Variability of DNA Methylation within Schizophrenia Risk Loci across Subregions of Human Hippocampus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Postmortem Human Hippocampus Tissue Samples
2.2. Sample Processing and DNA Methylation Measurement
2.3. Data Analysis
3. Results
3.1. Differentially Methylated Positions
3.2. Differentially Methylated Regions
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014, 511, 421–427. [Google Scholar]
- Ng, S.B.; Turner, E.H.; Robertson, P.D.; Flygare, S.D.; Bigham, A.W.; Lee, C.; Shaffer, T.; Wong, M.; Bhattacharjee, A.; Eichler, E.E.; et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature 2009, 461, 272–276. [Google Scholar] [CrossRef] [PubMed]
- Cnv and Schizophrenia Working Groups of the Psychiatric Genomics Consortium. Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat. Genet. 2017, 49, 27–35. [Google Scholar]
- Fatemi, S.H.; Folsom, T.D. The neurodevelopmental hypothesis of schizophrenia, revisited. Schizophr. Bull. 2009, 35, 528–548. [Google Scholar] [CrossRef] [PubMed]
- McCarty, R. Cross-fostering: Elucidating the effects of genexenvironment interactions on phenotypic development. Neurosci. Biobehav. Rev. 2017, 73, 219–254. [Google Scholar] [CrossRef] [PubMed]
- Raj, A.; Rifkin, S.A.; Andersen, E.; van Oudenaarden, A. Variability in gene expression underlies incomplete penetrance. Nature 2010, 463, 913–918. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, A.; Martins-de-Souza, D.; Akbarian, S.; Cassoli, J.S.; Ehrenreich, H.; Fischer, A.; Fonteh, A.; Gattaz, W.F.; Gawlik, M.; Gerlach, M.; et al. Consensus paper of the WFSBP Task Force on Biological Markers: Criteria for biomarkers and endophenotypes of schizophrenia, part III: Molecular mechanisms. World J. Biol. Psychiatry 2016. [Google Scholar] [CrossRef] [PubMed]
- Davies, M.N.; Volta, M.; Pidsley, R.; Lunnon, K.; Dixit, A.; Lovestone, S.; Coarfa, C.; Harris, R.A.; Milosavljevic, A.; Troakes, C.; et al. Functional annotation of the human brain methylome identifies tissue-specific epigenetic variation across brain and blood. Genome Biol. 2012. [Google Scholar] [CrossRef] [PubMed]
