MB0 and MBI Are Independent and Distinct Transactivation Domains in MYC that Are Essential for Transformation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture, Transfection, and Retroviral Infection
2.2. Plasmids and Expression Vectors
2.3. Reporter Assay
2.4. Immunoblotting
2.5. MYC/RAS Cotransformation
2.6. Proliferation and Apoptosis Assays
2.7. Quantitative Real-Time RT-PCR (qRT-PCR)
2.8. Data Analysis and Statistics
3. Results
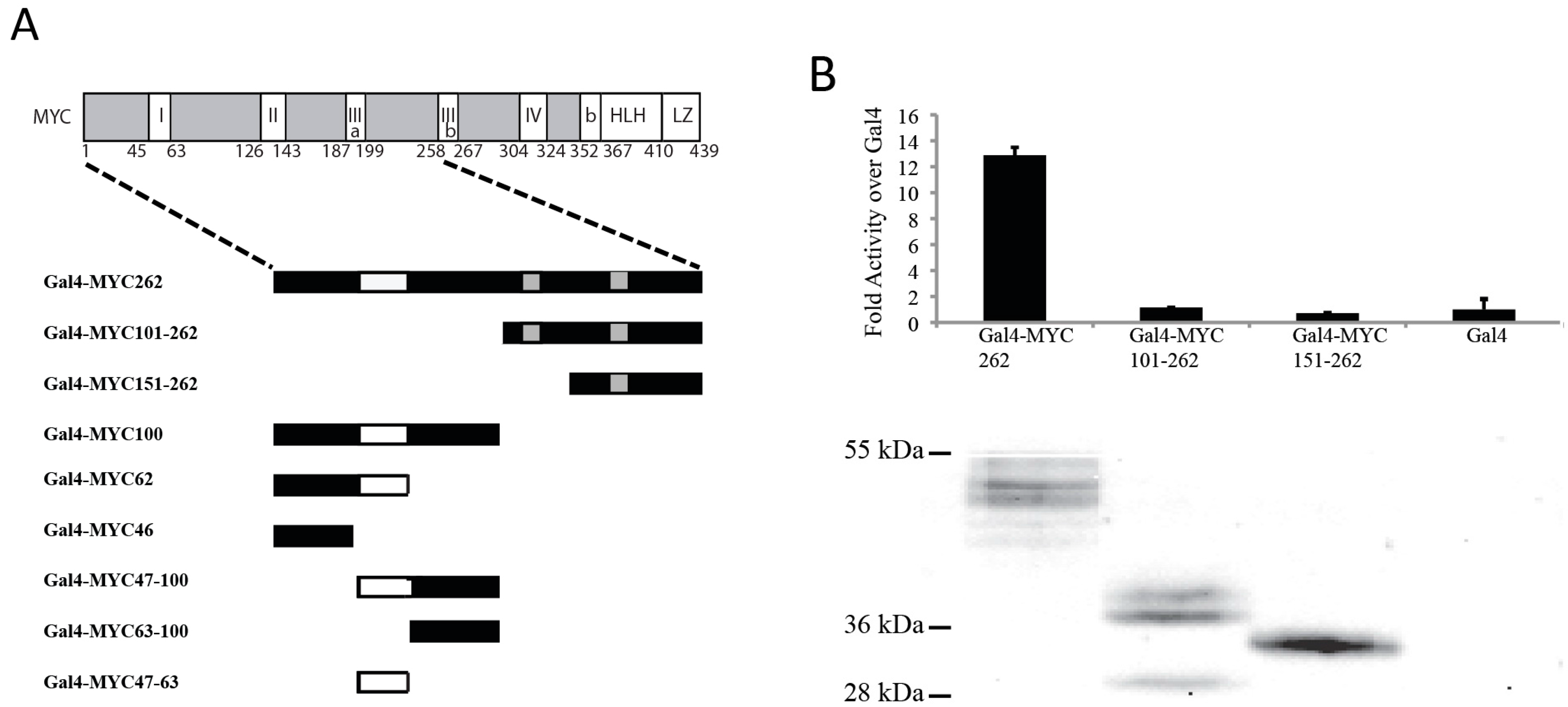
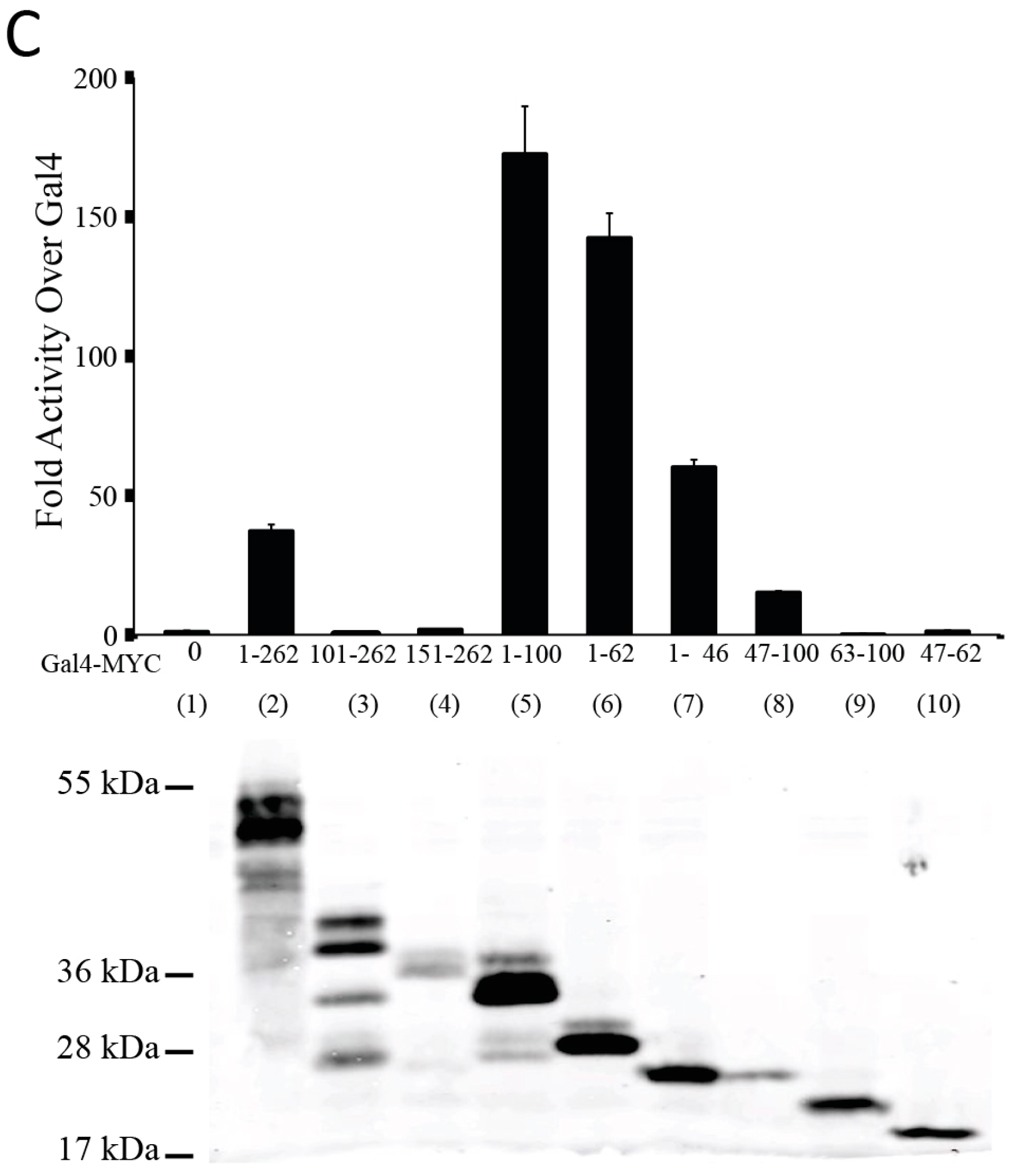
3.1. The N-Terminal 62 Amino Acids Represent the Transactivation Domain of MYC
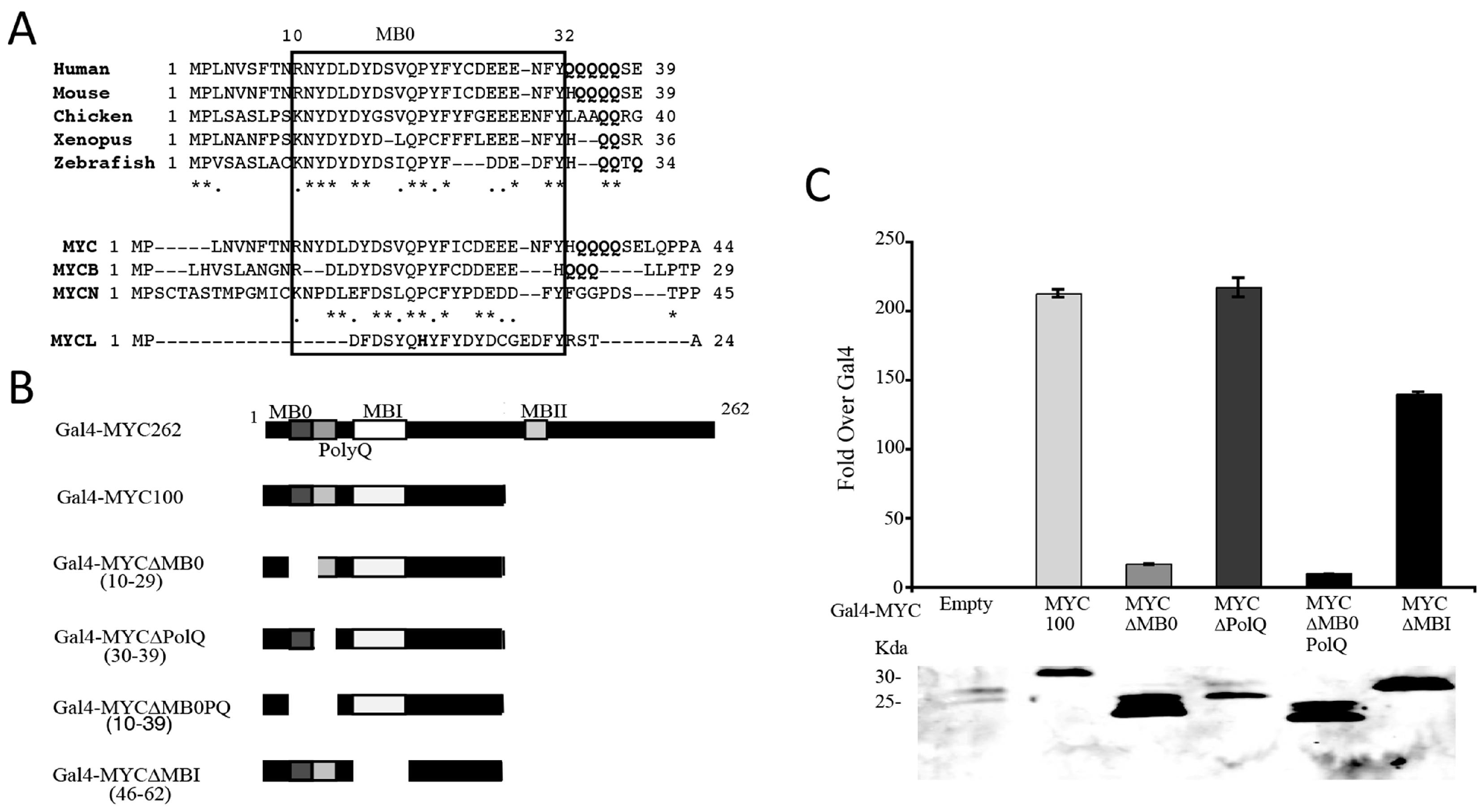
3.2. Identification of a Conserved MYC Transactivation Domain
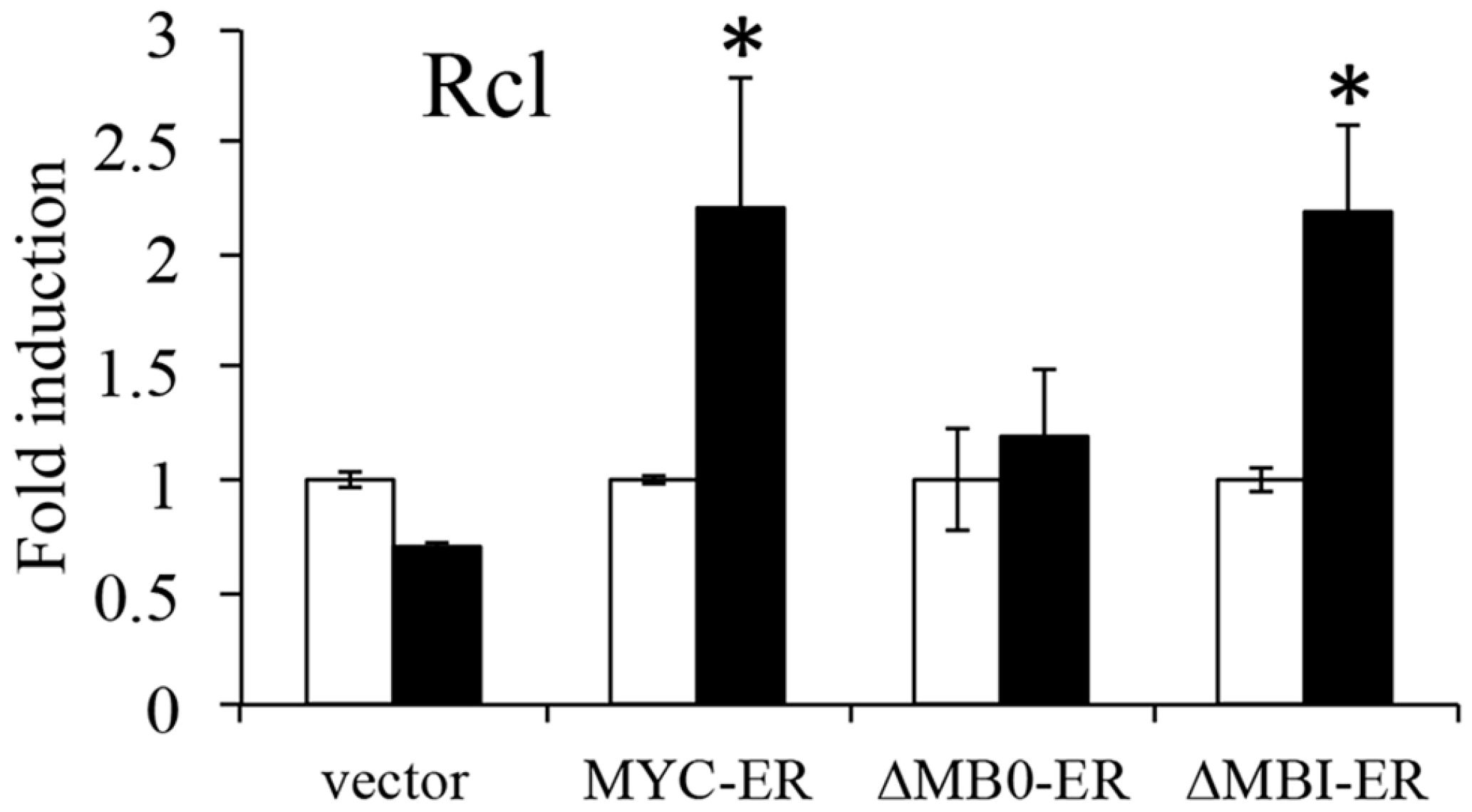
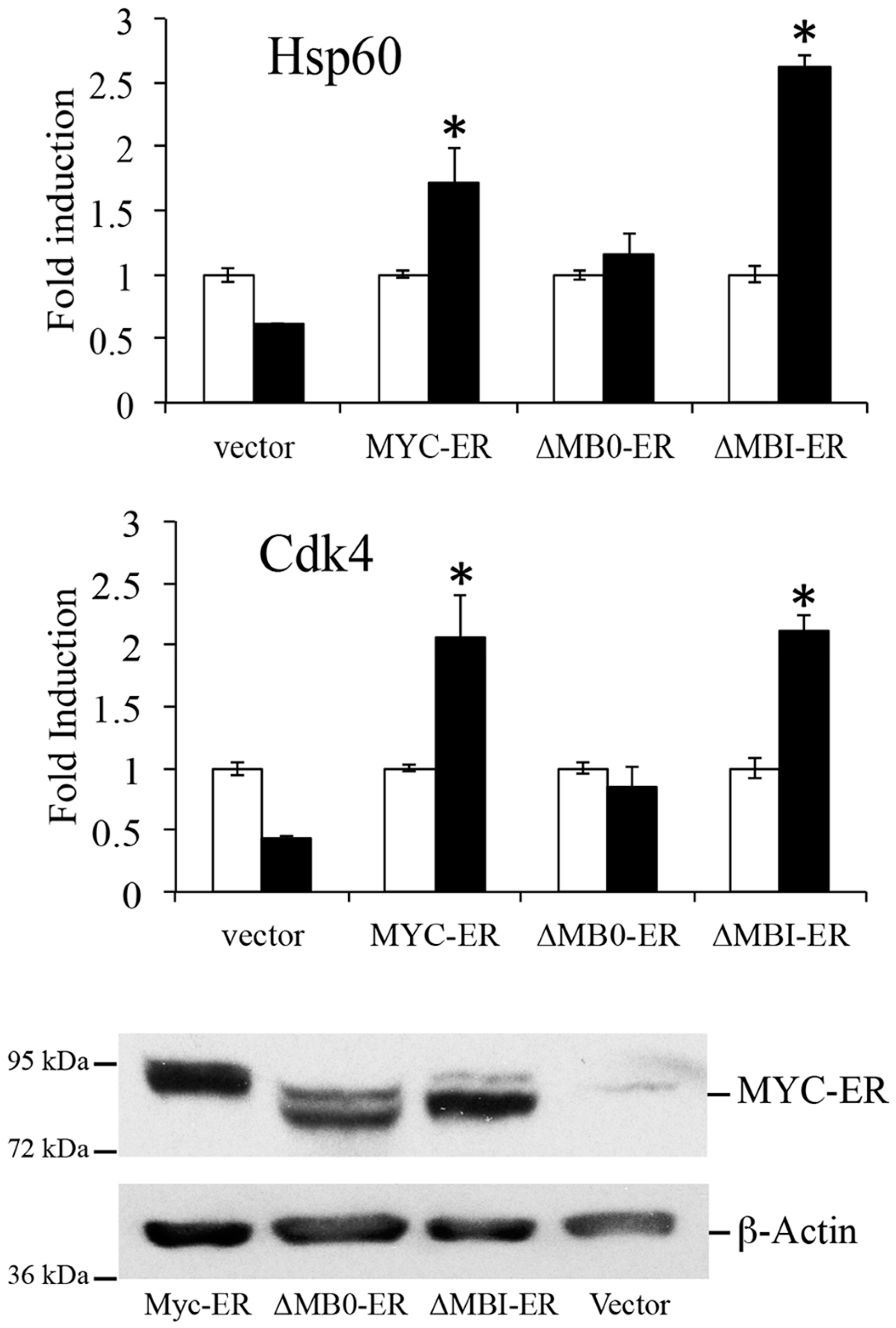
3.3. MB0 Is Necessary for Endogenous Target Gene Induction
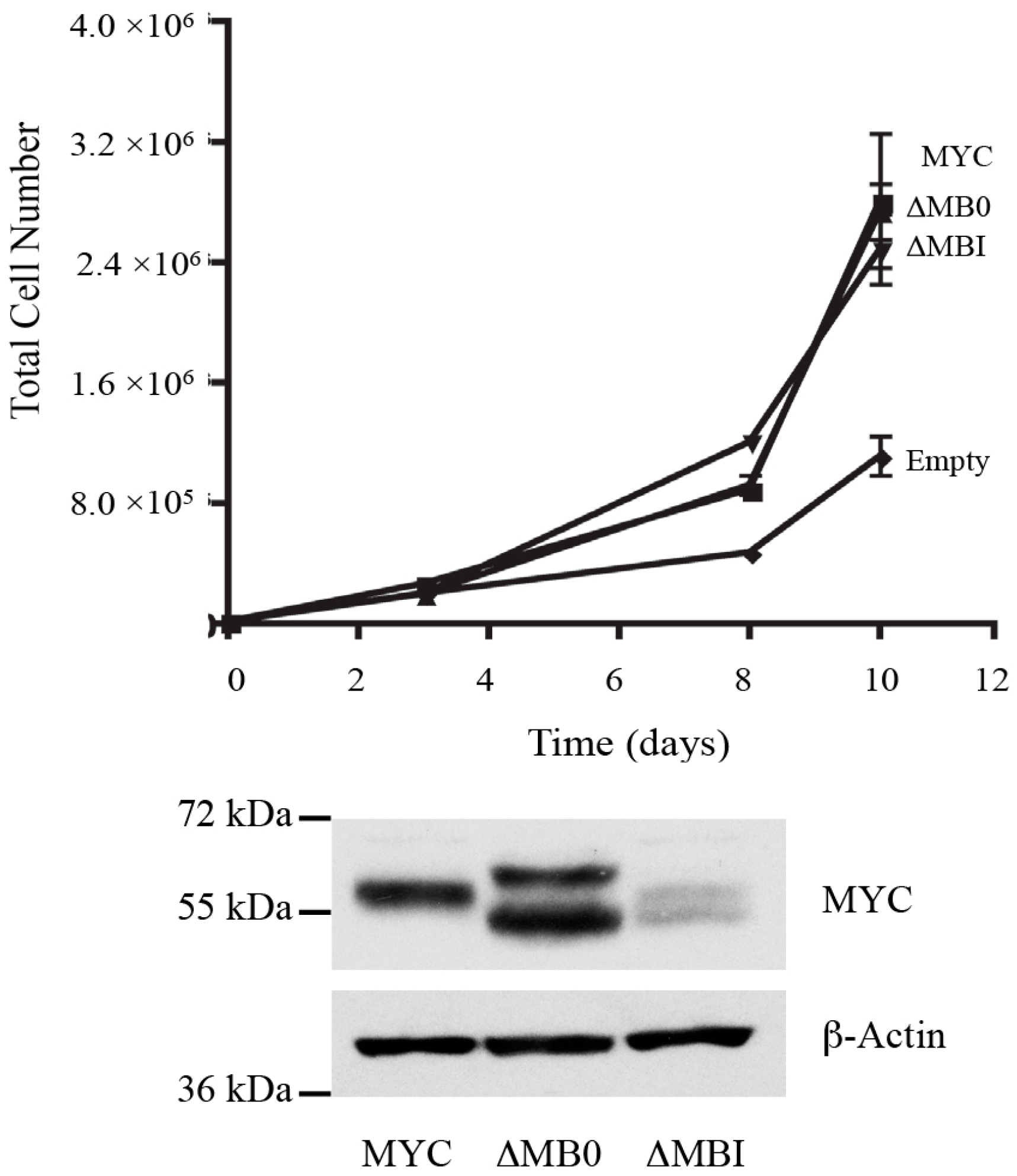
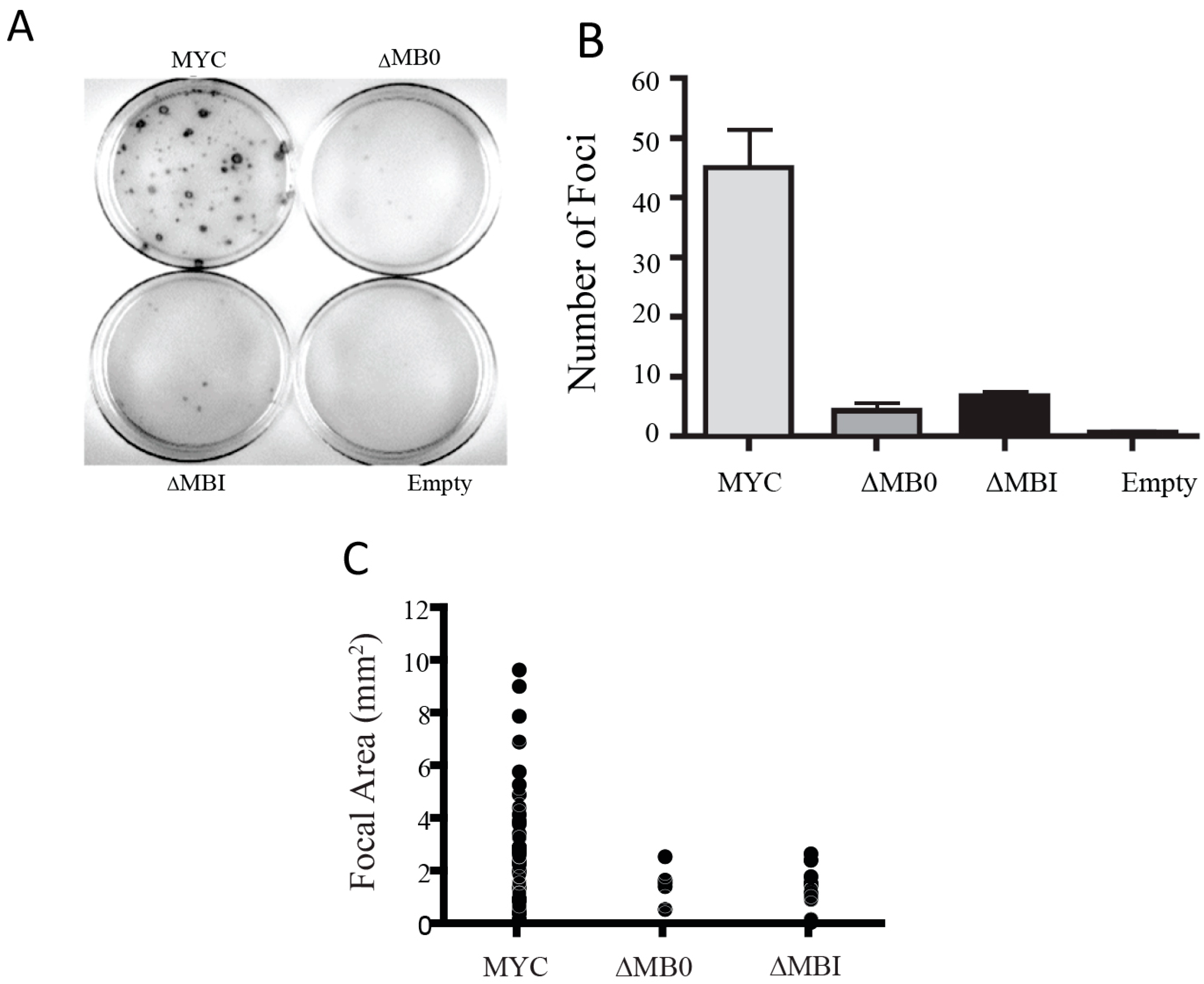
3.4. MB0 and MBI Are Essential for Cotransformation of Primary Cells with Activated RAS
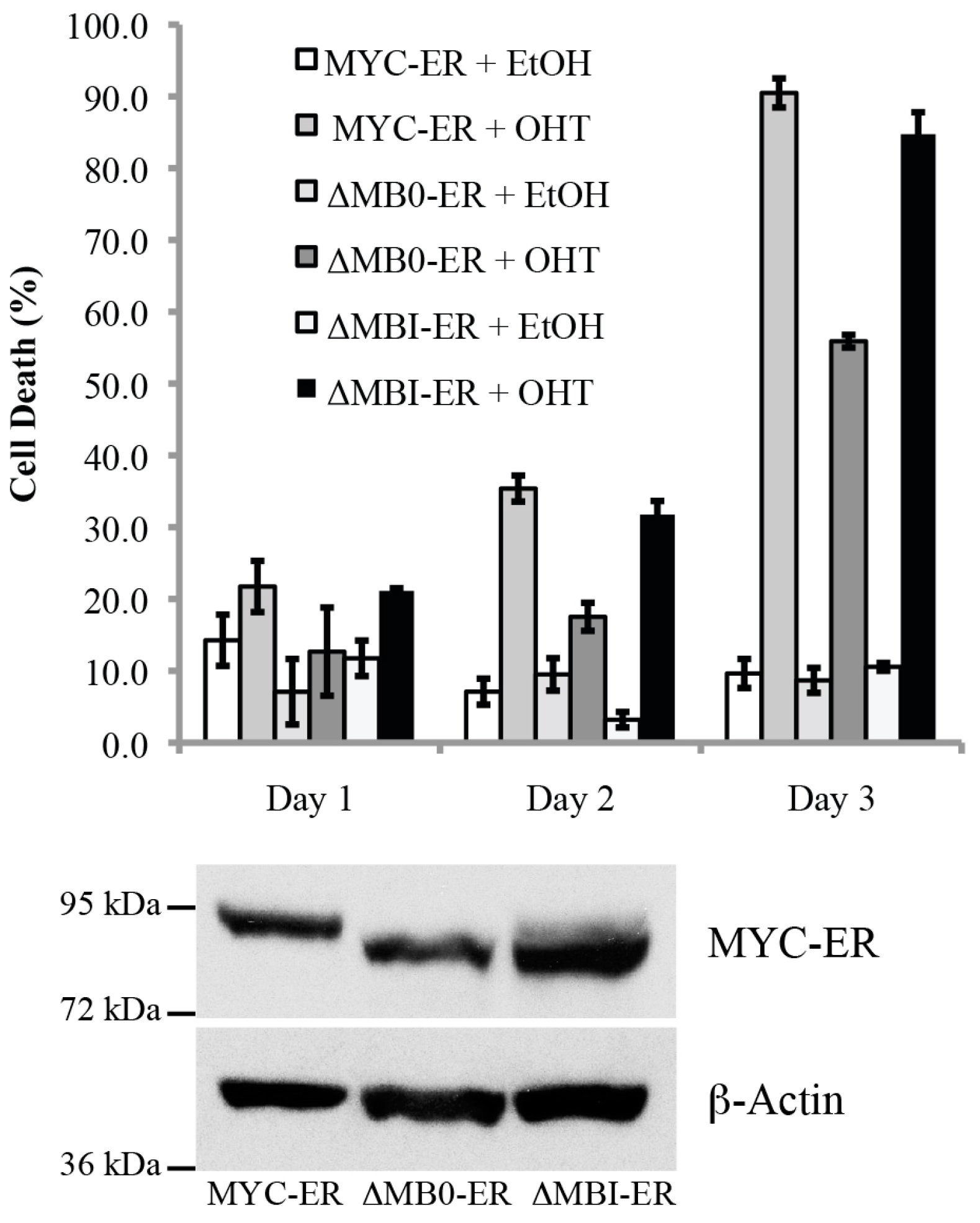
3.5. MB0, but Not MBI, Is Necessary for the Efficient Induction of p53-Independent Apoptosis
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Dang, C.V. MYC on the path to cancer. Cell 2012, 149, 22–35. [Google Scholar] [CrossRef] [PubMed]
- Meyer, N.; Penn, L.Z. Reflecting on 25 years with MYC. Nat. Rev. Cancer 2008, 8, 976–990. [Google Scholar] [CrossRef] [PubMed]
