Whang, I.; Lee, J. Molecular insights into a molluscan transferrin homolog identified from disk abalone (Haliotis discus discus) evidencing its detectable role in host antibacterial defense. Dev. Comp. Immunol. 2015, 53, 222–233. [Google Scholar] [CrossRef] [PubMed]
- Matozzo, V.; Gagné, F.; Marin, M.G.; Ricciardi, F.; Blaise, C. Vitellogenin as a biomarker of exposure to estrogenic compounds in aquatic invertebrates: A review. Environ. Int. 2008, 34, 531–545. [Google Scholar] [CrossRef] [PubMed]
- Osada, M.; Takamura, T.; Sato, H.; Mori, K. Vitellogenin synthesis in the ovary of scallop, Patinopecten yessoensis: Control by estradiol-17β and the central nervous system. J. Exp. Zool. A Comp. Exp. Biol. 2003, 299, 172–179. [Google Scholar] [CrossRef] [PubMed]
- Boutet, I.; Moraga, D.; Marinovic, L.; Obreque, J.; Chavez-Crooker, P. Characterization of reproduction-specific genes in a marine bivalve mollusc: Influence of maturation stage and sex on mRNA expression. Gene 2008, 407, 130–138. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, T.; Yamano, K.; Kitamura, M.; Hara, A. Ovarian follicle cells are the site of vitellogenin synthesis in the Pacific abalone Haliotis discus hannai. Comp. Biochem. Physiol. A: Mol. Integr. Physiol. 2008, 149, 293–298. [Google Scholar] [CrossRef] [PubMed]
- Corporeau, C.; Vanderplancke, G.; Boulais, M.; Suquet, M.; Quéré, C.; Boudry, P.; Huvet, A.; Madec, S. Proteomic identification of quality factors for oocytes in the Pacific oyster Crassostrea gigas. J. Proteom. 2012, 75, 5554–5563. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Zhang, Q.; Liu, H.; Liu, W.; Sun, Z.; Li, S.; Zhang, T. Cloning and expression of vitellogenin (Vg) gene and its correlations with total carotenoids content and total antioxidant capacity in noble scallop Chlamys nobilis (Bivalve: Pectinidae). Aquaculture 2012, 366, 46–53. [Google Scholar] [CrossRef]
- Ni, J.; Zeng, Z.; Kong, D.; Hou, L.; Huang, H.; Ke, C. Vitellogenin of Fujian oyster, Crassostrea angulata: Synthesized in the ovary and controlled by estradiol-17β. Gen. Comp. Endocrinol. 2014, 202, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Liu, Z.; Zhou, L.; Ji, G.; Yang, A. Molecular cloning, expression, purification and characterization of vitellogenin in scallop Patinopecten yessoensis with special emphasis on its antibacterial activity. Dev. Comp. Immunol. 2015, 49, 249–258. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-J.; Lee, N.; Woo, S.; Ryu, J.-C.; Yum, S. Transcriptomic change as evidence for cadmium-induced endocrine disruption in marine fish model of medaka, Oryzias javanicus. Mol. Cell. Toxicol. 2016, 12, 409–420. [Google Scholar] [CrossRef]
- Georges, A.; Auguste, A.; Bessière, L.; Vanet, A.; Todeschini, A.L.; Veitia, R.A. FOXL2: A central transcription factor of the ovary. J. Mol. Endocrinol. 2013, 52, 17–33. [Google Scholar] [CrossRef] [PubMed]
- Naimi, A.; Martinez, A.S.; Specq, M.L.; Diss, B.; Mathieu, M.; Sourdaine, P. Molecular cloning and gene expression of Cg-Foxl2 during the development and the adult gametogenetic cycle in the oyster Crassostrea gigas. Comp. Biochem. Physiol. B: Biochem. Mol. Biol. 2009, 154, 134–142. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.L.; Zhang, Z.F.; Shao, M.Y.; Liu, J.G.; Muhammad, F. Sexually dimorphic expression of foxl2 during gametogenesis in scallop Chlamys farreri, conserved with vertebrates. Dev. Genes Evol. 2012, 222, 279–286. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Xu, F.; Guo, X. Genomic analysis of the pacific oyster (Crassostrea gigas) reveals possible conservation of vertebrate sex determination in a mollusc. G3 2014, 4, 2207–2217. [Google Scholar] [CrossRef] [PubMed]
- Tong, Y.; Zhang, Y.; Huang, J.; Xiao, S.; Zhang, Y.; Li, J.; Chen, J.; Yu, Z. Transcriptomics analysis of Crassostrea hongkongensis for the discovery of reproduction-related genes. PLoS ONE 2015, 10, e0134280. [Google Scholar] [CrossRef] [PubMed]