- Hannon, E.; Lunnon, K.; Schalkwyk, L.; Mill, J. Interindividual methylomic variation across blood, cortex, and cerebellum: Implications for epigenetic studies of neurological and neuropsychiatric phenotypes. Epigenetics 2015, 10, 1024–1032. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, A.; Ziller, M.; Spengler, D. The Future is The Past: Methylation QTLs in Schizophrenia. Genes 2016, 7, 104. [Google Scholar] [CrossRef] [PubMed]
- Jaffe, A.E.; Gao, Y.; Deep-Soboslay, A.; Tao, R.; Hyde, T.M.; Weinberger, D.R.; Kleinman, J.E. Mapping DNA methylation across development, genotype and schizophrenia in the human frontal cortex. Nat. Neurosci. 2016, 19, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Hannon, E.; Spiers, H.; Viana, J.; Pidsley, R.; Burrage, J.; Murphy, T.M.; Troakes, C.; Turecki, G.; O’Donovan, M.C.; Schalkwyk, L.C.; et al. Methylation QTLs in the developing brain and their enrichment in schizophrenia risk loci. Nat. Neurosci. 2016, 19, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Ruzicka, W.B.; Subburaju, S.; Benes, F.M. Circuit- and Diagnosis-Specific DNA Methylation Changes at gamma-Aminobutyric Acid-Related Genes in Postmortem Human Hippocampus in Schizophrenia and Bipolar Disorder. JAMA Psychiatry 2015, 72, 541–551. [Google Scholar] [CrossRef] [PubMed]
- Freund, T.F.; Buzsaki, G. Interneurons of the hippocampus. Hippocampus 1996, 6, 347–470. [Google Scholar] [CrossRef]
- Aryee, M.J.; Jaffe, A.E.; Corrada-Bravo, H.; Ladd-Acosta, C.; Feinberg, A.P.; Hansen, K.D.; Irizarry, R.A. Minfi: A flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 2014, 30, 1363–1369. [Google Scholar] [CrossRef] [PubMed]
- Peters, T.J.; Buckley, M.J.; Statham, A.L.; Pidsley, R.; Samaras, K.; Lord, R.V.; Clark, S.J.; Molloy, P.L. De novo identification of differentially methylated regions in the human genome. Epigenet. Chromatin. 2015. [Google Scholar] [CrossRef]
- Dowell, R.D.; Ryan, O.; Jansen, A.; Cheung, D.; Agarwala, S.; Danford, T.; Bernstein, D.A.; Rolfe, P.A.; Heisler, L.E.; Chin, B.; et al. Genotype to phenotype: A complex problem. Science 2010, 328, 469. [Google Scholar] [CrossRef] [PubMed]
- Linder, R.A.; Seidl, F.; Ha, K.; Ehrenreich, I.M. The complex genetic and molecular basis of a model quantitative trait. Mol. Biol. Cell 2016, 27, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Illingworth, R.S.; Gruenewald-Schneider, U.; De Sousa, D.; Webb, S.; Merusi, C.; Kerr, A.R.; James, K.D.; Smith, C.; Walker, R.; Andrews, R.; et al. Inter-individual variability contrasts with regional homogeneity in the human brain DNA methylome. Nucleic Acids Res. 2015, 43, 732–744. [Google Scholar] [CrossRef] [PubMed]
- Viana, J.; Hannon, E.; Dempster, E.; Pidsley, R.; Macdonald, R.; Knox, O.; Spiers, H.; Troakes, C.; Al-Saraj, S.; Turecki, G.; et al. Schizophrenia-associated methylomic variation: Molecular signatures of disease and polygenic risk burden across multiple brain regions. Hum. Mol. Genet. 2017, 26, 210–225. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, J.R.; van der Brug, M.P.; Hernandez, D.G.; Traynor, B.J.; Nalls, M.A.; Lai, S.-L.; Arepalli, S.; Dillman, A.; Rafferty, L.P.; Troncoso, J.; et al. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genet. 2010, 6, e1000952. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.A. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012, 13, 484–492. [Google Scholar] [CrossRef] [PubMed]
- Benes, F.M.; Berretta, S. GABAergic interneurons: Implications for understanding schizophrenia and bipolar disorder. Neuropsychopharmacology 2001, 25, 1–27. [Google Scholar] [CrossRef]
- Benes, F.M.; Lim, B.; Matzilevich, D.; Walsh, J.P.; Subburaju, S.; Minns, M. Regulation of the GABA cell phenotype in hippocampus of schizophrenics and bipolars. Proc. Natl. Acad. Sci. USA 2007, 104, 10164–10169. [Google Scholar] [CrossRef] [PubMed]
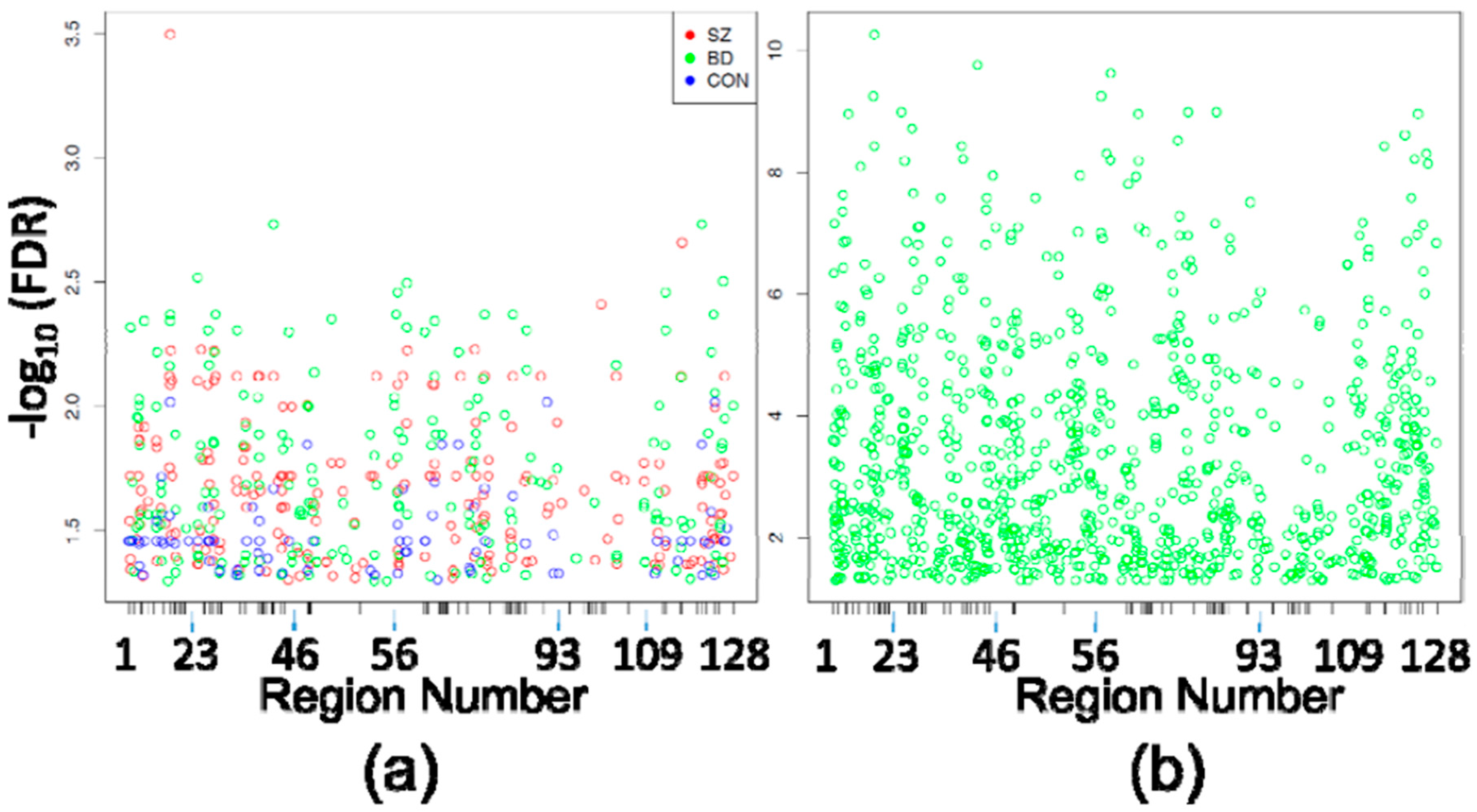
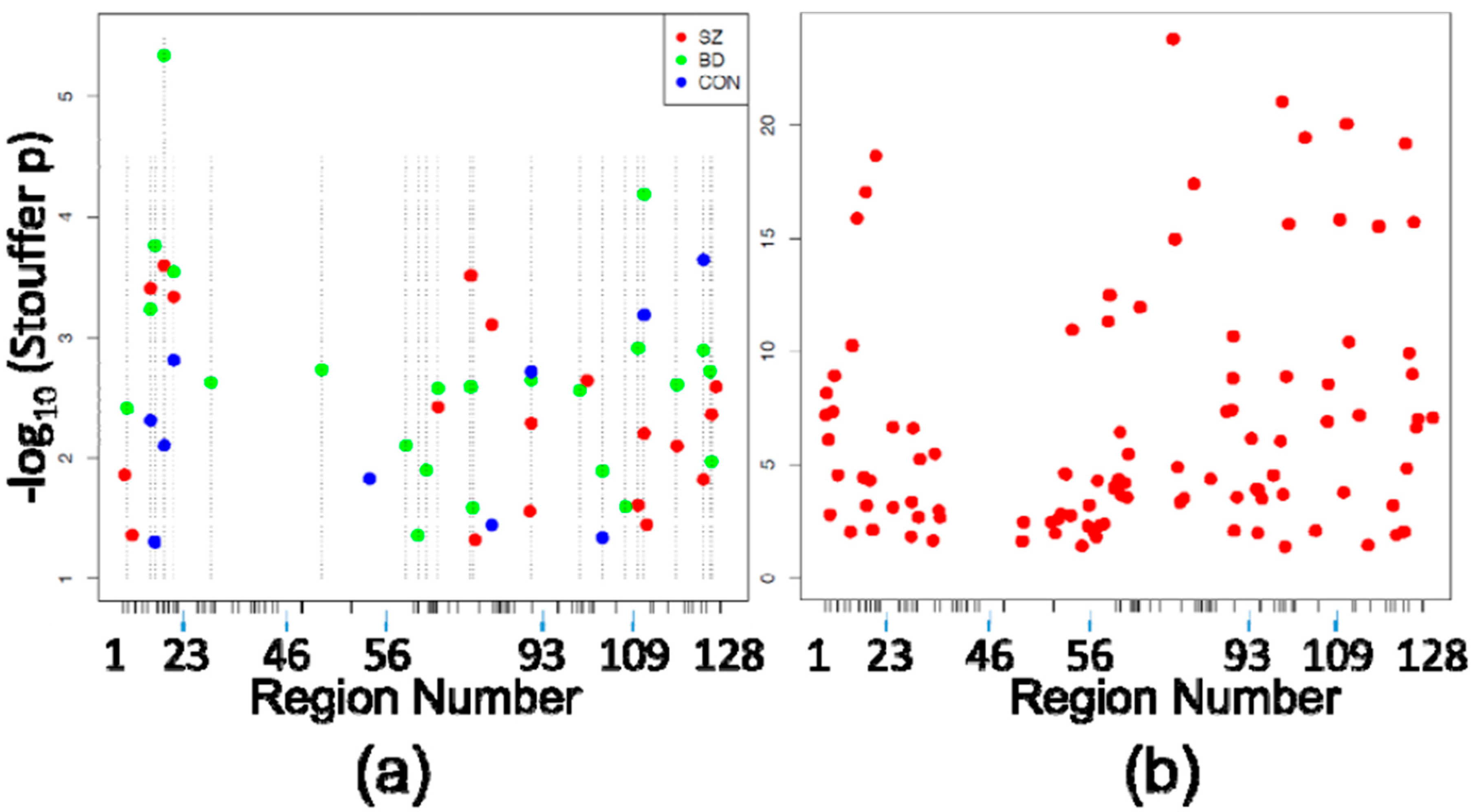
| Diagnosis | n | Gender | Age | PMI | pH |
|---|---|---|---|---|---|
| CON | 8 | 4M/4F | 64.1 ± 14.2 | 22.4 ± 5.7 | 6.4 ± 0.2 |
| SZ | 8 | 4M/4F | 67.9 ± 17.3 | 26.4 ± 10.3 | 6.4 ± 0.2 |
| BD | 8 | 3M/5F | 68.3 ± 11.7 | 25.0 ± 6.7 | 6.3 ± 0.2 |
| p | 0.86 | 0.82 | 0.58 | 0.46 |
| Comparison | # of DMPs | Mean Delta Beta | Mean FDR |
|---|---|---|---|
| SZ CA1 vs. SZ CA2/3 | 210 | 0.064 | 0.025 |
| BD CA1 vs. BD CA2/3 | 201 | 0.063 | 0.023 |
| CON CA1 vs. CON CA2/3 | 86 | 0.075 | 0.034 |
| Pooled Cases CA1 vs. CA2/3 | 966 | 0.035 | 0.0087 |
| SZ CA2/3 vs. CON CA2/3 | 1 | 0.035 | 0.0062 |
| SZ CA1 vs. CON CA1 | 0 | NA | NA |
| BD CA2/3 vs. CON CA2/3 | 0 | NA | NA |
| BD CA1 vs. CON CA1 | 0 | NA | NA |
| Comparison | # of DMPs | % of DMPs | |
| SZ vs. BD | 107 | 53.23 | |
| SZ vs. CON | 53 | 61.63 | |
| BD vs. CON | 59 | 68.6 | |
| All 3 Groups | 46 | 53.49 | |
| SZ vs. Pooled | 199 | 94.76 | |
| BD vs. Pooled | 195 | 97.01 | |
| CON vs. Pooled | 85 | 98.83 | |
| All vs. Pooled | 46 | 100 |
| Chr | Start | End | Description | SZ | BD | CON | Pooled |
|---|---|---|---|---|---|---|---|
| chr01 | 2381300 | 2381623 | 26 kb upstream of PLCH2 | 13 | 28 | ||
| chr01 | 2391242 | 2391347 | 16 kb upstream of PLCH2 | 16 | |||
| chr01 | 8484005 | 8484417 | RERE intron 12 | 18 | 24 | ||
| chr01 | 150123038 | 150123734 | PLEKH01 spans exon 2 | 3 | 5 | 5 | 21 |
| chr01 | 150229144 | 150230345 | CA14 TSS | 3 | 10 | 9 | |
| chr01 | 243584669 | 243584861 | SDCCAG8 intron #15 | 1 | 1 | 6 | 8 |
| chr02 | 72357938 | 72358024 | CYP26B1 3′ end | 4 | 4 | 3 | 6 |
| chr03 | 2553086 | 2553187 | CNTN4 intron 3 | 11 | 39 | ||
| chr06 | 28499677 | 28499825 | GPX5 exon 3 3′ boundary | 8 | 83 | ||
| chr07 | 1983170 | 1983503 | MADL1 intron 16 | 7 | 18 | ||
| chr07 | 2143508 | 2144767 | MADL1 intron 12 | 17 | 17 | ||
| chr07 | 86413439 | 86414302 | GRM3 intron 2 | 23 | 40 | ||
| chr07 | 104909431 | 104909815 | SRPK2 exon 3 5′ boundary | 19 | 45 | ||
| chr10 | 18689036 | 18689503 | CACNB2 exon 5 5′ boundary | 8 | 14 | 16 | |
| chr11 | 46365894 | 46367100 | DGKZ exon 1 | 2 | 13 | 1 | |
| chr11 | 46383032 | 46383209 | DGKZ exon 4 | 22 | 14 | ||
| chr11 | 46401423 | 46401447 | DGKZ exon 32 | 19 | 47 | ||
| chr11 | 57414402 | 57414908 | YPEL4 spans exon 2 | 5 | 8 | 7 | |
| chr12 | 57589254 | 57589740 | LRP1 exon 53 | 16 | 29 | ||
| chr12 | 57597137 | 57597238 | LRP1 exon 70 5′ boundary | 10 | 10 | 4 | 19 |
| chr14 | 104171260 | 104172224 | XRCC3 intron 6 | 15 | 2 | ||
| chr15 | 40599681 | 40600635 | PLCB2 TSS | 6 | 12 | ||
| chr15 | 91426668 | 91427884 | Overlaps FURIN 3′ end & FES TSS | 20 | 9 | 4 | |
| chr16 | 30124804 | 30124904 | GDPD3 3′ end | 21 | 36 | ||
| chr16 | 67918485 | 67919362 | NRN1L TSS | 15 | 6 | 10 | |
| chr16 | 67977866 | 67978450 | SLC12A4 exon 24 | 11 | 2 | 2 | 3 |
| chr16 | 68000764 | 68001415 | CLC12A4 exon 2 5′ boundary | 17 | 20 | ||
| chr17 | 18011514 | 18012134 | MYO15A TSS | 12 | 12 | 13 | |
| chr20 | 37464180 | 37464594 | PPP1R16B exon 2 | 14 | 7 | 1 | 5 |
| chr22 | 41613693 | 41613790 | L3MBTL2 intron 5 | 9 | 23 | ||
| chr22 | 41636942 | 41637617 | CHADL TSS | 9 | 18 | 11 | |
| chr22 | 42347907 | 42348061 | LINC00634 TSS | 7 | 35 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruzicka, W.B.; Subburaju, S.; Benes, F.M. Variability of DNA Methylation within Schizophrenia Risk Loci across Subregions of Human Hippocampus. Genes 2017, 8, 143. https://doi.org/10.3390/genes8050143
Ruzicka WB, Subburaju S, Benes FM. Variability of DNA Methylation within Schizophrenia Risk Loci across Subregions of Human Hippocampus. Genes. 2017; 8(5):143. https://doi.org/10.3390/genes8050143
Chicago/Turabian StyleRuzicka, W. Brad, Sivan Subburaju, and Francine M. Benes. 2017. "Variability of DNA Methylation within Schizophrenia Risk Loci across Subregions of Human Hippocampus" Genes 8, no. 5: 143. https://doi.org/10.3390/genes8050143





