DNA Methylation Targets Influenced by Bisphenol A and/or Genistein Are Associated with Survival Outcomes in Breast Cancer Patients
Abstract
:1. Introduction
2. Materials and Methods
2.1. BPA and GEN Exposures and Mammary Gland Procurement
2.2. MBD Cap-Seq Analysis
2.3. Bioinformatics Analysis
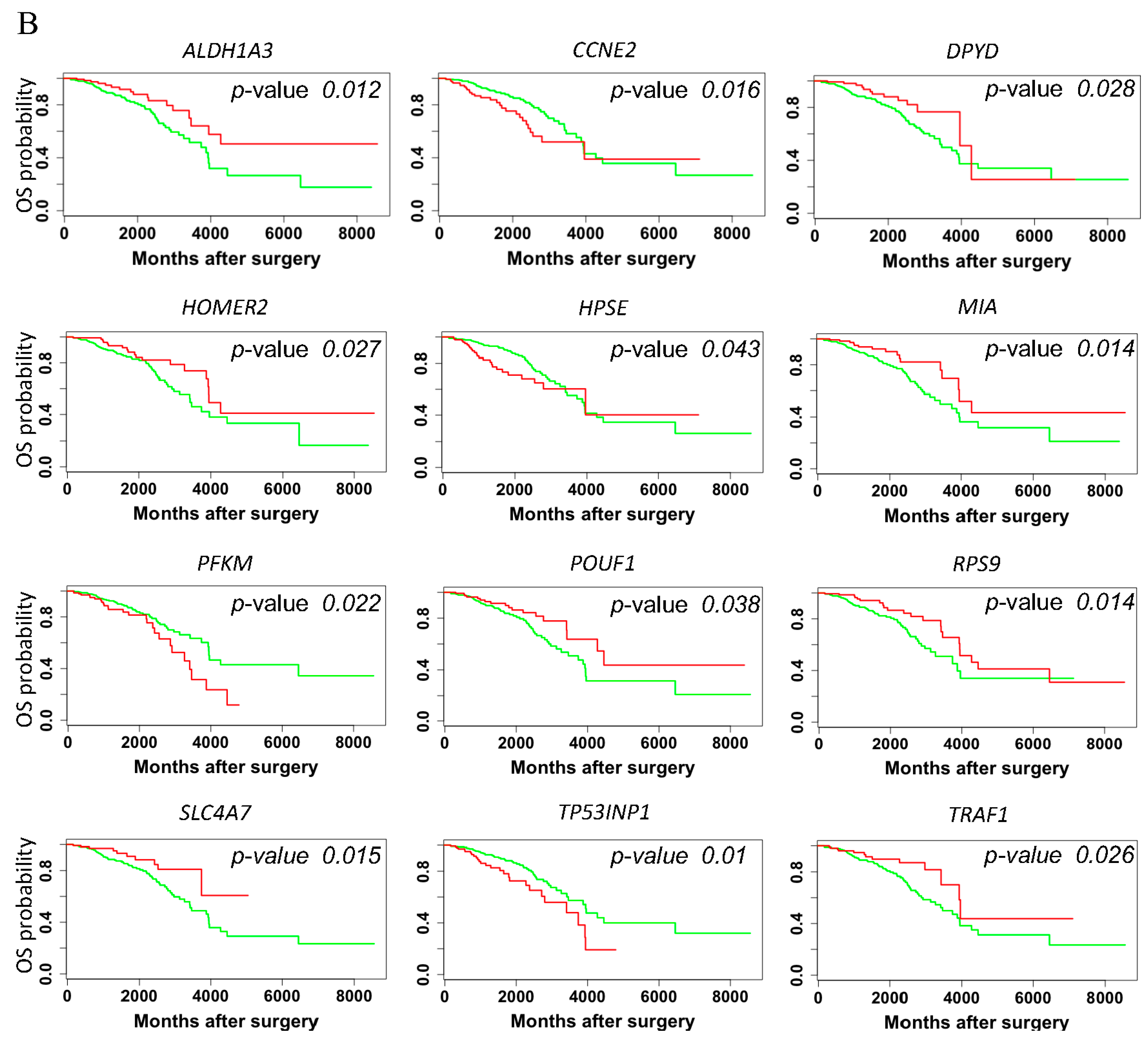
3. Results
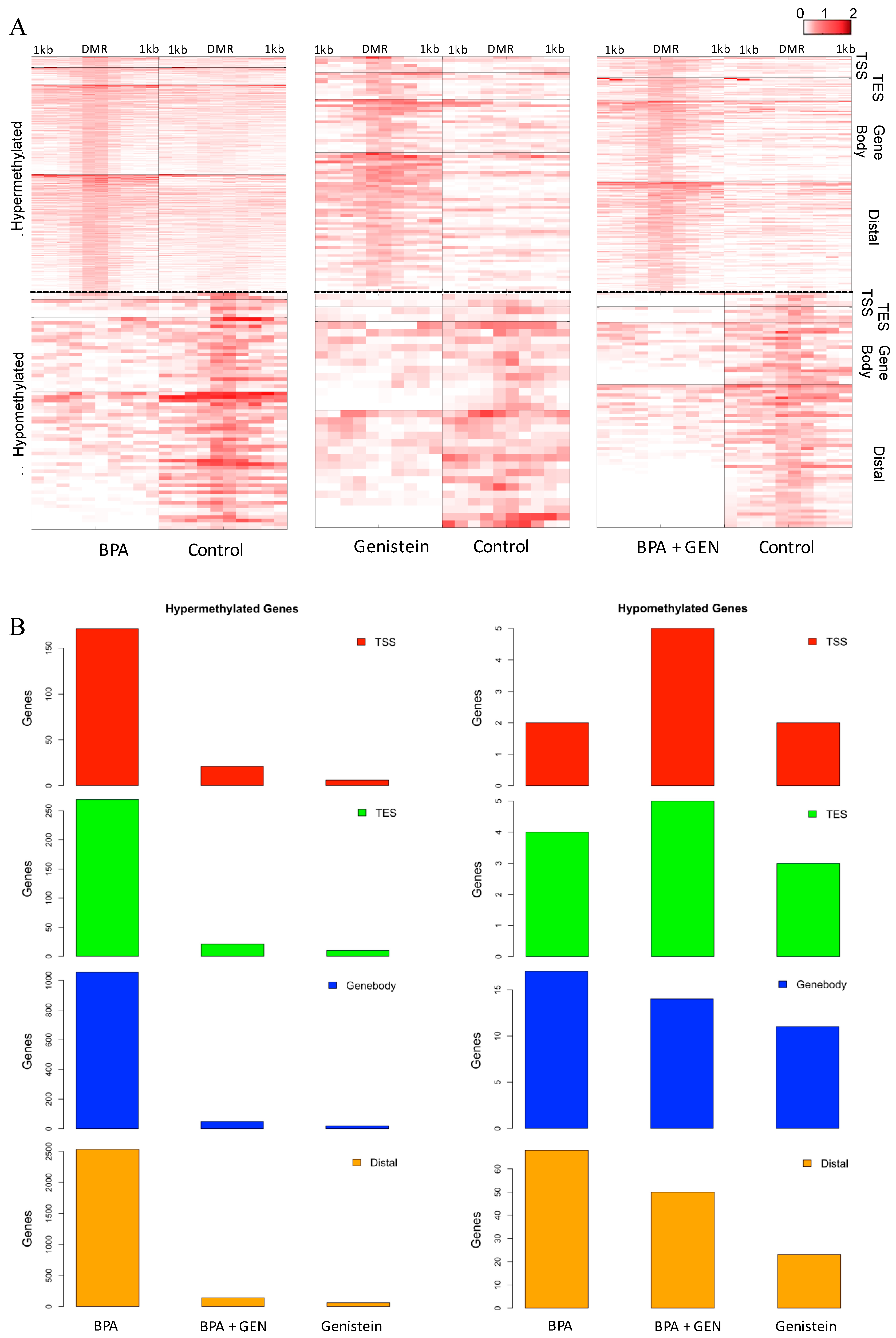
3.1. Identification of Differentially Methylated Loci Following Prepubertal Exposure to BPA and/or GEN in Rats
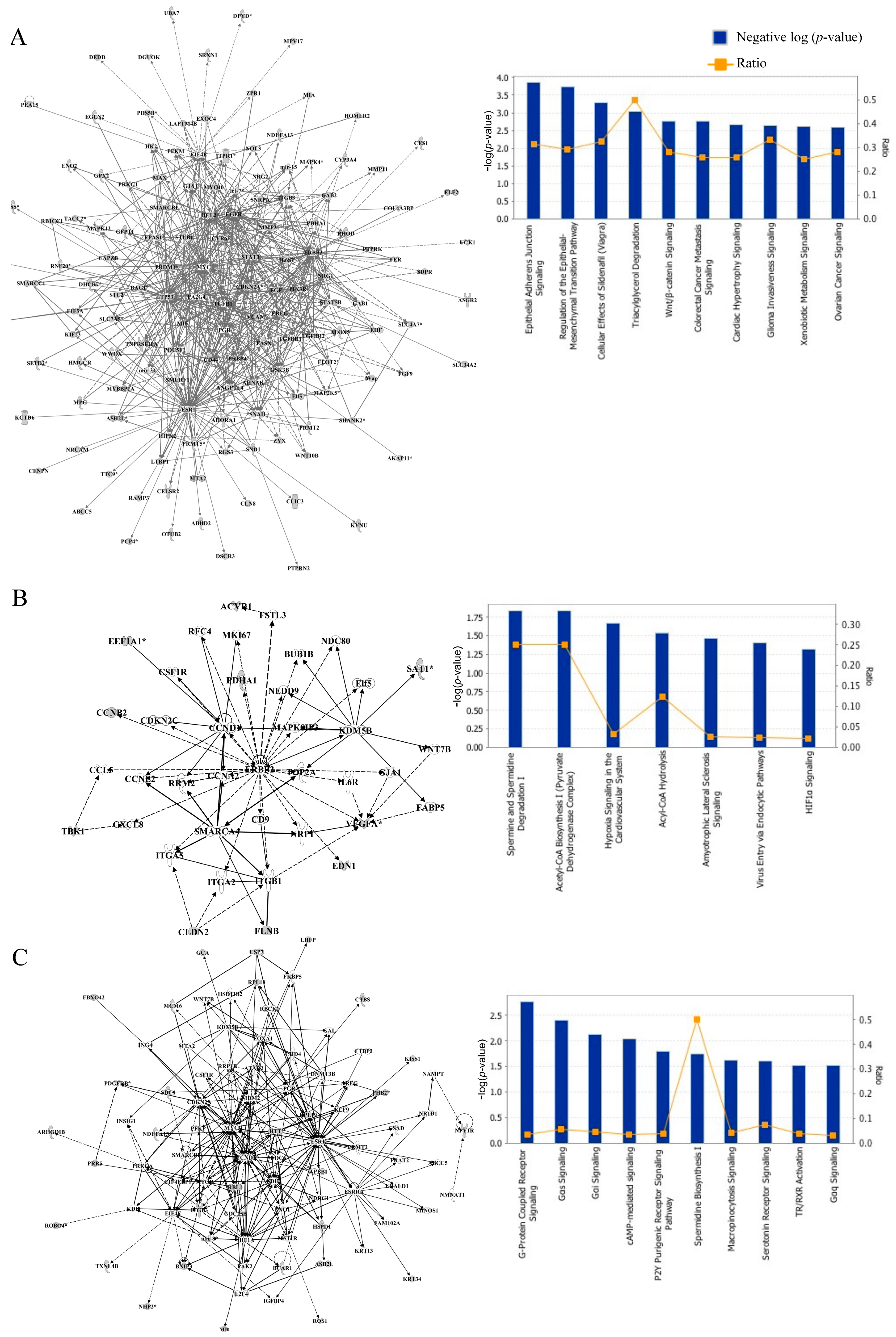
3.2. Network and Pathway Analyses of DNA Methylated Targets Mediated by Prepubertal BPA and/or GEN Exposures
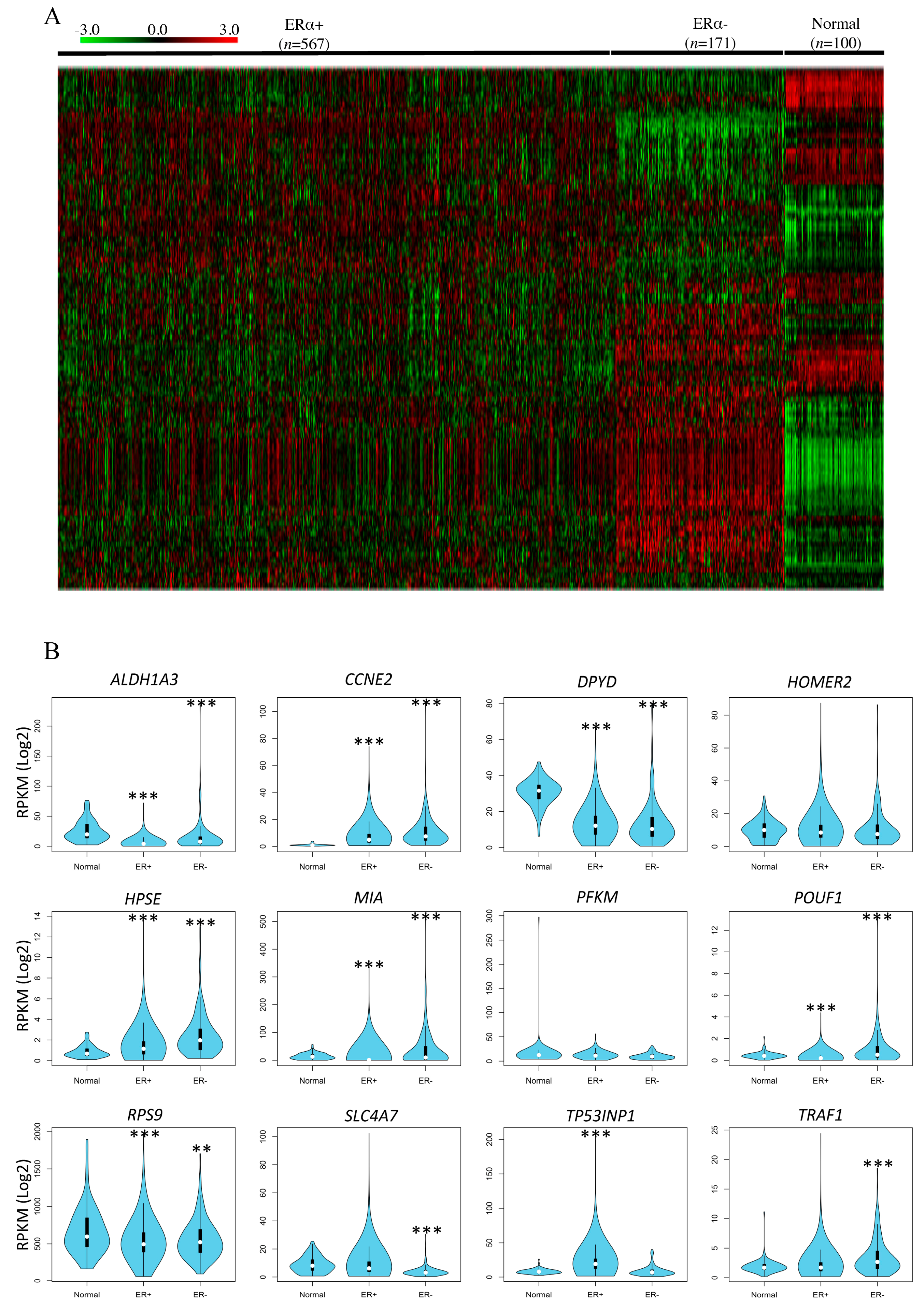
3.3. In Silico Correlation of Rat DNA Methylated Genes with Breast Cancer Patient Data from the Cancer Genome Atlas (TCGA)
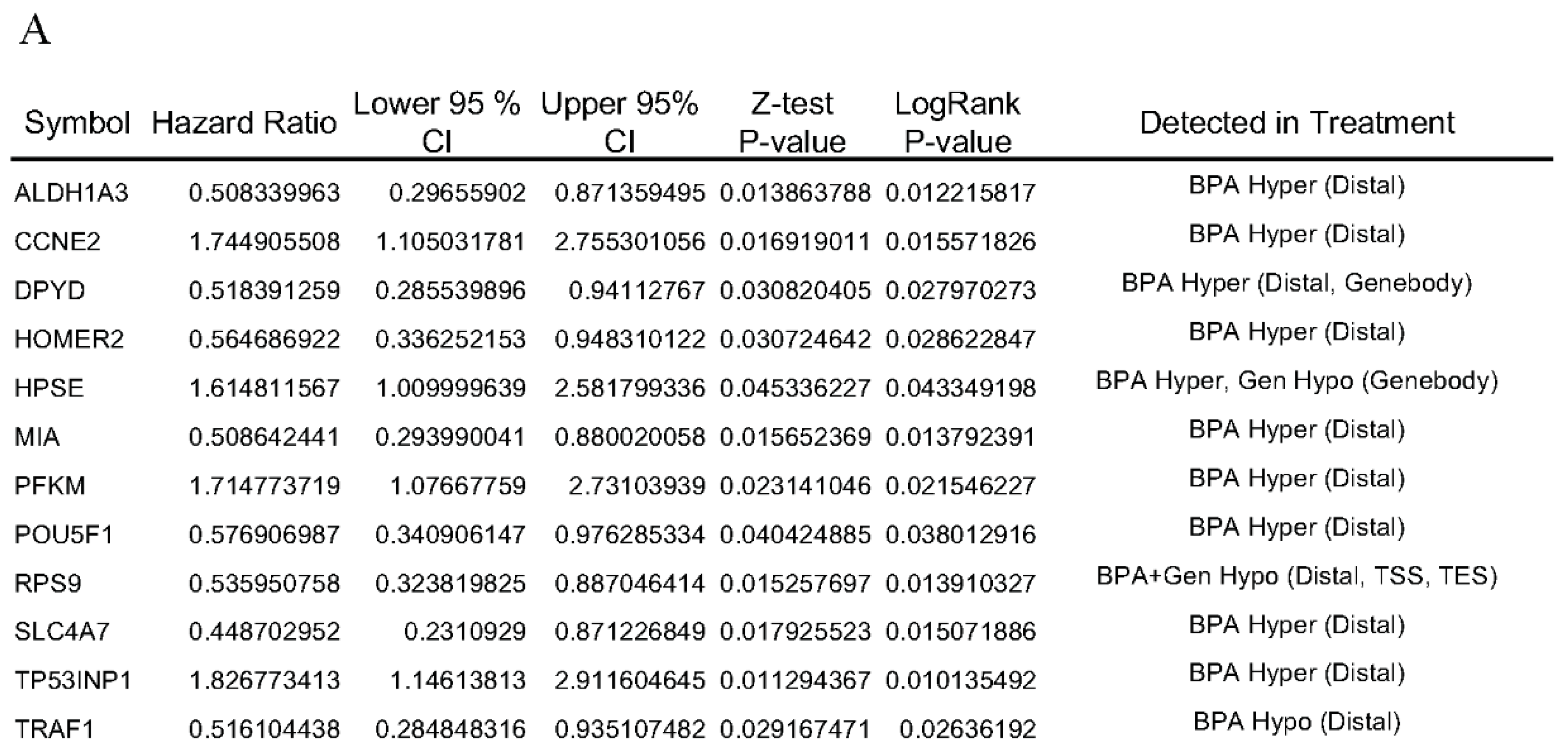
3.4. Hazard Ratio and Survival Analysis of rat DNA Methylation Regulated Targeted Genes in Breast Cancer Patients
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Data Access
References
- Ardies, C.M.; Dees, C. Xenoestrogens significantly enhance risk for breast cancer during growth and adolescence. Med. Hypotheses 1998, 50, 457–464. [Google Scholar] [CrossRef]
- Jenkins, S.; Betancourt, A.M.; Wang, J.; Lamartiniere, C.A. Endocrine-active chemicals in mammary cancer causation and prevention. J. Steroid Biochem. Mol. Biol. 2012, 129, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Jenkins, S.; Lamartiniere, C.A. Cell proliferation and apoptosis in rat mammary glands following combinational exposure to bisphenol A and genistein. BMC Cancer 2014, 14, 379. [Google Scholar] [CrossRef] [PubMed]
- Vandenberg, L.N.; Hauser, R.; Marcus, M.; Olea, N.; Welshons, W.V. Human exposure to bisphenol A (BPA). Reprod. Toxicol. 2007, 24, 139–177. [Google Scholar] [CrossRef] [PubMed]
- Calafat, A.M.; Ye, X.; Wong, L.Y.; Reidy, J.A.; Needham, L.L. Exposure of the U.S. Population to bisphenol A and 4-tertiary-octylphenol: 2003–2004. Environ. Health Perspect. 2008, 116, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Ikezuki, Y.; Tsutsumi, O.; Takai, Y.; Kamei, Y.; Taketani, Y. Determination of bisphenol A concentrations in human biological fluids reveals significant early prenatal exposure. Hum. Reprod. 2002, 17, 2839–2841. [Google Scholar] [CrossRef] [PubMed]
- Kuruto-Niwa, R.; Tateoka, Y.; Usuki, Y.; Nozawa, R. Measurement of bisphenol A concentrations in human colostrum. Chemosphere 2007, 66, 1160–1164. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, A.V.; Stathis, P.; Permuth, S.F.; Tokes, L.; Feldman, D. Bisphenol-A: An estrogenic substance is released from polycarbonate flasks during autoclaving. Endocrinology 1993, 132, 2279–2286. [Google Scholar] [CrossRef] [PubMed]
- Lapensee, E.W.; Tuttle, T.R.; Fox, S.R.; Ben-Jonathan, N. Bisphenol A at low nanomolar doses confers chemoresistance in estrogen receptor-alpha-positive and -negative breast cancer cells. Environ. Health Perspect. 2009, 117, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, S.; Raghuraman, N.; Eltoum, I.; Carpenter, M.; Russo, J.; Lamartiniere, C.A. Oral exposure to bisphenol A increases dimethylbenzanthracene-induced mammary cancer in rats. Environ. Health Perspect. 2009, 117, 910–915. [Google Scholar] [CrossRef] [PubMed]
- Lamartiniere, C.A.; Jenkins, S.; Betancourt, A.M.; Wang, J.; Russo, J. Exposure to the endocrine disruptor bisphenol A alters susceptibility for mammary cancer. Horm. Mol. Biol. Clin. Investig. 2011, 5, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, R.G.; Hoover, R.N.; Pike, M.C.; Hildesheim, A.; Nomura, A.M.; West, D.W.; Wu-Williams, A.H.; Kolonel, L.N.; Horn-Ross, P.L.; Rosenthal, J.F.; et al. Migration patterns and breast cancer risk in asian-american women. J. Natl. Cancer Inst. 1993, 85, 1819–1827. [Google Scholar] [CrossRef] [PubMed]
- Trock, B.J.; Hilakivi-Clarke, L.; Clarke, R. Meta-analysis of soy intake and breast cancer risk. J. Natl. Cancer Inst. 2006, 98, 459–471. [Google Scholar] [CrossRef] [PubMed]
- Shu, X.O.; Jin, F.; Dai, Q.; Wen, W.; Potter, J.D.; Kushi, L.H.; Ruan, Z.; Gao, Y.T.; Zheng, W. Soyfood intake during adolescence and subsequent risk of breast cancer among chinese women. Cancer Epidemiol. Biomark. Prev. 2001, 10, 483–488. [Google Scholar]
- Wu, A.H.; Wan, P.; Hankin, J.; Tseng, C.C.; Yu, M.C.; Pike, M.C. Adolescent and adult soy intake and risk of breast cancer in asian-americans. Carcinogenesis 2002, 23, 1491–1496. [Google Scholar] [CrossRef] [PubMed]
- Korde, L.A.; Wu, A.H.; Fears, T.; Nomura, A.M.; West, D.W.; Kolonel, L.N.; Pike, M.C.; Hoover, R.N.; Ziegler, R.G. Childhood soy intake and breast cancer risk in asian american women. Cancer Epidemiol. Biomark. Prev. 2009, 18, 1050–1059. [Google Scholar] [CrossRef] [PubMed]
- Thanos, J.; Cotterchio, M.; Boucher, B.A.; Kreiger, N.; Thompson, L.U. Adolescent dietary phytoestrogen intake and breast cancer risk (Canada). Cancer Causes Control 2006, 17, 1253–1261. [Google Scholar] [CrossRef] [PubMed]
- Murrill, W.B.; Brown, N.M.; Zhang, J.X.; Manzolillo, P.A.; Barnes, S.; Lamartiniere, C.A. Prepubertal genistein exposure suppresses mammary cancer and enhances gland differentiation in rats. Carcinogenesis 1996, 17, 1451–1457. [Google Scholar] [CrossRef] [PubMed]
- Lamartiniere, C.A.; Zhang, J.X.; Cotroneo, M.S. Genistein studies in rats: Potential for breast cancer prevention and reproductive and developmental toxicity. Am. J. Clin. Nutr. 1998, 68, 1400S–1405S. [Google Scholar] [PubMed]
- Fritz, W.A.; Coward, L.; Wang, J.; Lamartiniere, C.A. Dietary genistein: Perinatal mammary cancer prevention, bioavailability and toxicity testing in the rat. Carcinogenesis 1998, 19, 2151–2158. [Google Scholar] [CrossRef] [PubMed]
- Hilakivi-Clarke, L.; Onojafe, I.; Raygada, M.; Cho, E.; Skaar, T.; Russo, I.; Clarke, R. Prepubertal exposure to zearalenone or genistein reduces mammary tumorigenesis. Br. J. Cancer 1999, 80, 1682–1688. [Google Scholar] [CrossRef] [PubMed]
- Lamartiniere, C.A.; Cotroneo, M.S.; Fritz, W.A.; Wang, J.; Mentor-Marcel, R.; Elgavish, A. Genistein chemoprevention: Timing and mechanisms of action in murine mammary and prostate. J. Nutr. 2002, 132, 552S–558S. [Google Scholar] [PubMed]
- Setchell, K.D.; Zimmer-Nechemias, L.; Cai, J.; Heubi, J.E. Isoflavone content of infant formulas and the metabolic fate of these phytoestrogens in early life. Am. J. Clin. Nutr. 1998, 68, 1453S–1461S. [Google Scholar] [PubMed]
- Fukutake, M.; Takahashi, M.; Ishida, K.; Kawamura, H.; Sugimura, T.; Wakabayashi, K. Quantification of genistein and genistin in soybeans and soybean products. Food Chem. Toxicol. 1996, 34, 457–461. [Google Scholar] [CrossRef]
- Anderson, O.S.; Nahar, M.S.; Faulk, C.; Jones, T.R.; Liao, C.; Kannan, K.; Weinhouse, C.; Rozek, L.S.; Dolinoy, D.C. Epigenetic responses following maternal dietary exposure to physiologically relevant levels of bisphenol A. Environ. Mol. Mutagen. 2012, 53, 334–342. [Google Scholar] [CrossRef] [PubMed]
- Hanna, C.W.; Bloom, M.S.; Robinson, W.P.; Kim, D.; Parsons, P.J.; vom Saal, F.S.; Taylor, J.A.; Steuerwald, A.J.; Fujimoto, V.Y. DNA methylation changes in whole blood is associated with exposure to the environmental contaminants, mercury, lead, cadmium and bisphenol A, in women undergoing ovarian stimulation for IVF. Hum. Reprod. 2012, 27, 1401–1410. [Google Scholar] [CrossRef] [PubMed]
- Ho, S.M.; Tang, W.Y.; Belmonte de Frausto, J.; Prins, G.S. Developmental exposure to estradiol and bisphenol A increases susceptibility to prostate carcinogenesis and epigenetically regulates phosphodiesterase type 4 variant 4. Cancer Res. 2006, 66, 5624–5632. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.Y.; Hsu, H.K.; Singer, G.A.; Yan, P.S.; Rodriguez, B.A.; Liu, J.C.; Weng, Y.I.; Deatherage, D.E.; Chen, Z.; Pereira, J.S.; et al. Estrogen-mediated epigenetic repression of large chromosomal regions through DNA looping. Genome Res. 2010, 20, 733–744. [Google Scholar] [CrossRef] [PubMed]
- Prins, G.S.; Birch, L.; Tang, W.Y.; Ho, S.M. Developmental estrogen exposures predispose to prostate carcinogenesis with aging. Reprod. Toxicol. 2007, 23, 374–382. [Google Scholar] [CrossRef] [PubMed]
- Pudenz, M.; Roth, K.; Gerhauser, C. Impact of soy isoflavones on the epigenome in cancer prevention. Nutrients 2014, 6, 4218–4272. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Jadhav, R.R.; Liu, J.; Wilson, D.; Chen, Y.; Thompson, I.M.; Troyer, D.A.; Hernandez, J.; Shi, H.; Leach, R.J.; et al. Roles of distal and genic methylation in the development of prostate tumorigenesis revealed by genome-wide DNA methylation analysis. Sci. Rep. 2016, 6, 22051. [Google Scholar] [CrossRef] [PubMed]
- Yaoi, T.; Itoh, K.; Nakamura, K.; Ogi, H.; Fujiwara, Y.; Fushiki, S. Genome-wide analysis of epigenomic alterations in fetal mouse forebrain after exposure to low doses of bisphenol A. Biochem. Biophys. Res. Commun. 2008, 376, 563–567. [Google Scholar] [CrossRef] [PubMed]
- Lamartiniere, C.A.; Jenkins, S.B.; Wang, J. Genistein: Programming Against Breast Cancer. In Trends in Breast Cancer Prevention; Russo, J., Ed.; Springer International Publishing: Cham, Switzerland, 2016; pp. 23–50. [Google Scholar]
- Betancourt, A.M.; Wang, J.; Jenkins, S.; Mobley, J.; Russo, J.; Lamartiniere, C.A. Altered carcinogenesis and proteome in mammary glands of rats after prepubertal exposures to the hormonally active chemicals bisphenol A and genistein. J. Nutr. 2012, 142, 1382S–1388S. [Google Scholar] [CrossRef] [PubMed]
- Montales, M.T.; Rahal, O.M.; Kang, J.; Rogers, T.J.; Prior, R.L.; Wu, X.; Simmen, R.C. Repression of mammosphere formation of human breast cancer cells by soy isoflavone genistein and blueberry polyphenolic acids suggests diet-mediated targeting of cancer stem-like/progenitor cells. Carcinogenesis 2012, 33, 652–660. [Google Scholar] [CrossRef] [PubMed]
- Gu, F.; Doderer, M.S.; Huang, Y.W.; Roa, J.C.; Goodfellow, P.J.; Kizer, E.L.; Huang, T.H.; Chen, Y. Cms: A web-based system for visualization and analysis of genome-wide methylation data of human cancers. PLoS ONE 2013, 8, e60980. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Hsu, H.K.; Blattler, A.; Wang, Y.; Lan, X.; Wang, Y.; Hsu, P.Y.; Leu, Y.W.; Huang, T.H.; Farnham, P.J.; et al. Locating non-unique matched tags (LONUT) to improve the detection of the enriched regions for CHIP-seq data. PLoS ONE 2013, 8, e67788. [Google Scholar] [CrossRef] [PubMed]
- Jadhav, R.R.; Ye, Z.; Huang, R.L.; Liu, J.; Hsu, P.Y.; Huang, Y.W.; Rangel, L.B.; Lai, H.C.; Roa, J.C.; Kirma, N.B.; et al. Genome-wide DNA methylation analysis reveals estrogen-mediated epigenetic repression of metallothionein-1 gene cluster in breast cancer. Clin Epigenet. 2015, 7, 13. [Google Scholar] [CrossRef] [PubMed]
- Dhimolea, E.; Wadia, P.R.; Murray, T.J.; Settles, M.L.; Treitman, J.D.; Sonnenschein, C.; Shioda, T.; Soto, A.M. Prenatal exposure to BPA alters the epigenome of the rat mammary gland and increases the propensity to neoplastic development. PLoS ONE 2014, 9, e99800. [Google Scholar] [CrossRef] [PubMed]
- Dairkee, S.H.; Luciani-Torres, M.G.; Moore, D.H.; Goodson, W.H., 3rd. Bisphenol-a-induced inactivation of the p53 axis underlying deregulation of proliferation kinetics, and cell death in non-malignant human breast epithelial cells. Carcinogenesis 2013, 34, 703–712. [Google Scholar] [CrossRef] [PubMed]
- Dave, B.; Eason, R.R.; Till, S.R.; Geng, Y.; Velarde, M.C.; Badger, T.M.; Simmen, R.C. The soy isoflavone genistein promotes apoptosis in mammary epithelial cells by inducing the tumor suppressor PTEN. Carcinogenesis 2005, 26, 1793–1803. [Google Scholar] [CrossRef] [PubMed]
- The Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 2012, 490, 61–70. [Google Scholar]
- Gertz, J.; Reddy, T.E.; Varley, K.E.; Garabedian, M.J.; Myers, R.M. Genistein and bisphenol A exposure cause estrogen receptor 1 to bind thousands of sites in a cell type-specific manner. Genome Res. 2012, 22, 2153–2162. [Google Scholar] [CrossRef] [PubMed]
- Colt, J.S.; Blair, A. Parental occupational exposures and risk of childhood cancer. Environ. Health Perspect. 1998, 106, 909–925. [Google Scholar] [CrossRef] [PubMed]
- Doshi, T.; Mehta, S.S.; Dighe, V.; Balasinor, N.; Vanage, G. Hypermethylation of estrogen receptor promoter region in adult testis of rats exposed neonatally to bisphenol A. Toxicology 2011, 289, 74–82. [Google Scholar] [CrossRef] [PubMed]
- Mirza, S.; Sharma, G.; Parshad, R.; Gupta, S.D.; Pandya, P.; Ralhan, R. Expression of DNA methyltransferases in breast cancer patients and to analyze the effect of natural compounds on DNA methyltransferases and associated proteins. J. Breast Cancer 2013, 16, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Xie, Q.; Bai, Q.; Zou, L.Y.; Zhang, Q.Y.; Zhou, Y.; Chang, H.; Yi, L.; Zhu, J.D.; Mi, M.T. Genistein inhibits DNA methylation and increases expression of tumor suppressor genes in human breast cancer cells. Genes Chromosomes Cancer 2014, 53, 422–431. [Google Scholar] [CrossRef] [PubMed]
- Gudas, J.M.; Payton, M.; Thukral, S.; Chen, E.; Bass, M.; Robinson, M.O.; Coats, S. Cyclin e2, a novel G1 cyclin that binds Cdk2 and is aberrantly expressed in human cancers. Mol. Cell. Biol. 1999, 19, 612–622. [Google Scholar] [CrossRef] [PubMed]
- Ahsan, H.; Halpern, J.; Kibriya, M.G.; Pierce, B.L.; Tong, L.; Gamazon, E.; McGuire, V.; Felberg, A.; Shi, J.; Jasmine, F.; et al. A genome-wide association study of early-onset breast cancer identifies pfkm as a novel breast cancer gene and supports a common genetic spectrum for breast cancer at any age. Cancer Epidemiol. Biomark. Prev. 2014, 23, 658–669. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Ruan, J.; Zhao, P.; Chen, S.; Pan, L.; Liu, J. Heparanase is involved in proliferation and invasion of ovarian cancer cells. Cancer Biomark. 2015, 15, 525–534. [Google Scholar] [CrossRef] [PubMed]
- Seux, M.; Peuget, S.; Montero, M.P.; Siret, C.; Rigot, V.; Clerc, P.; Gigoux, V.; Pellegrino, E.; Pouyet, L.; N'Guessan, P.; et al. TP53INP1 decreases pancreatic cancer cell migration by regulating sparc expression. Oncogene 2011, 30, 3049–3061. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Han, H.; De Carvalho, D.D.; Lay, F.D.; Jones, P.A.; Liang, G. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell 2014, 26, 577–590. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zhang, Z.; Hill, D.L.; Chen, X.; Wang, H.; Zhang, R. Genistein, a dietary isoflavone, down-regulates the MDM2 oncogene at both transcriptional and posttranslational levels. Cancer Res. 2005, 65, 8200–8208. [Google Scholar] [CrossRef] [PubMed]
- Ismail, I.A.; Kang, K.S.; Lee, H.A.; Kim, J.W.; Sohn, Y.K. Genistein-induced neuronal apoptosis and g2/m cell cycle arrest is associated with MDC1 up-regulation and PLK1 down-regulation. Eur. J. Pharmacol. 2007, 575, 12–20. [Google Scholar] [CrossRef] [PubMed]
| Group Identification | Gavage Administered | Food Administered |
|---|---|---|
| (1) Control (SO) | Sesame Oil as Vehicle | AIN-76A |
| (2) Bisphenol A (BPA) | 250 µg BPA/kg BW | AIN-76A |
| (3) Genistein (GEN) | Sesame Oil as Vehicle | 250 mg genistein/kg AIN-76A diet |
| (4) BPA + GEN | 250 µg BPA/kg BW | 250 mg genistein/kg AIN-76A diet |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jadhav, R.R.; Santucci-Pereira, J.; Wang, Y.V.; Liu, J.; Nguyen, T.D.; Wang, J.; Jenkins, S.; Russo, J.; Huang, T.H.-M.; Jin, V.X.; et al. DNA Methylation Targets Influenced by Bisphenol A and/or Genistein Are Associated with Survival Outcomes in Breast Cancer Patients. Genes 2017, 8, 144. https://doi.org/10.3390/genes8050144
Jadhav RR, Santucci-Pereira J, Wang YV, Liu J, Nguyen TD, Wang J, Jenkins S, Russo J, Huang TH-M, Jin VX, et al. DNA Methylation Targets Influenced by Bisphenol A and/or Genistein Are Associated with Survival Outcomes in Breast Cancer Patients. Genes. 2017; 8(5):144. https://doi.org/10.3390/genes8050144
Chicago/Turabian StyleJadhav, Rohit R., Julia Santucci-Pereira, Yao V. Wang, Joseph Liu, Theresa D. Nguyen, Jun Wang, Sarah Jenkins, Jose Russo, Tim H.-M. Huang, Victor X. Jin, and et al. 2017. "DNA Methylation Targets Influenced by Bisphenol A and/or Genistein Are Associated with Survival Outcomes in Breast Cancer Patients" Genes 8, no. 5: 144. https://doi.org/10.3390/genes8050144