- Sapiro, R.; Kostetskii, I.; Olds-Clarke, P.; Gerton, G.L.; Radice, G.L.; Strauss, J.F. Male infertility, impaired sperm motility, and hydrocephalus in mice deficient in sperm-associated antigen 6. Mol. Cell. Biol. 2002, 22, 6298–6305. [Google Scholar] [CrossRef] [PubMed]
- Gaudet, J.; VanderElst, I.; Spence, A.M. Post-transcriptional regulation of sex determination in Caenorhabditis elegans: Widespread expression of the sex-determining gene fem-1 in both sexes. Mol. Biol. Cell 1996, 7, 1107–1121. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Boswell, R.; Wood, W.B. mag-1, a homolog of Drosophila mago nashi, regulates hermaphrodite germ-line sex determination in Caenorhabditis elegans. Dev. Biol. 2000, 218, 172–182. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, H.; Iguchi, N.; Toyama, Y.; Kitamura, K.; Takahashi, T.; Kaseda, K.; Maekawa, M.; Nishimune, Y. Mice deficient in the axonemal protein Tektin-t exhibit male infertility and immotile-cilium syndrome due to impaired inner arm dynein function. Mol. Cell. Biol. 2004, 24, 7958–7964. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Liu, Y.X.; Jiang, G.F. De novo assembly and characterization of the testis transcriptome and development of EST-SSR markers in the cockroach Periplaneta americana. Sci. Rep. 2015, 5, 11144. [Google Scholar] [CrossRef] [PubMed]
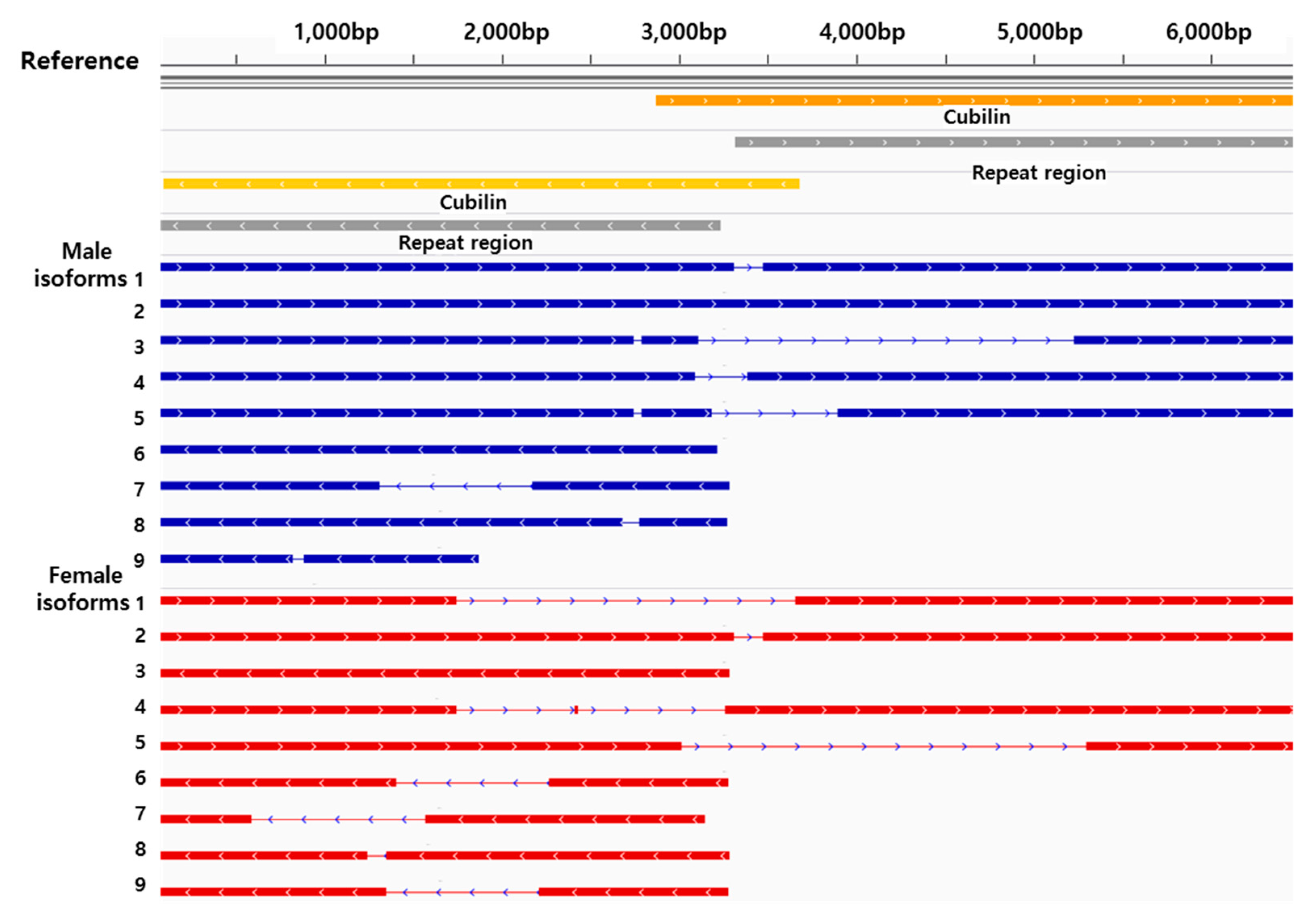
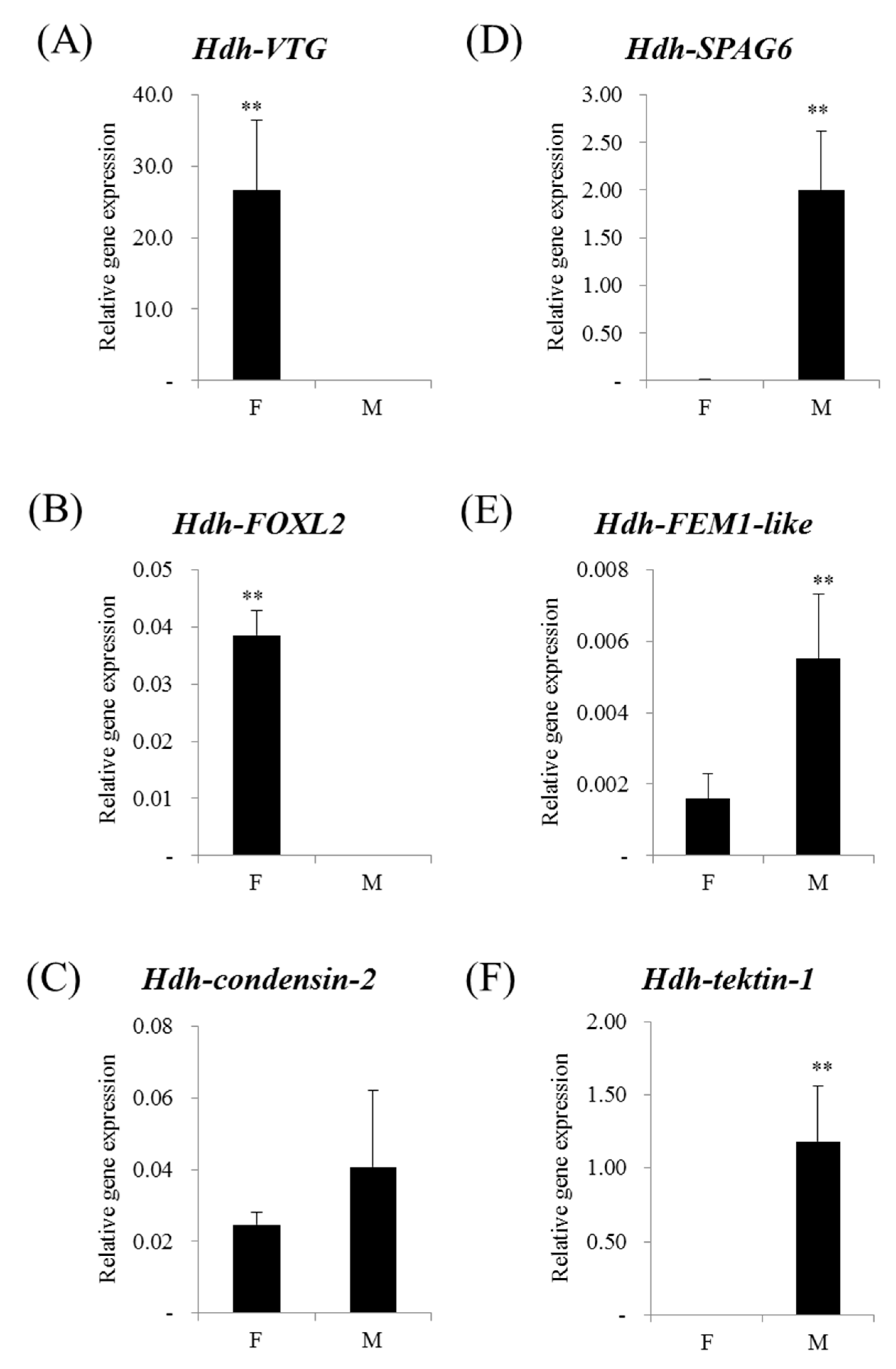
| Step | Data | Platform | Female | Male |
|---|---|---|---|---|
| 1 | High quality consensus sequence | RS_IsoSeq | 22,494 | 18,981 |
| 2 | Non-redundant representative sequence | CD-HIT | 18,692 | 15,271 |
| 3 | Reference isoforms | BLASTCLUST and TransDecoder | 15,363 | 12,409 |
| 4 | Final isoform transcriptome by combine representative sequence | GMAP and ToFU | 15,792 | 12,718 |
| 5 | Final gene set with representative isoforms | TransDecoder | 15,110 | 12,145 |
| Number of Total Orthologs | 13,057 |
|---|---|
| Average identity (%) | 99.33 |
| Average coverage of female (%) | 85.23 |
| Average coverage of male (%) | 90.63 |
| Number of 100% coverage orthologs | 502 |
| Number of 100% identity orthologs | 597 |