- Boone, D.N.; Qi, Y.; Li, Z.; Hann, S.R. Egr1 mediates p53-independent c-Myc-induced apoptosis via a noncanonical ARF-dependent transcriptional mechanism. Proc. Natl. Acad. Sci. USA 2011, 108, 632–637. [Google Scholar] [CrossRef] [PubMed]
- McMahon, S.B.; Van Buskirk, H.A.; Dugan, K.A.; Copeland, T.D.; Cole, M.D. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell 1998, 94, 363–374. [Google Scholar] [CrossRef]
- Cole, M.D.; Cowling, V.H. Transcription-independent functions of MYC: Regulation of translation and DNA replication. Nat. Rev. Mol. Cell Biol. 2008, 9, 810–815. [Google Scholar] [CrossRef] [PubMed]
- Dominguez-Sola, D.; Gautier, J. MYC and the control of DNA replication. Cold Spring Harb. Perspect. Med. 2014, 4, a014423. [Google Scholar] [CrossRef] [PubMed]
- Cowling, V.H.; Cole, M.D. Mechanism of transcriptional activation by the Myc oncoproteins. Semin. Cancer Biol. 2006, 16, 242–252. [Google Scholar] [CrossRef] [PubMed]
- Dang, C.V.; O’Donnell, K.A.; Zeller, K.I.; Nguyen, T.; Osthus, R.C.; Li, F. The c-Myc target gene network. Semin. Cancer Biol. 2006, 16, 253–264. [Google Scholar] [CrossRef] [PubMed]
- Blackwell, T.K.; Kretzner, L.; Blackwood, E.M.; Eisenman, R.N.; Weintraub, H. Sequence-specific DNA binding by the c-Myc protein. Science 1990, 250, 1149–1151. [Google Scholar] [CrossRef] [PubMed]
- Blackwood, E.M.; Eisenman, R.N. Max: A helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science 1991, 251, 1211–1217. [Google Scholar] [CrossRef] [PubMed]
- Blackwell, T.K.; Huang, J.; Ma, A.; Kretzner, L.; Alt, F.W.; Eisenman, R.N.; Weintraub, H. Binding of myc proteins to canonical and noncanonical DNA sequences. Mol. Cell. Biol. 1993, 13, 5216–5224. [Google Scholar] [CrossRef] [PubMed]
- Hann, S.R.; Dixit, M.; Sears, R.C.; Sealy, L. The alternatively initiated c-Myc proteins differentially regulate transcription through a noncanonical DNA-binding site. Genes Dev. 1994, 8, 2441–2452. [Google Scholar] [CrossRef] [PubMed]
- Tu, W.B.; Helander, S.; Pilstal, R.; Hickman, K.A.; Lourenco, C.; Jurisica, I.; Raught, B.; Wallner, B.; Sunnerhagen, M.; Penn, L.Z. Myc and its interactors take shape. Biochim. Biophys. Acta 2015, 1849, 469–483. [Google Scholar] [CrossRef] [PubMed]
- Farrell, S.; Simkovich, N.; Wu, Y.; Barberis, A.; Ptashne, M. Gene activation by recruitment of the RNA polymerase II holoenzyme. Genes Dev. 1996, 10, 2359–2367. [Google Scholar] [CrossRef] [PubMed]
- Nevado, J.; Gaudreau, L.; Adam, M.; Ptashne, M. Transcriptional activation by artificial recruitment in mammalian cells. Proc. Natl. Acad. Sci. USA 1999, 96, 2674–2677. [Google Scholar] [CrossRef] [PubMed]
- Kato, G.J.; Barrett, J.; Villa-Garcia, M.; Dang, C.V. An amino-terminal c-Myc domain required for neoplastic transformation activates transcription. Mol. Cell. Biol. 1990, 10, 5914–5920. [Google Scholar] [CrossRef] [PubMed]
- Spotts, G.D.; Patel, S.V.; Xiao, Q.; Hann, S.R. Identification of downstream-initiated c-Myc proteins which are dominant-negative inhibitors of transactivation by full-length c-Myc proteins. Mol. Cell. Biol. 1997, 17, 1459–1468. [Google Scholar] [CrossRef] [PubMed]
- Hirst, S.K.; Grandori, C. Differential activity of conditional MYC and its variant MYC-S in human mortal fibroblasts. Oncogene 2000, 19, 5189–5197. [Google Scholar] [CrossRef] [PubMed]
- Cowling, V.H.; Cole, M.D. An N-Myc truncation analogous to c-Myc-S induces cell proliferation independently of transactivation but dependent on Myc homology box II. Oncogene 2008, 27, 1327–1332. [Google Scholar] [CrossRef] [PubMed]
- Li, L.H.; Nerlov, C.; Prendergast, G.; MacGregor, D.; Ziff, E.B. c-Myc represses transcription in vivo by a novel mechanism dependent on the initiator element and Myc box II. EMBO J. 1994, 13, 4070–4079. [Google Scholar] [PubMed]
- Oster, S.K.; Mao, D.Y.; Kennedy, J.; Penn, L.Z. Functional analysis of the N-terminal domain of the Myc oncoprotein. Oncogene 2003, 22, 1998–2010. [Google Scholar] [CrossRef] [PubMed]
- Herbst, A.; Hemann, M.T.; Tworkowski, K.A.; Salghetti, S.E.; Lowe, S.W.; Tansey, W.P. A conserved element in Myc that negatively regulates its proapoptotic activity. EMBO Rep. 2005, 6, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, A.; Kume, A.; Nemoto, K.; Lee, S.Y.; Asami, Y.; Nemoto, F.; Nishimura, S.; Kuchino, Y. Isolation and characterization of s-myc, a member of the rat MYC gene family. Proc. Natl. Acad. Sci. USA 1989, 86, 9144–9148. [Google Scholar] [CrossRef] [PubMed]
- Benassayag, C.; Montero, L.; Colombie, N.; Gallant, P.; Cribbs, D.; Morello, D. Human c-Myc isoforms differentially regulate cell growth and apoptosis in Drosophila melanogaster. Mol. Cell. Biol. 2005, 25, 9897–9909. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Q.; Claassen, G.; Shi, J.; Adachi, S.; Sedivy, J.; Hann, S.R. Transactivation-defective c-MycS retains the ability to regulate proliferation and apoptosis. Genes Dev. 1998, 12, 3803–3808. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Gregory, M.A.; Li, Z.; Brousal, J.P.; West, K.; Hann, S.R. p19ARF directly and differentially controls the functions of c-Myc independently of p53. Nature 2004, 431, 712–717. [Google Scholar] [CrossRef] [PubMed]
- Legouy, E.; DePinho, R.; Zimmerman, K.; Collum, R.; Yancopoulos, G.; Mitsock, L.; Kriz, R.; Alt, F.W. Structure and expression of the murine L-myc gene. EMBO J. 1987, 6, 3359–3366. [Google Scholar] [PubMed]
- Helander, S.; Montecchio, M.; Pilstal, R.; Su, Y.; Kuruvilla, J.; Elven, M.; Ziauddin, J.M.; Anandapadamanaban, M.; Cristobal, S.; Lundstrom, P.; et al. Pre-Anchoring of Pin1 to Unphosphorylated c-Myc in a Fuzzy Complex Regulates c-Myc Activity. Structure 2015, 23, 2267–2279. [Google Scholar] [CrossRef] [PubMed]
- Facchini, L.M.; Chen, S.; Marhin, W.W.; Lear, J.N.; Penn, L.Z. The Myc negative autoregulation mechanism requires Myc-Max association and involves the c-myc P2 minimal promoter. Mol. Cell. Biol. 1997, 17, 100–114. [Google Scholar] [CrossRef] [PubMed]
- Littlewood, T.D.; Hancock, D.C.; Danielian, P.S.; Parker, M.G.; Evan, G.I. A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acids Res. 1995, 23, 1686–1690. [Google Scholar] [CrossRef] [PubMed]
- Resar, L.M.; Dolde, C.; Barrett, J.F.; Dang, C.V. B-Myc inhibits neoplastic transformation and transcriptional activation by c-Myc. Mol. Cell. Biol. 1993, 13, 1130–1136. [Google Scholar] [CrossRef] [PubMed]
- Barrett, J.; Birrer, M.J.; Kato, G.J.; Dosaka-Akita, H.; Dang, C.V. Activation domains of L-Myc and c-Myc determine their transforming potencies in rat embryo cells. Mol. Cell. Biol. 1992, 12, 3130–3137. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, M.; Takizawa, N.; Narita, M.; Ichisaka, T.; Yamanaka, S. Promotion of direct reprogramming by transformation-deficient Myc. Proc. Natl. Acad. Sci. USA 2010, 107, 14152–14157. [Google Scholar] [CrossRef] [PubMed]
- Andresen, C.; Helander, S.; Lemak, A.; Fares, C.; Csizmok, V.; Carlsson, J.; Penn, L.Z.; Forman-Kay, J.D.; Arrowsmith, C.H.; Lundstrom, P.; et al. Transient structure and dynamics in the disordered c-Myc transactivation domain affect Bin1 binding. Nucleic Acids Res. 2012, 40, 6353–6366. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Spears, E.; Boone, D.N.; Li, Z.; Gregory, M.A.; Hann, S.R. Domain-specific c-Myc ubiquitylation controls c-Myc transcriptional and apoptotic activity. Proc. Natl. Acad. Sci. USA 2013, 110, 978–983. [Google Scholar] [CrossRef] [PubMed]
- Boone, D.N.; Hann, S.R. The Myc-ARF-Egr1 pathway: Unleashing the apoptotic power of c-Myc. Cell Cycle 2011, 10, 2043–2044. [Google Scholar] [CrossRef] [PubMed]
- Hann, S.R. MYC cofactors: Molecular switches controlling diverse biological outcomes. Cold Spring Harb. Perspect. Med. 2014, 4, a014399. [Google Scholar] [CrossRef] [PubMed]
- Eberhardy, S.R.; Farnham, P.J. Myc recruits P-TEFb to mediate the final step in the transcriptional activation of the cad promoter. J. Biol. Chem. 2002, 277, 40156–40162. [Google Scholar] [CrossRef] [PubMed]
- Huang, A.; Ho, C.S.; Ponzielli, R.; Barsyte-Lovejoy, D.; Bouffet, E.; Picard, D.; Hawkins, C.E.; Penn, L.Z. Identification of a novel c-Myc protein interactor, JPO2, with transforming activity in medulloblastoma cells. Cancer Res. 2005, 65, 5607–5619. [Google Scholar] [CrossRef] [PubMed]
- Maertens, G.N.; Cherepanov, P.; Engelman, A. Transcriptional co-activator p75 binds and tethers the Myc-interacting protein JPO2 to chromatin. J. Cell Sci. 2006, 119, 2563–2571. [Google Scholar] [CrossRef] [PubMed]
- D’Artista, L.; Bisso, A.; Piontini, A.; Doni, M.; Verrecchia, A.; Kress, T.R.; Morelli, M.J.; Del Sal, G.; Amati, B.; Campaner, S. Pin1 is required for sustained B cell proliferation upon oncogenic activation of Myc. Oncotarget 2016, 7, 21786–21798. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Q.; West-Osterfield, K.; Spears, E.; Li, Z.; Panaccione, A.; Hann, S.R. MB0 and MBI Are Independent and Distinct Transactivation Domains in MYC that Are Essential for Transformation. Genes 2017, 8, 134. https://doi.org/10.3390/genes8050134
Zhang Q, West-Osterfield K, Spears E, Li Z, Panaccione A, Hann SR. MB0 and MBI Are Independent and Distinct Transactivation Domains in MYC that Are Essential for Transformation. Genes. 2017; 8(5):134. https://doi.org/10.3390/genes8050134
Chicago/Turabian StyleZhang, Qin, Kimberly West-Osterfield, Erick Spears, Zhaoliang Li, Alexander Panaccione, and Stephen R. Hann. 2017. "MB0 and MBI Are Independent and Distinct Transactivation Domains in MYC that Are Essential for Transformation" Genes 8, no. 5: 134. https://doi.org/10.3390/genes8050134





