1. Introduction
Dehydration is one of the major stresses that inhibits plant growth and can reduce crop productivity. Hence, drought resistance is a key target in helping to ensure global food supply. Plants respond to dehydration stress in three broad approaches: (1) Dehydration escape; (2) Dehydration avoidance; and (3) Dehydration tolerance. Such mechanisms are seen in a range of leguminous species, including the mung bean [
1] and pigeon pea [
2]. Dehydration escape is the ability of plants to complete their growth cycle and reach maturity with successful reproduction before the shortage of water reaches damaging levels [
3]. Mechanisms of avoidance include improved root traits for a greater extraction of soil moisture, stomatal closure, a decreased radiation absorption through leaf rolling, a decreased leaf area for reduced water loss, and the accumulation of osmoprotectants such as proline, trehalose, and dehydrins [
4]. Dehydration tolerance allows plants to survive through improved water-use efficiency, i.e., performing all of the biological, molecular, and cellular functions with minimal water. Numerous studies on the effects of dehydration stress on staple crops have been reported [
1,
2,
4,
5,
6,
7,
8,
9,
10].
Reduced water availability causes the production of abscisic acid (ABA), the phyto-hormone which initiates stomatal closure and influences other aspects of plant growth and physiology. It is responsible for regulating a broad range of genes during the dehydration response. The SNF1-related protein kinase, AREB (ABA-responsive element)/ABF are the key regulators of ABA signalling [
11]. Improving the dehydration tolerance has also been linked to a reduction in shoot growth, while root growth is maintained, leading to an altered partition between the root and shoot. This process is achieved by cell-wall synthesis and remodelling. The formation of reactive oxygen species (ROS) and lignin peroxidases are the key steps involved in cell wall thickening.
Stomatal closure limits the CO
2 uptake by leaves, which leads to a reduction in photosynthesis as the leaf’s internal CO
2 is depleted. Severe dehydration stress also limits photosynthesis by down-regulating the expression of ribulose-1, 5-bisphosphate carboxylase/oxygenase (Rubisco), fructose-1,6-bisphosphatase (FBPase), phosphoenolpyruvate carboxylase (PEPCase), pyruvate orthophosphate dikinase (PPDK), and NADP-malic enzyme (NADP-ME) [
12]. Plant responses to dehydration affect vegetative growth by reducing the leaf-area expansion and total dry matter, which in turn decreases light interception [
13]. Under dehydration stress, wheat (
Triticum dicoccoides) shows a reduction in the number of grains, grain yield, shoot dry weight, and harvest index [
8]. In soybean specimens (
Glycine max), the loss of seed yield was reported to be greatest when dehydration appeared during anthesis and the early reproductive stages [
6,
7,
8,
9].
A range of dehydration stress-related genes have been identified in
Arabidopsis thaliana, rice (
Oryza sativa), and other model plants [
14]. These can be classified into two main groups: (i) Effector proteins, whose role is to alleviate the effect of the stress (such as water channel proteins, detoxification enzymes, LEA proteins, chaperones, and osmoprotectants); and (ii) Regulatory proteins, which alter the expression or activity of effector genes and modify plant growth, such as the transcription factors DREB2 and AREB, and also protein kinases and phosphatases [
15].
In recent years, plant breeders have turned to landraces (i.e., locally adapted genetically mixed populations) for trait improvement in various crops, including barley [
16], sorghum [
17], sesame [
18], and soybean [
19]. An early attempt to investigate the use of landraces in addressing the problem of dehydration tolerance has been carried out in wheat [
20], although this did not delve into the specific genetics conferring the desirable traits. An alternative approach to identifying the genes conferring dehydration avoidance and tolerance is to study species that are already resilient under arid conditions. In this regard, bambara groundnut (
Vigna subterranea (L) Verdc.) is a potential candidate. It is an underutilised, drought-resistant African legume, which is mainly grown in sub-Saharan Africa [
5,
6,
7,
8,
9,
10,
11,
12,
13,
14,
15,
16,
17,
18,
19,
20,
21] and is sometimes used as an intercrop with major cereals, such as maize, because of its nitrogen fixing potential [
22]. Bambara groundnut is considered as a drought resistant crop with a reasonable protein content (18% to 22%), a high carbohydrate content (65%), and some level of lipids (6.5%) [
23]
, with a similar overall composition to chickpea. A number of bambara groundnut landraces have well-developed tap roots which grow up to a height of 30–35 cm [
24].
From the results of Mabhaudhi et al. [
25], bambara groundnut has been shown to adopt dehydration-escape mechanisms, including a shortened vegetative growth period, early flowering, a reduced duration of the reproductive stage, and early maturity under dehydration stress. Such responses are likely to be employed where the initial plant growth is based on stored soil water, but further rain is unlikely. It has been reported that bambara groundnut responds to dehydration stress by partitioning more assimilate into the root, relative to the shoots, so that a greater soil volume can be exploited [
26,
27]. Nyamudeza [
27] also observed that bambara groundnut allocated a greater fraction of its total dry weight to the roots than the groundnut, irrespective of the available soil moisture. This would suggest that bambara groundnut commits a greater supply of assimilates to root growth, irrespective of the soil moisture status. This strategy may have clear advantages when water subsequently becomes limited, but there could be a trade-off with the yield under benign environments. A greater root dry-weight was also reported when the bambara landrace, Burkina, was subjected to dehydration stress [
28]. Dehydration-avoidance traits have also been observed, especially the accumulation of proline [
21] and a reduced leaf area [
29].
This study aims to investigate the effects of dehydration on gene expression in this reportedly drought-resistant species. The transcriptomes of two genotypes (DipC and Tiga Nicuru (TN)) were sampled, to identify what is common and how they differ in their response to a prolonged, but slowly intensifying, dehydration treatment. The climatic conditions in their native regions (Botswana and Mali, respectively) suggest that they are likely to have evolved in regions which would select for drought resistance, while potentially exhibiting some variation in the mechanisms employed to deal with dehydration, as they are morphologically and phenologically distinct [
30]. Chai et al. [
30,
31] reported that transgressive segregation was observed in the segregating F
5 population derived from the TNxDipC cross. The contrast between the two parental lines for a number of traits such as the days-to-maturity, stomatal conductance, 100-seed weight, leaf area, internode length, peduncle length, pod number per plant, and leaf carbon (delta C
13) isotope analysis, suggest that some of these mechanisms for adaptation to dehydration could be non-identical in the two genotypes. For example, delta C
13 was associated with a higher yield as observed in DipC, compared to TN [
30]. In addition, the results showed that there were lines in the segregating population that performed better in terms of the ability to produce higher yields under drought conditions than the parental genotypes. Hence, evaluating the transcriptome of the two parental lines under dehydration stress could be a good indicator to investigate the molecular mechanism occurring in the two genotypes and its relationship to phenology and phenotype.
As a complete genome sequence is not available and microarray tools are still to be developed in this species, cross-species hybridisation with the Affymetrix Glycine-max microarray was investigated to test if this approach is acceptable for bambara groundnut transcriptomics, as it has been successful for other species [
32,
33,
34].
4. Discussion
Landraces are a potentially valuable resource for finding genes conferring useful agricultural and processing traits. Bambara groundnut is an underutilised African legume whose landraces are adapted, in many cases, to arid conditions. We have developed single genotypes derived from landraces for analysis. There have been several dehydration studies carried out on bambara groundnut, but the molecular mechanisms of how the crop responds and adapts to dehydration stress are still under investigation. This study has carried out transcriptomic comparisons in two genotypes of bambara groundnut, DipC and TN, in an attempt to identify potential genes conferring advantageous traits for crop growth and yields in marginal environments.
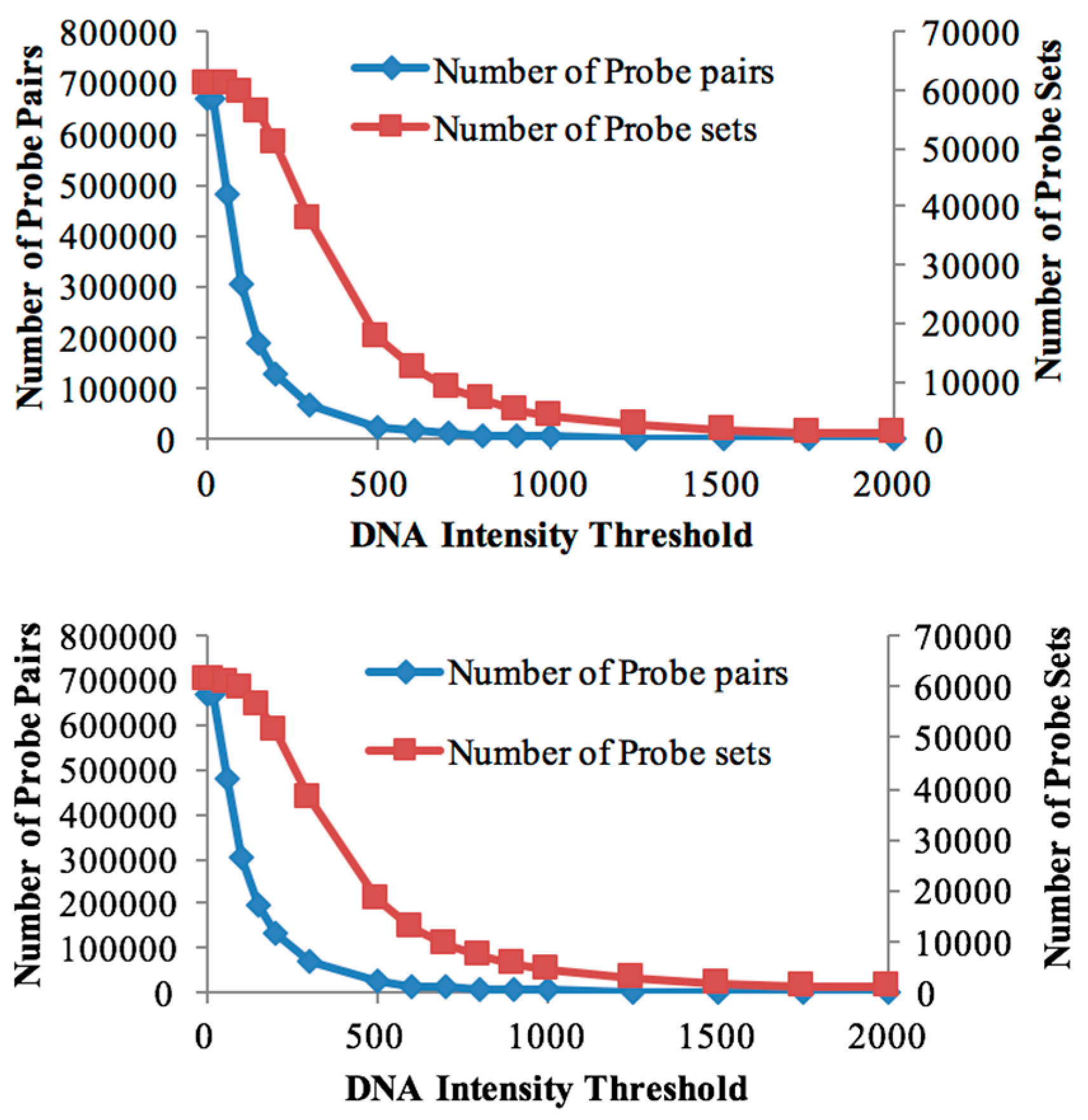
Cross-species hybridisation to the soybean microarray has been shown to be informative for investigating the bambara groundnut transcriptome, as good gene (probe-set) retention was observed at high gDNA hybridisation thresholds. In support of the results, Bonthala et al. [
44], reported a high correlation between cross-species microarray data and RNA-sequencing approaches for detecting differentially expressed genes under a cold temperature stress experiment in bambara groundnut. Probe-sets retained by the mask after genomic hybridisation are almost identical (>99%), suggesting that, at this level of resolution, the two genotypes are highly similar at the sequence level. Four known dehydration-associated genes, seen to be differentially expressed in these data, were subjected to qPCR, and supported the notion that the observed trends in the microarray data are valid.
The 26,496 probe sets common between the two genotypes, under irrigated conditions, (with a RMA cut-off of 0.97), include some sixty dehydration- and ABA-related genes. The latter include genes for producing osmoprotectants. They might provide two components of the dehydration avoidance capability of these genotypes, by retaining normal cell functioning when water access becomes limited. Clearly, if the plant has already activated part of the dehydration response, it could have multiple effects. The presence of osmoprotectants might draw in even more water than otherwise might be the case, and there will be a greater proportion of biomass devoted to root growth, resulting in even deeper roots that are better able to survive dehydration later on. Bambara groundnut is known to allocate a greater fraction of its dry weight to the roots than to the shoots, irrespective of the soil moisture status [
27]. This strategy may have clear advantages when water subsequently becomes limited, suggesting an adaptation to harsh environments and a decision to prioritise survival. In addition, as bambara groundnut is grown in harsh environments and has not undergone intensive breeding for the yield and above ground biomass, this suggests that it still allocates more effort to developing root architecture to handle dehydration when it happens. Moreover, Nayamudeza [
27] also stated that the fraction of total dry weight allocated to the roots in bambara groundnut is greater than that allocated to the groundnut. In addition, a relatively higher expression of dehydration-associated genes in both genotypes under water-sufficient treatment including
ABI1 (ABA Insensitive 1),
ABF1 (ABRE binding factor 1),
ERD4 (Early responsive to dehydration 4), and
RD19 (Response to dehydration 19), compared to other species such as Soybean [
76] (see
Figure S5), suggest that bambara groundnut could at least be in a partially ready state for dehydration, even in the absence of dehydration stress. However, further research is needed to validate this hypothesis.
Given that 59,782 and 59,835 probe-sets were used to evaluate the transcriptome changes after probe-masking in DipC and TN, respectively, there were only very small numbers of genes significantly differentially expressed (189 in DipC and 81 in TN) under water-limited treatment. It could be speculated that the slow and progressive dehydration stress might not cause significant shock to the plants.
The upregulated genes in both genotypes were subdivided into ~75% dehydration responsive (with expression levels returning to normal after recovery) and ~25% dehydration perturbed (where the expression levels remained altered). In the case of downregulated genes, 80–85% of the expression levels returned to being comparable with the non-stressed state. The dehydration-perturbed expression levels might be caused by changes at the chromatin level, through DNA methylation or histone modification, and it is therefore interesting to note that a protein-lysine demethylase is repressed by dehydration.
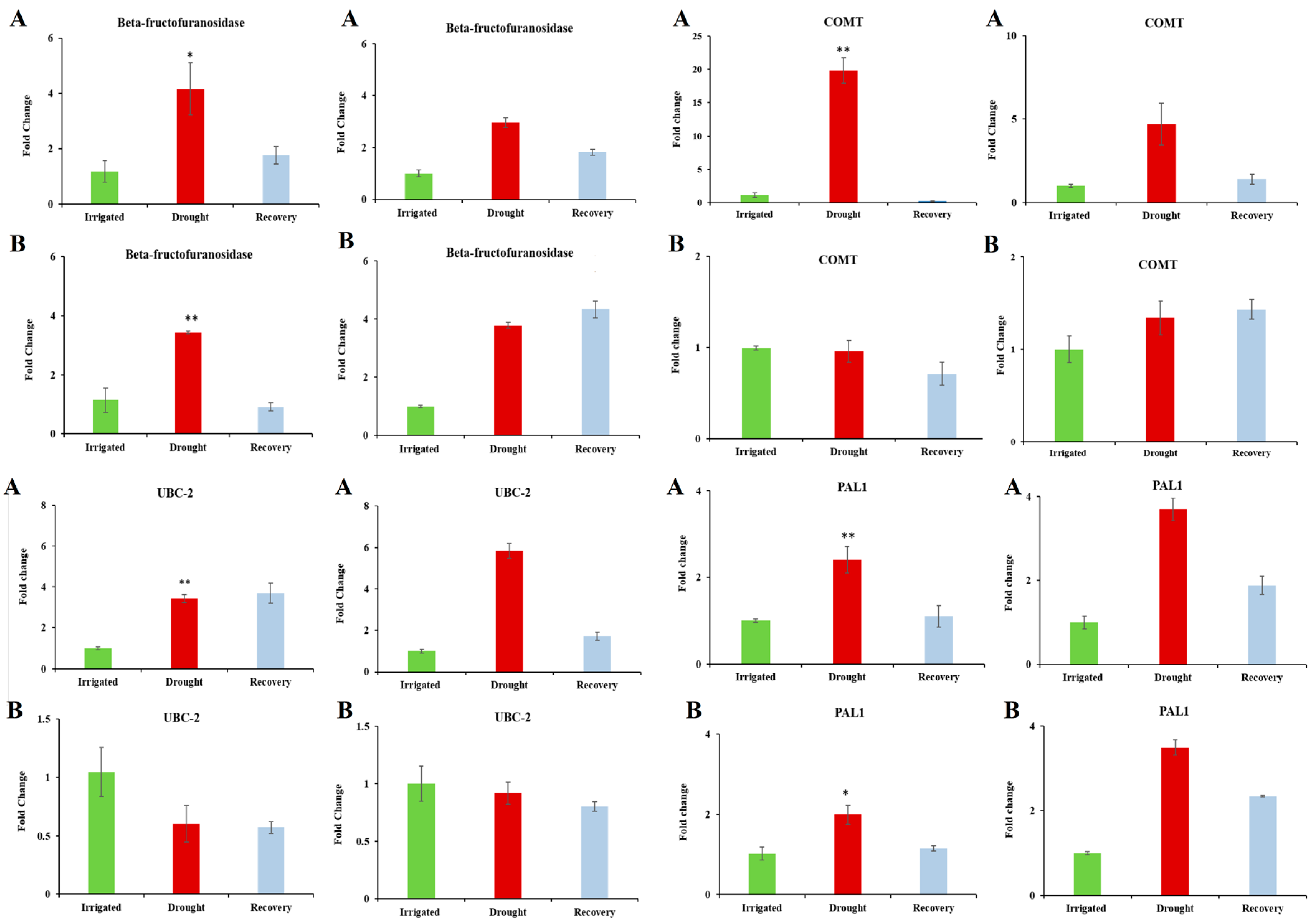
The above observations show that the two genotypes appear to be very similar in terms of their genotype (validating the comparability of the transcriptome data compared using the microarray), while exhibiting differences in their general transcriptional behaviour in water-sufficient conditions and in response to dehydration stress. However, when the sets of differentially expressed genes are compared, there is almost no overlap. Out of 189 and 91 genes differentially expressed in DipC and TN, respectively, only nine were common between the two genotypes, suggesting that some of the mechanisms for adaption to dehydration are substantially different in the two genotypes. Of these, Beta-fructofuranosidase contributes to osmoprotection [
46,
47], an MYB gene is associated with the stomatal opening in Arabidopsis thaliana [
50],
BRH1 affects the stomatal density [
51], and bsd2 affects photosynthesis in maize [
52], while
JMJD5 plays an epigenetic role [
49], as mentioned above.
Figure 4 illustrates how two genotypes with very similar genomes may have adapted to achieve dehydration response traits (transcriptional and hormone signalling to affect cell-wall modification, lignin synthesis, photosynthesis, transporters, hormone signalling, osmoprotection, oxidative stress) through largely different sets of effector genes.
Several transcription factors that seem likely to play a role in the bambara groundnut dehydration response and which are common to both genotypes are
BRH1 and an MYB transcription factor, which are known to affect the stomata in
Arabidopsis thaliana [
50], and
JMJD5. DipC shows a more significant response, with changes to
WRKY40, and is of particular interest. It is a well-known member of plant dehydration-response networks [
67] and is the most highly linked TF node in the co-expression networks. For DipC, the network also reveals the importance of
PRR7, a core circadian clock component known to play a complex role in abiotic stresses [
77]. It is somewhat surprising that TN does not show a >2-fold change in the expression of
WRKY40, but it may have roles for CONSTANS-like 1 (another clock-related gene associated with flowering in rice that may be associated with abiotic stress in bambara groundnut [
78]) and
MYB60, which affect stomatal closure in
A. thaliana [
79], and
AGL-83, a MADS-Box protein with an uncertain role.