| Contig data | Female | Male |
|---|---|---|
| Total genes | 15,110 | 12,145 |
| Genes with no isoforms | 14,591 | 11,754 |
| Genes with at least two isoforms | 519 | 391 |
| Total length of genes with isoforms (bp) | 1,599,611 | 1,166,159 |
| Average length (bp) | 3082 | 2982 |
| Maximum length (bp) | 8315 | 8058 |
| Minimum length (bp) | 741 | 847 |
| Cluster ID | Length (bp) | #Isoform | Description | Matched Species | GenBank No. |
|---|---|---|---|---|---|
| F_Cluster00018 | 6565 | 27 | deleted in malignant brain tumors one protein | Columba livia | ZDB-GENE-060228-6 |
| F_Cluster11205 | 2162 | 11 | - | - | - |
| F_Cluster00024 | 6380 | 9 | PREDICTED: cubilin | Haplochromis burtoni | ZDB-GENE-060228-6 |
| F_Cluster00089 | 4372 | 9 | PREDICTED: cubilin-like | Lingula anatina | ZDB-GENE-060228-6 |
| F_Cluster13261 | 1675 | 9 | - | - | - |
| F_Cluster00812 | 3837 | 8 | PREDICTED: cyclin-L1-like | Crassostrea gigas | H2U6Q2 |
| F_Cluster00002 | 8315 | 7 | PREDICTED: LOW QUALITY PROTEIN: sushi, von Willebrand factor type A, epidermal growth factor (EGF) and pentraxin domain-containing protein 1 | Callorhinchus milii | F1MNH3 |
| F_Cluster00829 | 3833 | 6 | - | - | - |
| F_Cluster03162 | 3343 | 6 | PREDICTED: serine/arginine-rich splicing factor 6-like isoform X1 | Crassostrea gigas | A0A0D9SEM4 |
| F_Cluster05356 | 3088 | 6 | heterogeneous nuclear ribonucleoprotein L, partial | Aplysia californica | R4GHI6 |
| F_Cluster11593 | 2114 | 6 | - | - | - |
| F_Cluster00004 | 7437 | 5 | hypothetical protein LOTGIDRAFT_214098 | Lottia gigantea | NP_001116989.1 |
| F_Cluster00011 | 6791 | 5 | hypothetical protein AC249_AIPGENE2795 | Exaiptasia pallida | F1NX90 |
| F_Cluster10316 | 2268 | 5 | PREDICTED: Na(+)/H(+) exchange regulatory cofactor NHE-RF1-like | Biomphalaria glabrata | XP_414851.3 |
| F_Cluster12757 | 1916 | 5 | - | - | - |
| Cluster ID | Length (bp) | #Isoform | Description | Matched species | GenBank No. |
|---|---|---|---|---|---|
| M_Cluster00016 | 6579 | 52 | hypothetical protein cypCar_00021969, partial | Cyprinus carpio | ZDB-GENE-060228-6 |
| M_Cluster00705 | 3598 | 11 | PREDICTED: cyclin-L1-like | Crassostrea gigas | H2U6Q2 |
| M_Cluster00017 | 6523 | 9 | PREDICTED: cubilin | Oreochromis niloticus | ZDB-GENE-060228-6 |
| M_Cluster01458 | 3359 | 8 | PREDICTED: mesocentin-like | Aplysia californica | - |
| M_Cluster09253 | 1770 | 8 | - | - | - |
| M_Cluster09585 | 1697 | 8 | - | - | - |
| M_Cluster00226 | 3901 | 7 | hypothetical protein LOTGIDRAFT_115468 | Lottia gigantea | XP_002415964.1 |
| M_Cluster00908 | 3513 | 7 | serine-arginine protein 55 | Melipona quadrifasciata | E1C270 |
| M_Cluster00059 | 4569 | 5 | hypothetical protein LOTGIDRAFT_200884 | Lottia gigantea | NP_001040037.1 |
| M_Cluster00931 | 3505 | 5 | - | - | - |
| M_Cluster01425 | 3366 | 5 | - | - | - |
| M_Cluster01740 | 3296 | 5 | putative splicing factor, arginine/serine-rich 7 | Crassostrea gigas | NP_064477.1 |
| M_Cluster01748 | 3295 | 5 | - | - | - |
| M_Cluster06621 | 2255 | 5 | PREDICTED: tryptophan 2,3-dioxygenase-like | Lingula anatina | M3X838 |
| M_Cluster09868 | 1649 | 5 | - | - | - |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, M.A.; Rhee, J.-S.; Kim, T.H.; Lee, J.S.; Choi, A.-Y.; Choi, B.-S.; Choi, I.-Y.; Sohn, Y.C. Alternative Splicing Profile and Sex-Preferential Gene Expression in the Female and Male Pacific Abalone Haliotis discus hannai. Genes 2017, 8, 99. https://doi.org/10.3390/genes8030099
Kim MA, Rhee J-S, Kim TH, Lee JS, Choi A-Y, Choi B-S, Choi I-Y, Sohn YC. Alternative Splicing Profile and Sex-Preferential Gene Expression in the Female and Male Pacific Abalone Haliotis discus hannai. Genes. 2017; 8(3):99. https://doi.org/10.3390/genes8030099
Chicago/Turabian StyleKim, Mi Ae, Jae-Sung Rhee, Tae Ha Kim, Jung Sick Lee, Ah-Young Choi, Beom-Soon Choi, Ik-Young Choi, and Young Chang Sohn. 2017. "Alternative Splicing Profile and Sex-Preferential Gene Expression in the Female and Male Pacific Abalone Haliotis discus hannai" Genes 8, no. 3: 99. https://doi.org/10.3390/genes8030099






