The Dual Roles of MYC in Genomic Instability and Cancer Chemoresistance
Abstract
:1. Introduction: MYC as a Transcription Factor
2. MYC in Cancer Development and Cancer Stemness
3. Oncogenic Addiction to MYC as an “Achilles’ Heel” of MYC-Driven Cancer Cells
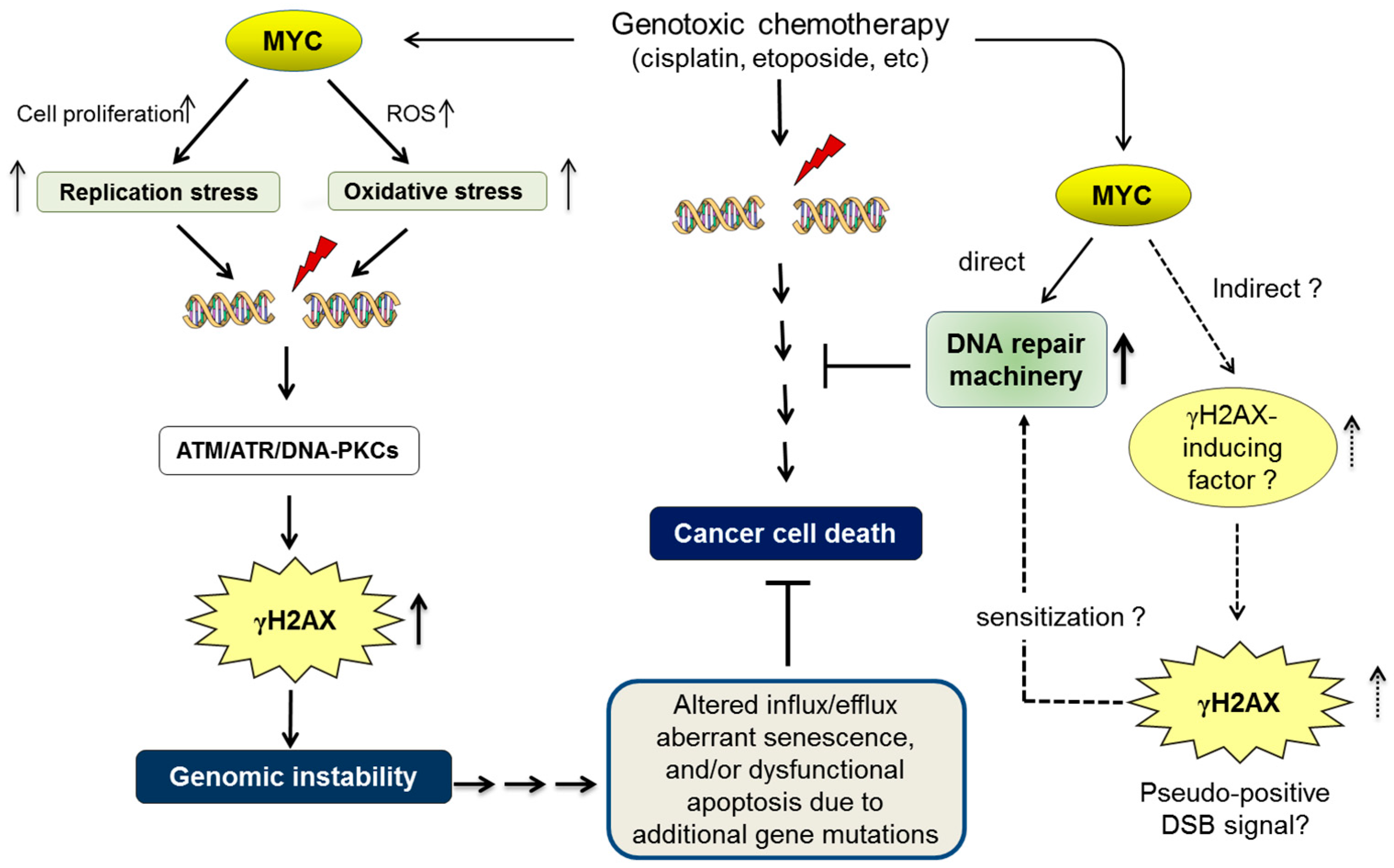
4. MYC-Induced Replication Stress, Genomic Instability, and Oncogenic Transformation
5. MYC in DNA Repair and Cancer Chemoresistance
6. Conclusions
Acknowledgments
Conflicts of Interest
References
- Eilers, M.; Schirm, S.; Bishop, J.M. The MYC protein activates transcription of the alpha-prothymosin gene. EMBO J. 1991, 10, 133–141. [Google Scholar] [PubMed]
- Shim, H.; Dolde, C.; Lewis, B.C.; Wu, C.S.; Dang, G.; Jungmann, R.A.; Dalla-Favera, R.; Dang, C.V. c-Myc transactivation of LDH-A: Implications for tumor metabolism and growth. Proc. Natl. Acad. Sci. USA 1997, 94, 6658–6663. [Google Scholar] [CrossRef] [PubMed]
- Evan, G.I.; Wyllie, A.H.; Gilbert, C.S.; Littlewood, T.D.; Land, H.; Brooks, M.; Waters, C.M.; Penn, L.Z.; Hancock, D.C. Induction of apoptosis in fibroblasts by c-MYC protein. Cell 1992, 69, 119–128. [Google Scholar] [CrossRef]
- Freytag, S.O.; Geddes, T.J. Reciprocal regulation of adipogenesis by Myc and C/EBPalpha. Science 1992, 256, 379–382. [Google Scholar] [CrossRef] [PubMed]
- Dang, C.V. MYC on the Path to Cancer. Cell 2012, 149, 22–35. [Google Scholar] [CrossRef] [PubMed]
- Gabay, M.; Li, Y.; Felsher, D.W. MYC Activation Is a Hallmark of Cancer Initiation and Maintenance. Cold Spring Harb. Perspect. Med. 2014, 4, a014241. [Google Scholar] [CrossRef] [PubMed]
- Tansey, W.P. Mammalian MYC Proteins and Cancer. New J. Sci. 2014, 2014, 1–27. [Google Scholar] [CrossRef]
- Nisen, P.D.; Zimmerman, K.A.; Cotter, S.V.; Gilbert, F.; Alt, F.W. Enhanced expression of the N-myc gene in Wilm’s tumors. Cancer Res. 1986, 46, 6217–6222. [Google Scholar] [PubMed]
- Zimmerman, K.A.; Yancopoulos, G.D.; Collum, R.G.; Smith, R.K.; Kohl, N.E.; Denis, K.A.; Nau, M.M.; Witte, O.N.; Toran-Allerand, D.; Gee, C.E. Differential expression of myc family genes during murine development. Nature 1986, 319, 780–783. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, B.; Morgenbesser, D.; DePinho, R. Myc family oncoproteins function through a common pathway to transform normal cells in culture: Cross-interference by Max and trans-acting dominant mutants. Genes Dev. 1992, 6, 1480–1492. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef]
- Evan, G.I.; Littlewood, T.D. The role of c-Myc in cell growth. Curr. Opin. Genet. Dev. 1993, 3, 44–49. [Google Scholar] [CrossRef]
- Henriksson, M.; Luscher, B. Proteins of the Myc network: Essential regulators of cell growth and differentiation. Adv. Cancer Res. 1996, 68, 109–182. [Google Scholar] [PubMed]
- Huang, M.; Weiss, W.A. Neuroblastoma and MYCN. Cold Spring Harb. Perspect. Med. 2013, 3, a014415. [Google Scholar] [CrossRef] [PubMed]
- Beltran, H. The N-myc Oncogene: Maximizing its Targets, Regulation, and Therapeutic Potential. Mol. Cancer Res. 2014, 12, 815–822. [Google Scholar] [CrossRef] [PubMed]
- Bush, A.; Mateyak, M.; Dugan, K.; Obaya, A.; Adachi1, S.; Sedivy, J.; Cole, M. c-myc null cells misregulate cad and gadd45 but not other proposed c-Myc targets. Genes Dev. 1998, 12, 3797–3802. [Google Scholar] [CrossRef] [PubMed]
- Malynn, B.A.; Alboran, I.M.; O’Hagan, R.C.; Bronson, R.; Davidson, L.; DePinho, R.A.; Alt, F.W. N-myc can functionally replace c-myc in murine development, cellular growth, and differentiation. Genes Dev. 2000, 14, 1390–1399. [Google Scholar] [PubMed]
- McMahon, S.B.; Van Buskirk, H.A.; Dugan, K.A.; Copeland, T.D.; Cole, M.D. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell 1998, 94, 363–374. [Google Scholar] [CrossRef]
- Gu, W.; Bhatia, K.; Magrath, I.T.; Dang, C.V.; Dalla-Favera, R. Binding and suppression of the Myc transcriptional activation domain by p107. Science 1994, 264, 251–254. [Google Scholar] [CrossRef] [PubMed]
- Sakamuro, D.; Elliott, K.J.; Wechsler-Reya, R.; Prendergast, G.C. BIN1 is a novel MYC-interacting protein with features of a tumor suppressor. Nat. Genet. 1996, 14, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Sakamuro, D.; Prendergast, G.C. New Myc-interacting proteins: A second Myc network emerges. Oncogene 1999, 18, 2942–2954. [Google Scholar] [CrossRef] [PubMed]
- Barrett, J.; Birrer, M.J.; Kato, G.J.; Dosaka-Akita, H.; Dang, C.V. Activation domains of L-Myc and c-Myc determine their transforming potencies in rat embryo cells. Mol. Cell. Biol. 1992, 12, 3130–3137. [Google Scholar] [CrossRef] [PubMed]
- Prendergast, G.C.; Ziff, E.B. A new bind for Myc. Trends Genet. 1992, 8, 91–96. [Google Scholar] [CrossRef]
- Amati, B.; Brooks, M.W.; Levy, N.; Littlewood, T.D.; Evan, G.I.; Land, H. Oncogenic activity of the c-Myc protein requires dimerization with Max. Cell 1993, 72, 233–245. [Google Scholar] [CrossRef]
- Hopewell, R.; Ziff, E.B. The nerve growth factor-responsive PC12 cell line does not express the Myc dimerization partner. Max. Mol. Cell. Biol. 1995, 15, 3470–3478. [Google Scholar] [CrossRef] [PubMed]
- Steiger, D.; Furrer, M.; Schwinkendorf, D.; Gallant, P. Max independent functions of Myc in Drosophila melanogaster. Nat. Genet. 2008, 40, 1084–1091. [Google Scholar] [CrossRef] [PubMed]
- Wert, M.; Kennedy, S.; Palfrey, H.C.; Hay, N. Myc drives apoptosis in PC12 cells in the absence of Max. Oncogene 2001, 20, 3746–3750. [Google Scholar] [CrossRef] [PubMed]
- Cowling, V.H.; Cole, M.D. The Myc transactivation domain promotes global phosphorylation of the RNA polymerase II carboxy-terminal domain independently of direct DNA binding. Mol. Cell. Biol. 2007, 27, 2059–2073. [Google Scholar] [CrossRef] [PubMed]
- Peukert, K.; Staller, P.; Schneider, A.; Carmichael, G.; Hänel, F.; Eilers, M. An alternative pathway for gene regulation by Myc. EMBO J. 1997, 16, 5672–5686. [Google Scholar] [CrossRef] [PubMed]
- Javahery, R.; Khachi, A.; Lo, K.; Zenzie-Gregory, B.; Smale, S.T. DNA sequence requirements for transcriptional initiator activity in mammalian cells. Mol. Cell. Biol. 1994, 14, 116–127. [Google Scholar] [CrossRef] [PubMed]
- Staller, P.; Peukert, K.; Kiermaier, A.; Seoane, J.; Lukas, J.; Karsunky, H.; Möröy, T.; Bartek, J.; Massagué, J.; Hänel, F.; et al. Repression of p15INK4b expression by Myc through association with Miz-1. Nat. Cell Biol. 2001, 3, 392–399. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Cetinkaya, C.; Munoz-Alonso, M.J.; von der Lehr, N.; Bahram, F.; Beuger, V.; Eilers, M.; Leon, J.; Larsson, L.G. Myc represses differentiation-induced p21CIP1 expression via Miz-1-dependent interaction with the p21 core promoter. Oncogene 2003, 22, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Pyndiah, S.; Tanida, S.; Ahmed, K.M.; Cassimere, E.K.; Choe, C.; Sakamuro, D. c-MYC suppresses BIN1 to release poly (ADP-ribose) polymerase 1: A mechanism by which cancer cells acquire cisplatin resistance. Sci. Signal. 2011, 4, Ra19. [Google Scholar] [CrossRef] [PubMed]
- Cole, M.D.; Cowling, V.H. Transcription-Independent Functions of MYC: Regulation of Translation and DNA Replication. Nat. Rev. Mol. Cell Biol. 2008, 9, 810–815. [Google Scholar] [CrossRef] [PubMed]
- Kelly, K.; Cochran, B.H.; Stiles, C.D.; Leder, P. Cell-specific regulation of the c-myc gene by lymphocyte mitogens and platelet-derived growth factor. Cell 1983, 35, 603–610. [Google Scholar] [CrossRef]
- Campisi, J.; Gray, H.E.; Pardee, A.B.; Dean, M.; Sonenshein, G.E. Cell-cycle control of c-myc but not c-ras expression is lost following chemical transformation. Cell 1984, 36, 241–247. [Google Scholar] [CrossRef]
- Mateyak, M.K.; Obaya, A.J.; Adachi, S.; Sedivy, J.M. Phenotypes of c-Myc-deficient rat fibroblasts isolated by targeted homologous recombination. Cell Growth Differ. 1997, 8, 1039–1048. [Google Scholar] [PubMed]
- Pfeifer-Ohlsson, S.; Rydnert, J.; Goustin, A.S.; Larsson, E.; Bethsholtz, C.; Ohlsson, R. Cell-type-specific pattern of myc protooncogene expression in developing human embryos. Proc. Natl. Acad. Sci. USA 1985, 82, 5050–5054. [Google Scholar] [CrossRef] [PubMed]
- Land, H.; Parada, L.F.; Weinberg, R.A. Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes. Nature 1983, 304, 596–602. [Google Scholar] [CrossRef] [PubMed]
- Adams, J.M.; Harris, A.W.; Pinkert, C.A.; Corcoran, L.M.; Alexander, W.S.; Cory, S.; Palmiter, R.D.; Brinster, R.L. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature 1985, 318, 533–538. [Google Scholar] [CrossRef] [PubMed]
- Chesi, M.; Robbiani, D.F.; Sebag, M.; Chng, W.J.; Affer, M.; Tiedemann, R.; Valdez, R.; Palmer, S.E.; Haas, S.S.; Stewart, A.K.; et al. AID-dependent activation of a MYC transgene induces multiple myeloma in a conditional mouse model of post-germinal center malignancies. Cancer Cell 2008, 13, 167–180. [Google Scholar] [CrossRef] [PubMed]
- Leder, A.; Pattengale, P.K.; Kuo, A.; Stewart, T.A.; Leder, P. Consequences of widespread deregulation of the c-myc gene in transgenic mice: Multiple neoplasms and normal development. Cell 1986, 45, 485–495. [Google Scholar] [CrossRef]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Laurenti, E.; Wilson, A.; Trumpp, A. Myc’s other life: Stem cells and beyond. Curr. Opin. Cell Biol. 2009, 21, 844–854. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.M.; Dalton, S. The cell cycle and Myc intersect with mechanisms that regulate pluripotency and reprogramming. Cell Stem Cell 2009, 5, 141–149. [Google Scholar] [CrossRef] [PubMed]
- Ben-David, U.; Benvenisty, N. The tumorigenicity of human embryonic and induced pluripotent stem cells. Nat. Rev. Cancer 2011, 11, 268–277. [Google Scholar] [CrossRef] [PubMed]
- Riggs, J.W.; Barrilleaux, B.L.; Varlakhanova, N.; Bush, K.M.; Chan, V.; Knoepfler, P.S. Induced pluripotency and oncogenic transformation are related processes. Stem Cells Dev. 2013, 22, 37–50. [Google Scholar] [CrossRef] [PubMed]
- Wolfera, A.; Wittnera, B.S.; Irimiab, D.; Flavinb, R.J.; Lupienb, M.; Gunawardaneb, R.N.; Meyerg, C.A.; Lightcaph, E.S.; Tamayo, P.; Mesirov, J.P.; et al. MYC regulation of a “poor-prognosis” metastatic cancer cell state. Proc. Natl. Acad. Sci. USA 2010, 107, 3698–3703. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.N.; Singh, A.M.; Dalton, S. Myc sustains pluripotency by repressing primitive endoderm specification. Cell Stem Cell 2010, 7, 343–354. [Google Scholar] [CrossRef] [PubMed]
- Laurenti, E.; Varnum-Finney, B.; Wilson, A.; Ferrero, I.; Blanco-Bose, W.E.; Ehninger, A.; Knoepfler, P.S.; Cheng, P.F.; MacDonald, H.R.; Eisenman, R.N.; et al. Hematopoietic stem cell function and survival depend on c-Myc and N-Myc activity. Cell Stem Cell 2008, 3, 611–624. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.; Murphy, M.J.; Oskarsson, T.; Kaloulis, K.; Bettess, M.D.; Oser, G.M.; Pasche, A.C.; Knabenhans, C.; Macdonald, H.R.; Trumpp, A. c-Myc controls the balance between hematopoietic stem cell self-renewal and differentiation. Genes Dev. 2004, 18, 2747–2763. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, I.B. Disorders in cell circuitry during multistage carcinogenesis: The role of homeostasis. Carcinogenesis 2000, 21, 857–864. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, I.B. Addiction to oncogenes—The Achilles’ heal of cancer. Science 2002, 297, 63–64. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, K.; Imamoto, F. Transcriptional control of the endogenous MYC protooncogene by antisense RNA. Proc. Natl. Acad. Sci. USA 1987, 84, 7363–7367. [Google Scholar] [CrossRef] [PubMed]
- Loke, S.L.; Stein, C.; Zhang, X.; Avigan, M.; Cohen, J.; Neckers, L.M. Delivery of c-myc antisense phosphorothioate oligodeoxynucleotides to hematopoietic cells in culture by liposome fusion: Specific reduction in c-myc protein expression correlates with inhibition of cell growth and DNA synthesis. Curr. Top. Microbiol. Immunol. 1988, 141, 282–289. [Google Scholar] [PubMed]
- Felsher, D.W.; Bishop, J.M. Reversible tumorigenesis by MYC in hematopoietic lineages. Mol. Cell. 1999, 4, 199–207. [Google Scholar] [CrossRef]
- Pelengaris, S.; Khan, M.; Evan, G.I. Suppression of Myc-induced apoptosis in beta cells exposes multiple oncogenic properties of Myc and triggers carcinogenic progression. Cell 2002, 109, 321–334. [Google Scholar] [CrossRef]
- Jain, M.; Arvanitis, C.; Chu, K.; Dewey, W.; Leonhardt, E.; Trinh, M.; Sundberg, C.D.; Bishop, J.M.; Felsher, D.W. Sustained loss of a neoplastic phenotype by brief inactivation of MYC. Science 2002, 297, 102–104. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, I.B.; Joe, A.K. Mechanisms of disease: Oncogene addiction—A rationale for molecular targeting in cancer therapy. Nat. Clin. Pract. Oncol. 2006, 3, 448–457. [Google Scholar] [CrossRef] [PubMed]
- Hoelder, S.; Clarke, P.A.; Workman, P. Discovery of small molecule cancer drugs: Successes, challenges and opportunities. Mol. Oncol. 2012, 6, 155–176. [Google Scholar] [CrossRef] [PubMed]
- Horiuchi, D.; Anderton, B.; Goga, A. Taking on Challenging Targets: Making MYC Druggable. Am. Soc. Clin. Oncol. Educ. Book 2014, e497–e502. [Google Scholar] [CrossRef] [PubMed]
- Darmon, E.; Leach, D.R.F. Bacterial Genome Instability. Mol. Biol. Rev. 2014, 78, 1–39. [Google Scholar] [CrossRef] [PubMed]
- Popescu, N.C.; Zimonjic, D.B. Chromosome-mediated alterations of the MYC gene in human cancer. J. Cell. Mol. Med. 2002, 6, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Lengauer, C.; Kinzler, K.W.; Vogelstein, B. Genetic instabilities in human cancers. Nature 1998, 396, 643–649. [Google Scholar] [CrossRef] [PubMed]
- Negrini, S.; Gorgoulis, V.G.; Halazonetis, T.D. Genomic instability—An evolving hallmark of cancer. Nat. Rev. Mol. Cell Biol. 2010, 11, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Loeb, L.A. Mutator phenotype may be required for multistage carcinogenesis. Cancer Res. 1991, 51, 3075–3079. [Google Scholar] [PubMed]
- Hartwell, L. Defects in a cell cycle checkpoint may be responsible for the genomic instability of cancer cells. Cell 1992, 71, 543–546. [Google Scholar] [CrossRef]
- Juan, J.; Muraguchi, T.; Iezza, G.; Sears, R.C.; McMahon, M. Diminished WNT → β-catenin → c-MYC signaling is a barrier for malignant progression of BRAFV600E-induced lung tumors. Genes Dev. 2014, 28, 561–575. [Google Scholar] [CrossRef] [PubMed]
- Barone, M.V.; Courtneidge, S.A. Myc but not Fos rescue of PDGF signalling block caused by kinase-inactive Src. Nature 1995, 378, 509–512. [Google Scholar] [CrossRef] [PubMed]
- Taub, A.; Kirsch, I.; Morton, C.; Lenoir, G.M.; Swan, D.; Tronick, S.; Aaronson, S.; Leder, P. Translocation of the c-myc gene into the immunoglobulin heavy chain locus in human Burkitt lymphoma and murine plasmacytoma cells. Proc. Natl. Acad. Sci. USA 1982, 79, 7837–7841. [Google Scholar] [CrossRef] [PubMed]
- Shen-Ong, G.L.C.; Keath, E.J.; Piccoli, S.P.; Cole, M.D. Novel myc oncogene RNA from abortive immunoglobulin-gene recombination in mouse plasmacytomas. Cell 1982, 31, 443–480. [Google Scholar] [CrossRef]
- Pear, W.S.; Wahlstrom, G.; Nelson, S.F.; Axelson, H.; Szeles, A.; Wiener, F.; Bazin, H.; Klein, G.; Sumegi, J. 6;7 chromosomal translocation in spontaneously arising rat immunocytomas Evidence for c-myc breakpoint clustering and correlation between isotype expression and the c-myc target. Mol. Cell. Biol. 1988, 8, 441–451. [Google Scholar] [CrossRef] [PubMed]
- Koh, C.M.; Bieberich, C.J.; Dang, C.V.; Nelson, N.; Yegnasubramanian, S.; De Marzo, A. MYC and Prostate Cancer. Genes Cancer 2010, 1, 617–628. [Google Scholar] [CrossRef] [PubMed]
- Felsher, D.W.; Bishop, J.M. Transient excess of MYC activity can elicit genomic instability and tumorigenesis. Proc. Natl. Acad. Sci. USA 1999, 96, 3940–3944. [Google Scholar] [CrossRef] [PubMed]
- Kuzyk, A.; Mai, S. c-MYC-Induced Genomic Instability. Cold Spring Harb. Perspect. Med. 2014, 4, a014373. [Google Scholar] [CrossRef] [PubMed]
- Kuttler, F.; Mai, S. Formation of non-random extrachromosomal elements during development, differentiation and oncogenesis. Semin. Cancer Biol. 2007, 17, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Mai, S.; Fluri, M.; Siwarski, D.; Huppi, K. Genomic instability in MycER-activated Rat1A MycER cells. Chromosome Res. 1996, 4, 365–371. [Google Scholar] [CrossRef] [PubMed]
- Rockwood, L.D.; Torrey, T.A.; Kim, J.S.; Coleman, A.E.; Kovalchuk, A.L.; Xiang, S.; Ried, T.; Morse, H.C., III; Janz, S. Genomic instability in mouse Burkitt lymphoma is dominated by illegitimate genetic recombinations, not point mutations. Oncogene 2002, 21, 7235–7240. [Google Scholar] [CrossRef] [PubMed]
- Silva, A.G.; Graves, H.A.; Guffei, A.; Ricca, T.I.; Mortara, R.A.; Jasiulionis, M.G.; Mai, S. Telomere-centromere-driven genomic instability contributes to karyotype evolution in a mouse model of melanoma. Neoplasia 2010, 12, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Wade, M.; Wahl, G.M. c-Myc, Genome Instability, and Tumorigenesis: The Devil Is in the Details. CTMI 2006, 302, 169–203. [Google Scholar]
- Mai, S. Overexpression of c-myc precedes amplification of the gene encoding dihydrofolate reductase. Gene 1994, 148, 253–260. [Google Scholar] [CrossRef]
- Mai, S.; Hanley-Hyde, J.; Fluri, M. c-Myc overexpression associated DHFR gene amplification in hamster, rat, mouse and human cell lines. Oncogene 1996, 12, 277–288. [Google Scholar] [PubMed]
- Mai, S.; Hanley-Hyde, J.; Rainey, G.J.; Kuschak, T.I.; Paul, J.T.; Littlewood, T.D.; Mischak, H.; Stevens, L.M.; Henderson, D.W.; Mushinski, J.F. Chromosomal and extrachromosomal instability of the cyclin D2 gene is induced by Myc overexpression. Neoplasia 1999, 1, 241–252. [Google Scholar] [CrossRef] [PubMed]
- Kuschak, T.I.; Taylor, C.; McMillan-Ward, E.; Israels, S.; Henderson, D.W.; Mushinski, J.F.; Wright, J.A.; Mai, S. The ribonucleotide reductase R2 gene is a non-transcribed target of c-Myc-induced genomic instability. Gene 1999, 238, 351–365. [Google Scholar] [CrossRef]
- Guijon, F.B.; Greulich-Bode, K.; Paraskevas, M.; Baker, P.; Mai, S. Premalignant cervical lesions are characterized by dihydrofolate reductase gene amplification and c-Myc overexpression: Possible biomarkers. J. Low. Genit. Tract. Dis. 2007, 11, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Rogakou, E.P.; Pilch, D.R.; Orr, A.H.; Ivanova, V.S.; Bonner, W.M. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998, 273, 5858–5868. [Google Scholar] [CrossRef] [PubMed]
- Sedelnikova, O.A.; Pilch, D.R.; Redon, C.; Bonner, W.M. Histone H2AX in DNA damage and repair. Cancer Biol. Ther. 2003, 2, 233–235. [Google Scholar] [PubMed]
- Karlsson, A.; Deb-Basu, D.; Cherry, A.; Turner, S.; Ford, J.; Felsher, D.W. Defective double-strand DNA break repair and chromosomal translocations by MYC overexpression. Proc. Natl. Acad. Sci. USA 2003, 100, 9974–9979. [Google Scholar] [CrossRef] [PubMed]
- Khanna, K.K.; Jackson, S.P. DNA double-strand breaks: Signaling, repair and the cancer connection. Nat. Genet. 2001, 27, 247–254. [Google Scholar] [CrossRef] [PubMed]
- Vafa, O.; Wade, M.; Kern, S.; Beeche, M.; Pandita, T.K.; Hampton, G.M.; Wahl, G.M. c-Myc can induce DNA damage, increase reactive oxygen species, and mitigate p53 function: A mechanism for oncogene-induced genetic instability. Mol. Cell. 2002, 9, 1031–1044. [Google Scholar] [CrossRef]
- Ray, S.; Atkuri, K.R.; Deb-Basu, D.; Adler, A.S.; Chang, H.Y.; Herzenberg, L.A.; Felsher, D.W. MYC can induce DNA breaks in vivo and in vitro independent of reactive oxygen species. Cancer Res. 2006, 66, 6598–6605. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Owonikoko, T.; Ramalingam, S.S.; Doetsch, P.W.; Xiao, Z.; Khuri, F.; Curran, W.; Deng, X. c-Myc suppression of DNA double-strand break repair. Neoplasia 2012, 14, 1190–1202. [Google Scholar] [CrossRef] [PubMed]
- Ambrosio, S.; Amente, S.; Napolitano, G.; Di Palo, G.; Lania, L.; Majello, B. MYC impairs resolution of site-specific DNA double-strand breaks repair. Mutat. Res. 2015, 774, 6–13. [Google Scholar] [CrossRef] [PubMed]
- Classon, M.; Henriksson, M.; Sümegi, J.; Klein, G.; Hammarskjöld, M.L. Elevated c-myc expression facilitates the replication of SV40 DNA in human lymphoma cells. Nature 1987, 330, 272–274. [Google Scholar] [CrossRef] [PubMed]
- Dominguez-Sola, D.; Ying, C.Y.; Grandori, C.; Ruggiero, L.; Chen, B.; Li, M.; Galloway, D.A.; Gu, W.; Gautier, J.; Dalla-Favera, R. Non-transcriptional control of DNA replication by c-Myc. Nature 2007, 448, 445–451. [Google Scholar] [CrossRef] [PubMed]
- Campaner, S.; Amati, B. Two sides of the Myc-induced DNA damage response: From tumor suppression to tumor maintenance. Cell Div. 2012, 7, 6. [Google Scholar] [CrossRef] [PubMed]
- Bartkova, J.; Rezaei, N.; Liontos, M.; Karakaidos, P.; Kletsas, D.; Issaeva, N.; Vassiliou, L.V.; Kolettas, E.; Niforou, K.; Zoumpourlis, V.C.; et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature 2006, 444, 633–637. [Google Scholar] [CrossRef] [PubMed]
- Di Micco, R.; Fumagalli, M.; Cicalese, A.; Piccinin, S.; Gasparini, P.; Luise, C.; Schurra, C.; Garre’, M.; Nuciforo, P.G.; Bensimon, A.; et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature 2006, 444, 638–642. [Google Scholar] [CrossRef] [PubMed]
- Serrano, M.; Lin, A.W.; McCurrach, M.E.; Beach, D.; Lowe, S.W. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 1997, 88, 593–602. [Google Scholar] [CrossRef]
- Wajapeyee, N.; Serra, R.W.; Zhu, X.; Mahalingam, M.; Green, M.R. Oncogenic BRAF induces senescence and apoptosis through pathways mediated by the secreted protein IGFBP7. Cell 2008, 132, 363–374. [Google Scholar] [CrossRef] [PubMed]
- Ruley, H.E. Adenovirus early region 1A enables viral and cellular transforming genes to transform primary cells in culture. Nature 1983, 304, 602–606. [Google Scholar] [CrossRef] [PubMed]
- Malumbres, M.; Barbacid, M. RAS oncogenes: The first 30 years. Nat. Rev. Cancer 2003, 3, 459–465. [Google Scholar] [CrossRef] [PubMed]
- Kerbel, R.S.; Kobayashi, H.; Graham, C.H. Intrinsic or acquired drug resistance and metastasis: Are they linked phenotypes? J. Cell. Biochem. 1994, 56, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Dijt, F.J.; Fichtinger-Schepman, A.M.; Berends, F.; Reedijk, J. Formation and repair of cisplatin-induced adducts to DNA in cultured normal and repair-deficient human fibroblasts. Cancer Res. 1988, 48, 6058–6062. [Google Scholar] [PubMed]
- Dabholkar, M.; Bostick-Bruton, F.; Weber, C.; Bohr, V.A.; Egwuagu, C.; Reed, E. ERCC1 and ERCC2 expression in malignant tissues from ovarian cancer patients. J. Natl. Cancer Inst. 1992, 84, 1512–1517. [Google Scholar] [CrossRef] [PubMed]
- Sheridan, E.; Silcocks, P.; Smith, J.; Hancock, B.W.; Goyns, M.H. P53 mutation in a series of epithelial ovarian cancer from the U.K. and its prognostic significance. Eur. J. Cancer 1994, 30A, 1701–1704. [Google Scholar] [CrossRef]
- Perego, P.; Giarola, M.; Righetti, S.C.; Supino, R.; Caserini, C.; Delia, D.; Pierotti, M.A.; Miyashita, T.; Reed, J.C.; Zunino, F. Association between cisplatin resistance and mutation of p53 gene and reduced Bax expression in ovarian carcinoma cell systems. Cancer Res. 1996, 56, 556–562. [Google Scholar] [PubMed]
- Agarwal, R.; Kaye, S.B. Ovarian cancer: Strategies for overcoming resistance to chemotherapy. Nat. Rev. Cancer 2003, 3, 502–516. [Google Scholar] [CrossRef] [PubMed]
- Borst, P.; Evers, R.; Kool, M.; Wijnholds, J. A family of drug transporters: The multidrug resistance-associated proteins. J. Natl. Cancer Inst. 2000, 92, 1295–1302. [Google Scholar] [CrossRef] [PubMed]
- Longley, D.B.; Johnston, P.G. Molecular mechanisms of drug resistance. J. Pathol. 2005, 205, 275–292. [Google Scholar] [CrossRef] [PubMed]
- Aguilera, A.; Gómez-González, B. Genome instability: A mechanistic view of its causes and consequences. Nat. Rev. Genet. 2008, 9, 204–217. [Google Scholar] [CrossRef] [PubMed]
- Rebucci, M.; Michiels, C. Molecular aspects of cancer cell resistance to chemotherapy. Biochem. Pharmacol. 2013, 85, 1219–1226. [Google Scholar] [CrossRef] [PubMed]
- Walker, T.L.; White, J.D.; Esdale, W.J.; Burton, M.A.; DeCruz, E.E. Tumour cells surviving in vivo cisplatin chemotherapy display elevated c-myc expression. Br. J. Cancer 1996, 73, 610–614. [Google Scholar] [CrossRef] [PubMed]
- Leonetti, C.; Biroccio, A.; Benassi, B.; Stringaro, A.; Stoppacciaro, A.; Semple, S.C.; Zupi, G. Encapsulation of c-myc antisense oligodeoxynucleotides in lipid particles improves antitumoral efficacy in vivo in a human melanoma line. Cancer Gene Ther. 2001, 8, 459–468. [Google Scholar] [CrossRef] [PubMed]
- Knapp, D.C.; Mata, J.E.; Reddy, M.T.; Devi, G.R.; