Role of Archaeal HerA Protein in the Biology of the Bacterium Thermus thermophilus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Growth Conditions
2.2. Isolation of Mutants
2.3. HepA Inmunodetection
2.4. Protein Purification and ATPase Assays
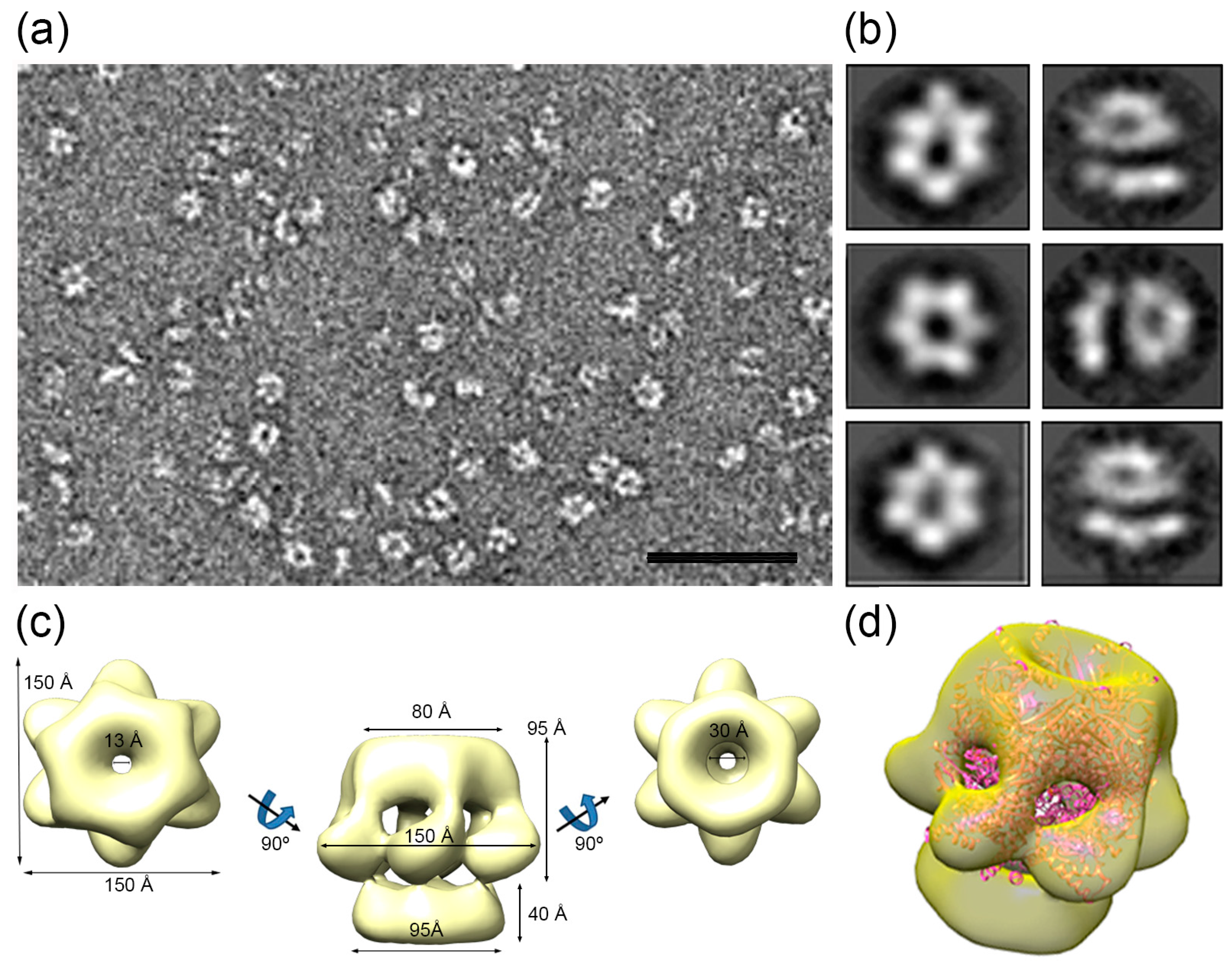
2.5. Single-Particle Electron Microscopy and Image Processing
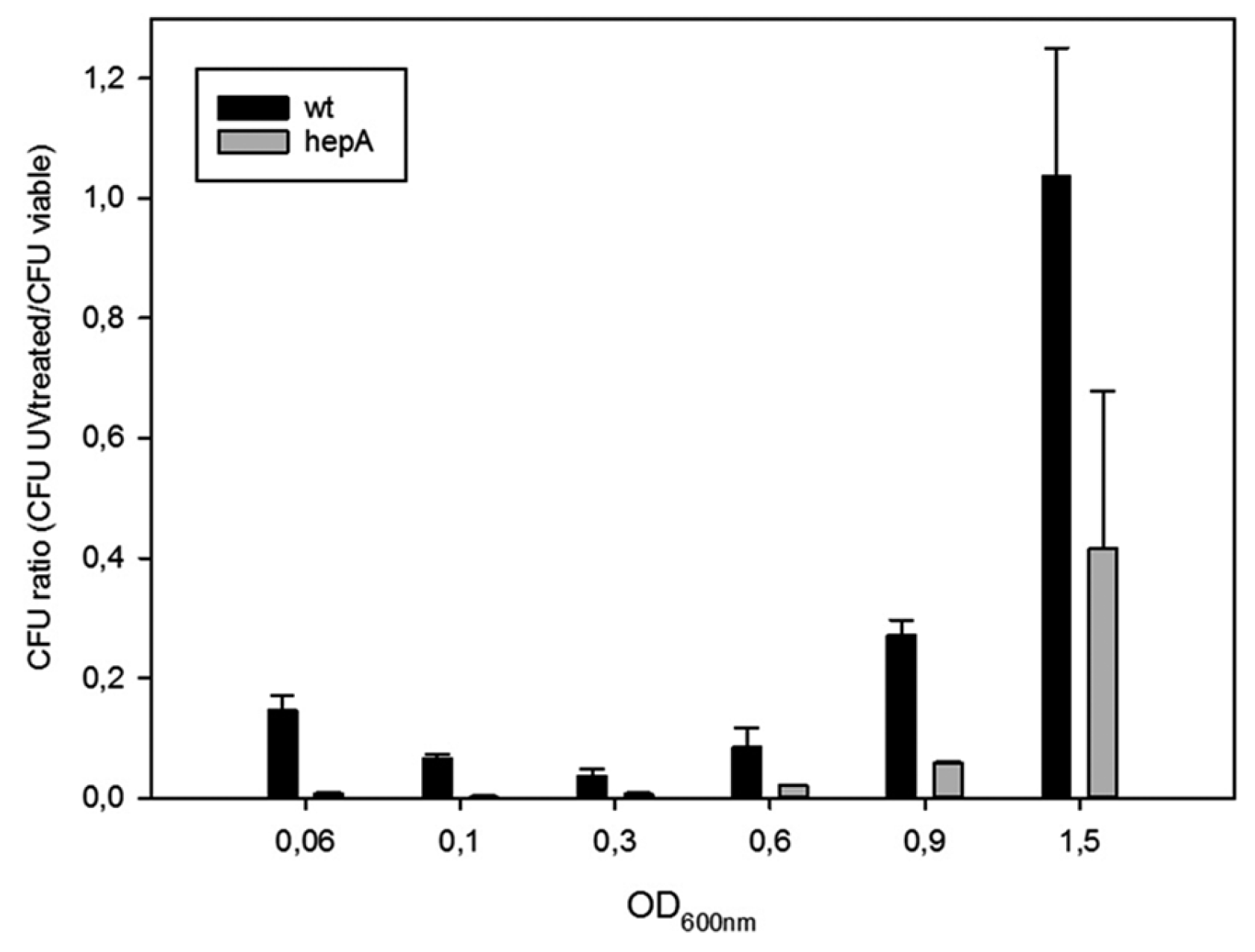
2.6. UV and Temperature Resistance Assays
2.7. Confocal Microscopy
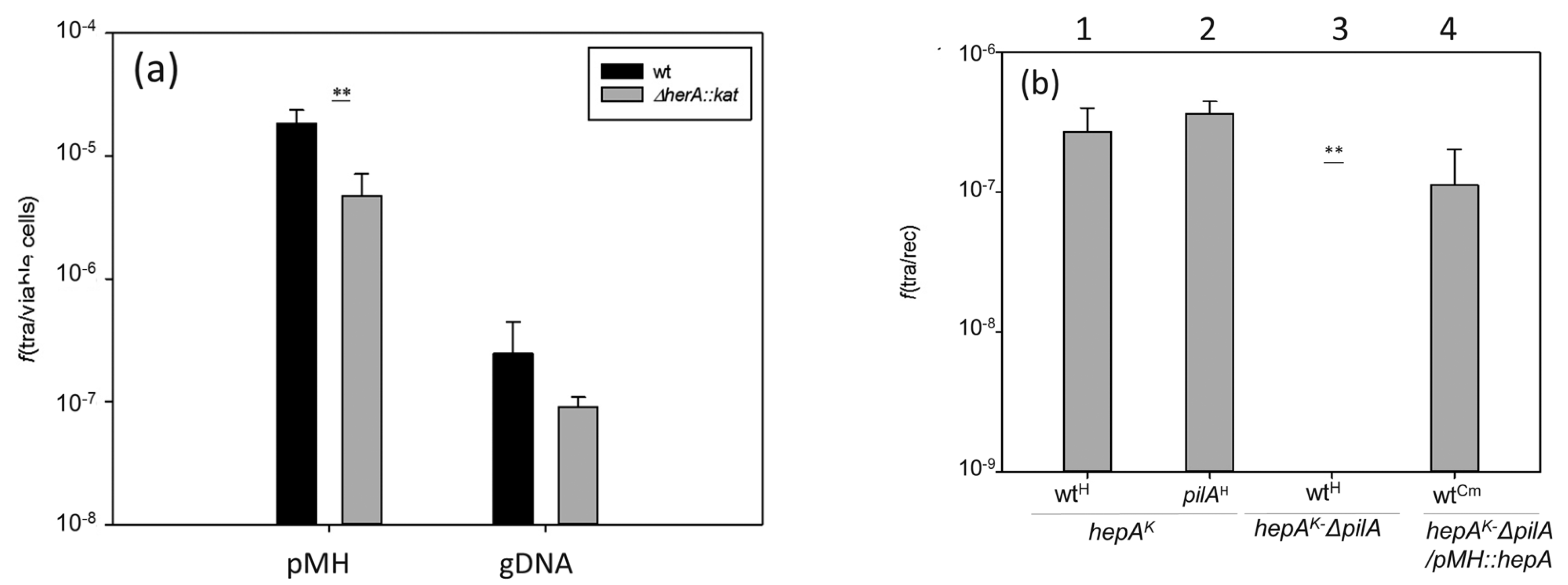
2.8. Transformation and Transjugation Assays
3. Results
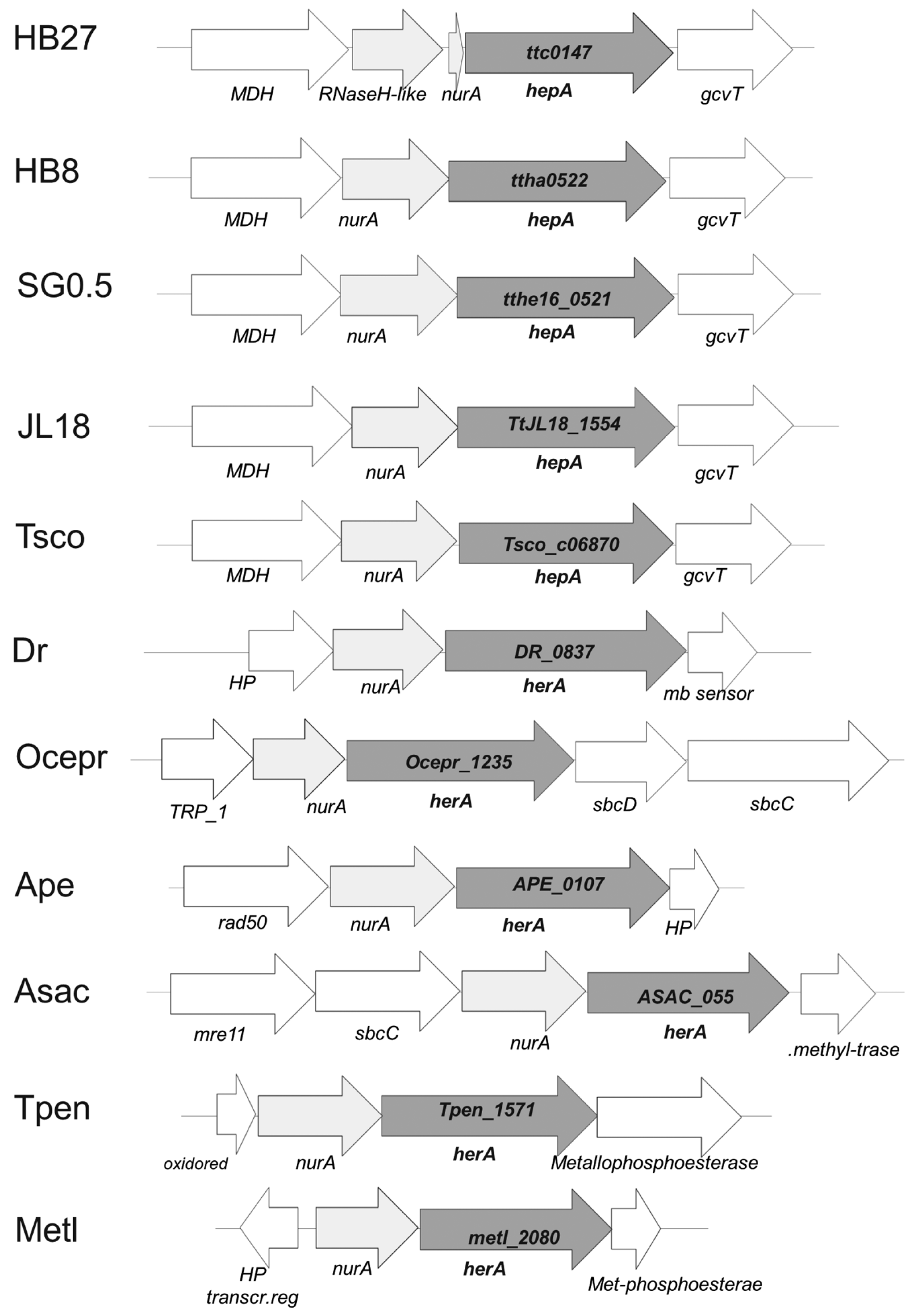
3.1. Presence of HerA Homologs in Thermus
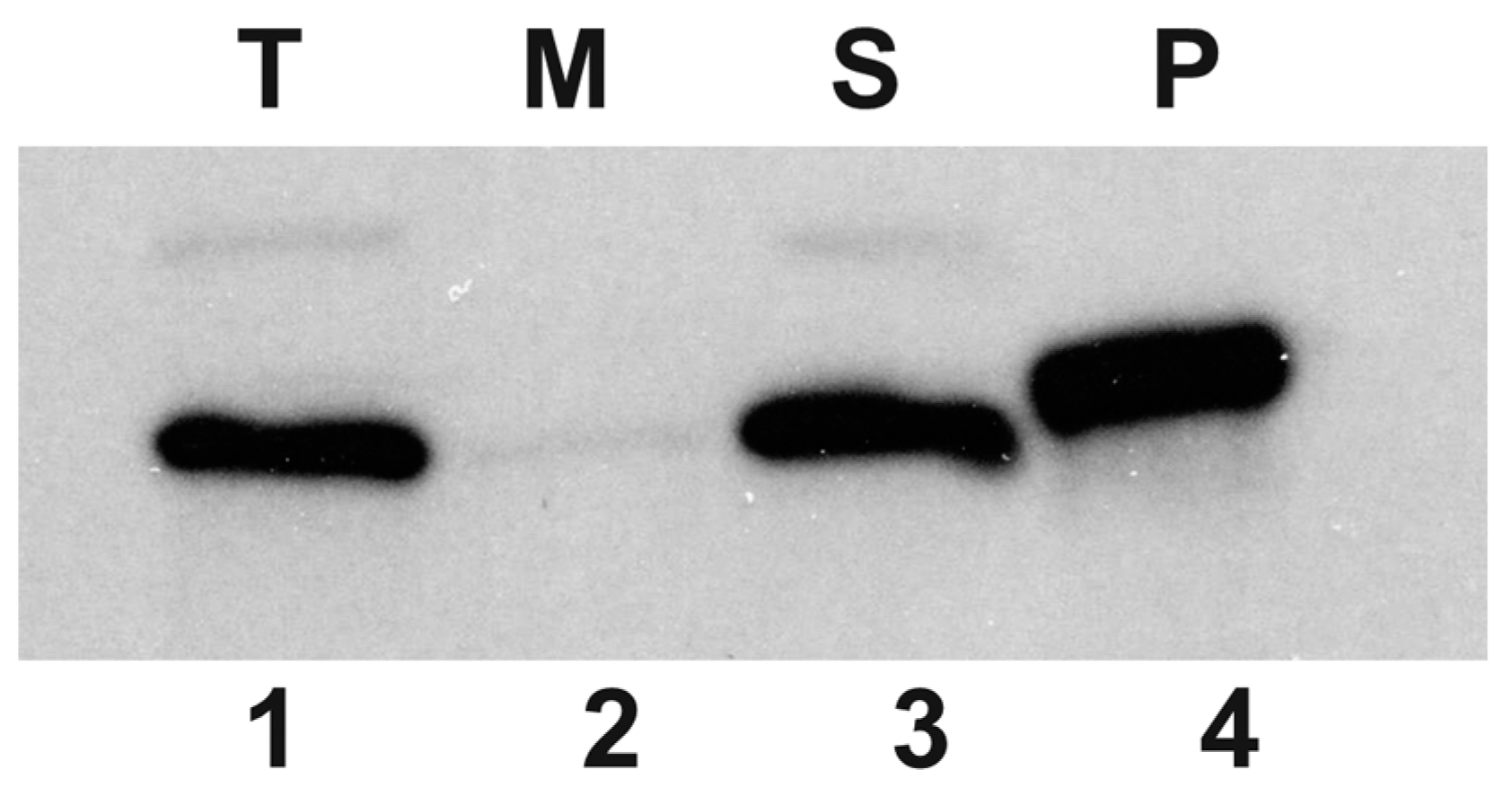
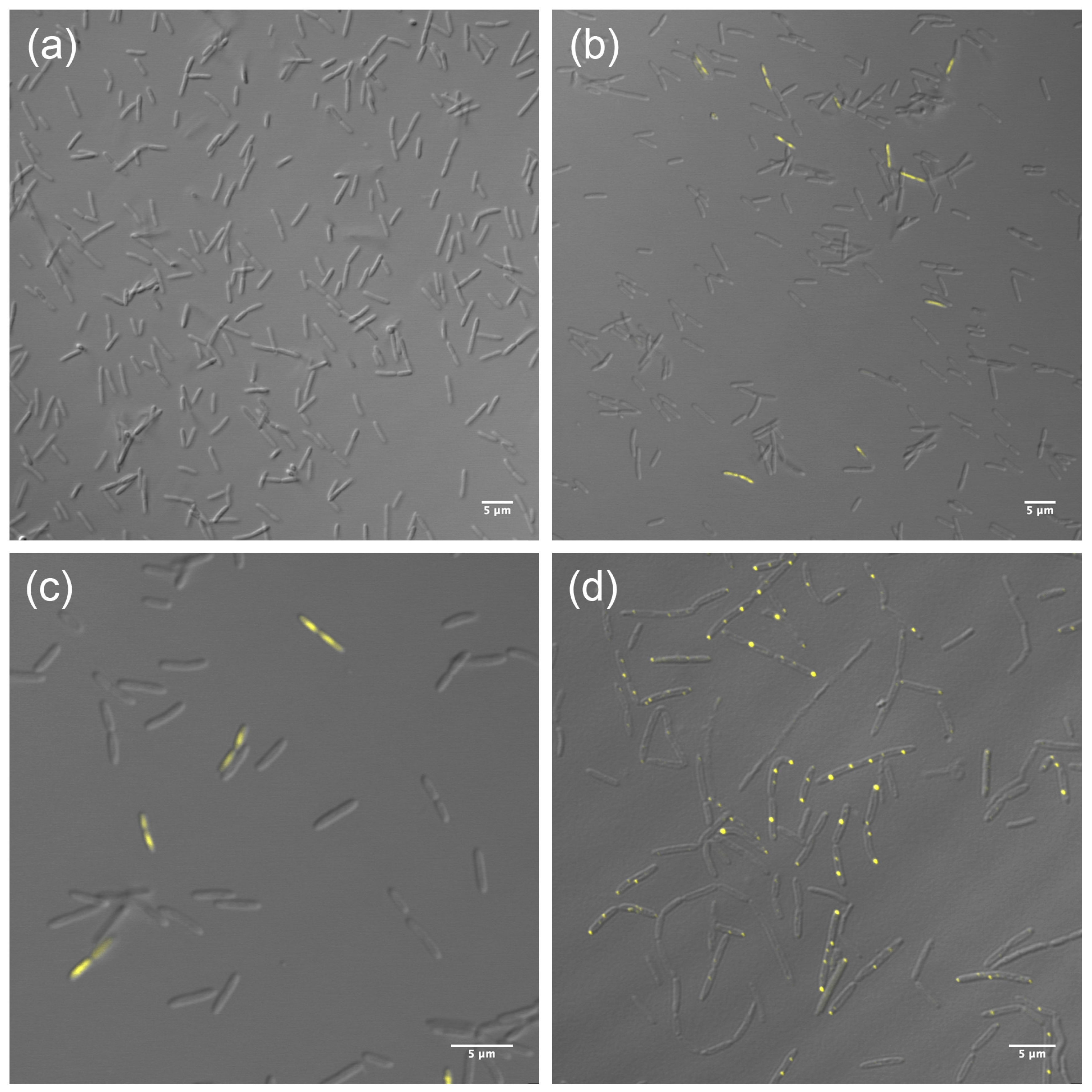
3.2. Localization of HepA
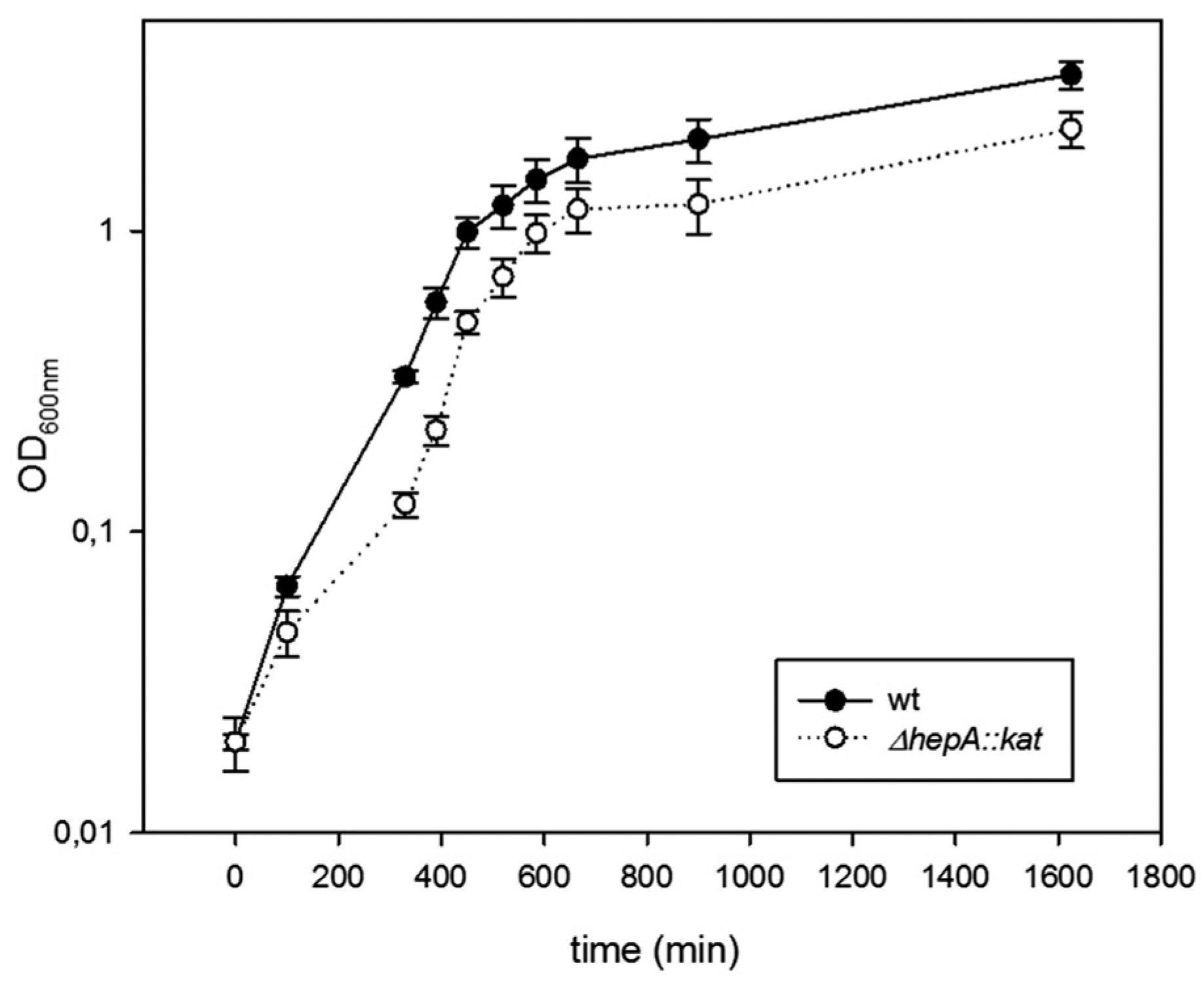
3.3. The Role of HepA
3.4. HepA is Required for DNA Donation in Transjugation
3.5. HepA Shows Low ATPase Activity
3.6. HepA Is a Complex Hexameric ATPase
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Wu, D.; Hugenholtz, P.; Mavromatis, K.; Pukall, R.; Dalin, E.; Ivanova, N.N.; Kunin, V.; Goodwin, L.; Wu, M.; Tindall, B.J.; et al. A phylogeny-driven genomic encyclopaedia of bacteria and archaea. Nature 2009, 462, 1056–1060. [Google Scholar] [CrossRef] [PubMed]
- Hug, L.A.; Baker, B.J.; Anantharaman, K.; Brown, C.T.; Probst, A.J.; Castelle, C.J.; Butterfield, C.N.; Hernsdorf, A.W.; Amano, Y.; Ise, K.; et al. A new view of the tree of life. Nat. Microbiol. 2016, 1, 16048. [Google Scholar] [CrossRef] [PubMed]
- Omelchenko, M.V.; Wolf, Y.I.; Gaidamakova, E.K.; Matrosova, V.Y.; Vasilenko, A.; Zhai, M.; Daly, M.J.; Koonin, E.V.; Makarova, K.S. Comparative genomics of thermus thermophilus and deinococcus radiodurans: Divergent routes of adaptation to thermophily and radiation resistance. BMC Evol. Biol. 2005, 5, 57. [Google Scholar] [CrossRef] [PubMed]
- Bruggemann, H.; Chen, C. Comparative genomics of Thermus thermophilus: Plasticity of the megaplasmid and its contribution to a thermophilic lifestyle. J. Biotechnol. 2006, 124, 654–661. [Google Scholar] [CrossRef] [PubMed]
- Iyer, L.M.; Makarova, K.S.; Koonin, E.V.; Aravind, L. Comparative genomics of the ftsk—HerA superfamily of pumping atpases: Implications for the origins of chromosome segregation, cell division and viral capsid packaging. Nucleic Acids Res. 2004, 32, 5260–5279. [Google Scholar] [CrossRef] [PubMed]
- Rzechorzek, N.J.; Blackwood, J.K.; Bray, S.M.; Maman, J.D.; Pellegrini, L.; Robinson, N.P. Structure of the hexameric HerA ATPase reveals a mechanism of translocation-coupled DNA-end processing in archaea. Nat. Commun. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Cheng, K.; Chen, X.; Xu, G.; Wang, L.; Xu, H.; Yang, S.; Zhao, Y.; Hua, Y. Biochemical and functional characterization of the NurA-HerA complex from Deinococcus radiodurans. J. Bacteriol. 2015, 197, 2048–2061. [Google Scholar] [CrossRef] [PubMed]
- Klostermeier, D. Rearranging RNA structures at 75 °C? Toward the molecular mechanism and physiological function of the thermus thermophilus dead-box helicase HerA. Biopolymers 2013, 99, 1137–1146. [Google Scholar] [PubMed]
- Ramirez-Arcos, S.; Fernandez-Herrero, L.A.; Berenguer, J. A thermophilic nitrate reductase is responsible for the strain specific anaerobic growth of Thermus thermophilus HB8. Biochim. Biophys. Acta 1998, 1396, 215–227. [Google Scholar] [CrossRef]
- Hanahan, D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983, 166, 557–580. [Google Scholar] [CrossRef]
- Rosenberg, A.H.; Lade, B.N.; Dao-shan, C.; Lin, S.-W.; Dunn, J.J.; Studier, F.W. Vectors for selective expression of cloned DNAs by T7 RNApolymerase. Gene 1987, 56, 125–135. [Google Scholar] [CrossRef]
- Cava, F.; Zafra, O.; Magalon, A.; Blasco, F.; Berenguer, J. A new type of NADH dehydrogenase specific for nitrate respiration in the extreme thermophile Thermus thermophilus. J. Biol. Chem. 2004, 279, 45369–45378. [Google Scholar] [CrossRef] [PubMed]
- Swarts, D.C.J.; Matthijs, M.; Westra, E.R.; Zhu, Y.; Janssen, J.H.; Snijders, A.P.; Wang, Y.; Patel, D.J.; Berenguer, J.; Brouns, S.J.; et al. DNA-guided DNA interference by a prokaryotic arognaute. Nature 2014, 507, 258–261. [Google Scholar] [CrossRef] [PubMed]
- Blesa, A.; César, C.E.; Averhoff, B.; Berenguer, J. Non canonical cell-to-cell DNA transfer in Thermus spp. Is insensitive to argonaute-mediated interference. J. Bacteriol. 2014. [Google Scholar] [CrossRef]
- Blesa, A.; Baquedano, I.; Quintáns, N.G.; Mata, C.P.; Castón, J.R.; Berenguer, J. The transjugation machinery of Thermus thermophilus: Identification of TdtA, an ATPase involved in DNA donation. PLoS Genet. 2017, 13, e1006669. [Google Scholar] [CrossRef] [PubMed]
- Vieira, J.; Messing, J. The pUC plasmids, an m13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene 1982, 19, 259–268. [Google Scholar] [CrossRef]
- Marabini, R.; Masegosa, I.M.; San Martin, M.C.; Marco, S.; Fernandez, J.J.; de la Fraga, L.G.; Vaquerizo, C.; Carazo, J.M. Xmipp: An image processing package for electron microscopy. J. Struct. Biol. 1996, 116, 237–240. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Mindell, J.A.; Grigorieff, N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 2003, 142, 334–347. [Google Scholar] [CrossRef]
- Sorzano, C.O.; Bilbao-Castro, J.R.; Shkolnisky, Y.; Alcorlo, M.; Melero, R.; Caffarena-Fernandez, G.; Li, M.; Xu, G.; Marabini, R.; Carazo, J.M. A clustering approach to multireference alignment of single-particle projections in electron microscopy. J. Struct. Biol. 2010, 171, 197–206. [Google Scholar] [CrossRef] [PubMed]
- Ludtke, S.J.; Baldwin, P.R.; Chiu, W. Eman: Semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 1999, 128, 82–97. [Google Scholar] [CrossRef] [PubMed]
- Scheres, S.H.; Nunez-Ramirez, R.; Sorzano, C.O.; Carazo, J.M.; Marabini, R. Image processing for electron microscopy single-particle analysis using Xmipp. Nat. Protoc. 2008, 3, 977–990. [Google Scholar] [CrossRef] [PubMed]
- Phosphate-buffered saline (PBS). Available online: http://cshprotocols.cshlp.org/content/2006/1/pdb.rec8247 (accessed on 26 April 2017).
- Constantinesco, F.; Forterre, P.; Koonin, E.V.; Aravind, L.; Elie, C. A bipolar DNA helicase gene, herA, clusters with rad50, mre11 and nurA genes in thermophilic archaea. Nucleic Acids Res. 2004, 32, 1439–1447. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, B.B.; Paull, T.T. The p. Furiosus mre11/rad50 complex promotes 5′ strand resection at a DNA double-strand break. Cell 2008, 135, 250–260. [Google Scholar] [CrossRef] [PubMed]
- Chae, J.; Kim, Y.C.; Cho, Y. Crystal structure of the NurA-dAMP-Mn2+ complex. Nucleic Acids Res. 2012, 40, 2258–2270. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Tian, B.; Li, S.; Ao, X.; Dalgaard, K.; Gokce, S.; Liang, Y.; She, Q. Genetic manipulation in Sulfolobus islandicus and functional analysis of DNA repair genes. Biochem. Soc. Trans. 2013, 41, 405–410. [Google Scholar] [CrossRef] [PubMed]
- Henne, A.; Bruggemann, H.; Raasch, C.; Wiezer, A.; Hartsch, T.; Liesegang, H.; Johann, A.; Lienard, T.; Gohl, O.; Martinez-Arias, R.; et al. The genome sequence of the extreme thermophile Thermus thermophilus. Nat. Biotechnol. 2004, 22, 547–553. [Google Scholar] [CrossRef] [PubMed]
- Castan, P.; Casares, L.; Barbe, J.; Berenguer, J. Temperature-dependent hypermutational phenotype in recA mutants of Thermus thermophilus HB27. J. Bacteriol. 2003, 185, 4901–4907. [Google Scholar] [CrossRef] [PubMed]
- Ohtani, N.; Tomita, M.; Itaya, M. An extreme thermophile, Thermus thermophilus, is a polyploid bacterium. J. Bacteriol. 2010, 192, 5499–5505. [Google Scholar] [CrossRef] [PubMed]

| Strain | Genotype | Source |
|---|---|---|
| E. coli DH5α | supE44 ΔlacU169 (Φ80 lacZΔM15) hsdR17, recA1, endA1, gyrA96, thi-1 relA1 | [10] |
| E. coli BL21 (DE3) | F- ompT gal dcm lon HsdSB (rB-mB-) λ(DE3 [lacI lacUV5-T7 gene 1 ind1 sam7 nin5]) | [11] |
| T. thermophilus HB27 | ATCC BAA-163/DSM7039 | Y. Koyama |
| T. thermophilus NAR1 | [pTT27::nar] | [12] |
| T. thermophilus HB8 | ATCC 27634 | Y. Koyama |
| T. thermophilus HB27EC | HB27 ago::agoISTth7 | [13] |
| T. thermophilus HB27Cm | This work | |
| T. thermophilus ∆pilA4 | HB27EC ∆pilA4 | [14] |
| T. thermophilus HB27 ∆pilA4::hyg | HB27EC ΔTTC0858::hyg | [14] |
| T. thermophilus HB27H | HB27ECΔTTC0313::hyg | [14] |
| T. thermophilus HB27 gdh::kat | HB27 [∆TTC1211::kat] | [12] |
| T. thermophilus HB27 hepA::pk | HB27EC TTC0147::pK18 | This work |
| T. thermophilus HB27 ∆hepA::kat | HB27EC ∆TTC0147::kat | This work |
| T. thermophilus HB27 ∆hepA::hyg | HB27EC ∆TTC0147::hyg, | This work |
| T. thermophilus HB27 ∆hepA::kat pilA4 | HB27EC ∆TTC0147::kat, ∆pilA4 | This work |
| T. thermophilus HB27 hepAYFPph | HB27EC [TTC0147-sYFP:pH118] | This work |
| T. thermophilus HB27 ∆hepA, ∆pilA4 PMH | HB27EC ∆TTC0147::kat, ∆pilA4 [pMH::TTC0147] | This work |
| Primer (Use) | Sequence 5′–> 3′ |
|---|---|
| TTC0313dir | CTTTACGAGGCCCTCTTGGAG |
| TTC0313rev | CCACCGCTCGGGGAC |
| AB92 (check pilA4 deletion) | AAATGCTGAAGCTTGGCGGCAAC |
| AB93 (check pilA4 deletion) | AAAAGAATTCGGGAGTTAGGCTTGGGATTGTG |
| AB219 (check hepA deletion) | CTACCTGAAGAACTCCCGGCGCAG |
| AB220 (check hepA deletion) | GTGAAGCGTATCGGCGTGGTCTTG |
| AB221 (insertion hepA) | aaagaattcGAGGTGGCCTACCTCAACCTG |
| AB222 (insertion hepA) | aaactgcagGAGCTCGTCCAGGACGATGAAG |
| AB231 (hepA-YFP fusion) | actagtCCTGAAGAACTCCCGGCGCAG |
| AB232 (hepA-YFP fusion) | ccatggGTGAAGCGTATCGGCGTG |
| AB249 (deletion hepA) | aaagaattcGTAATCTAGGGGCATGGCCTG |
| AB250 (deletion hepA) | aaatctagaCTTCACCTCGCACCTCCCA |
| AB251 (deletion hepA) | aaatctagaGATGGCCTCGCCATGAAGA |
| AB252 (deletion hepA) | aaactgcagCAGGGGCGAATAC |
| AB247 (overexpression hepA) | aaacatatgGTGAAGCGTATCGGCGTG |
| AB248 (overexpression hepA) | aaaaagcttCTACCCGAAGAACTCCC |
| Plasmid | Description/Use | Reference |
|---|---|---|
| pET28b(+) | Recombinant expression of proteins in E. coli | Novagen |
| pUC19/18 | Cloning vector | [16] |
| pUC19::kat | pUC19 with the thermostable kat resistance gene cassette | This work |
| pUC19::hyg | pUC19 with the Hyg resistance gene cassette | This work |
| pK118 | Suicide vector for T. thermophilus, Thermostable Km resistance | [12] |
| pH118 | Suicide vector for T. thermophilus, Thermostable Hyg resistance | [15] |
| pMK184 | Cloning vector for T. thermophilus. Km resistance | [15] |
| pMH184 | Cloning vector for T. thermophilus. Hyg resistance | [15] |
| pMHPnqosYFP | Expression of sYFP in T. thermophilus | [12] |
| pAB135 | pET28b derivative for overexpression of N-terminal His-tagged HepA | This work |
| pAB207 | pUC18::hepA::kat. For the isolationon of ΔhepA::kat mutants | This work |
| pAB219 | pH118:hepA::sYFP. pH118 derivative to generate chromosomal fusions of HepA to sYFP | This work |
| pAB175 | Derivative of pMHPnqosYFP for the ectopic expression of the HepA-sYFP fusion in T. thermophilus | This work |
| pAB150 | Plasmid for the isolation of Hyg resistant single insertion hepA mutants | This work |
| pAB151 | Plasmid for the isolation of Km resistant single insertion hepA mutants | This work |
| pAB141 | Hyg-resistant plasmid for the hepA mutants | This work |
| Viable CFUs Ratio | 60 °C | 70 °C | 75 °C |
|---|---|---|---|
| wt (gdh) | 18.7 ± 1.7 × 107 | 40.9 ± 8 × 107 | 16.30 ± 1 × 107 |
| ΔhepA | 20.1 ± 9.3 × 107 | 33.3 ± 1 × 106 | 12.8 ± 3.9 × 104 |
| ΔhepA/wt | 1.1 ± 0.05 | 8.16 ± 1.3 × 10−2 | 7.8 ± 0.004 × 10−4 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Blesa, A.; Quintans, N.G.; Baquedano, I.; Mata, C.P.; Castón, J.R.; Berenguer, J. Role of Archaeal HerA Protein in the Biology of the Bacterium Thermus thermophilus. Genes 2017, 8, 130. https://doi.org/10.3390/genes8050130
Blesa A, Quintans NG, Baquedano I, Mata CP, Castón JR, Berenguer J. Role of Archaeal HerA Protein in the Biology of the Bacterium Thermus thermophilus. Genes. 2017; 8(5):130. https://doi.org/10.3390/genes8050130
Chicago/Turabian StyleBlesa, Alba, Nieves G. Quintans, Ignacio Baquedano, Carlos P. Mata, José R. Castón, and José Berenguer. 2017. "Role of Archaeal HerA Protein in the Biology of the Bacterium Thermus thermophilus" Genes 8, no. 5: 130. https://doi.org/10.3390/genes8050130






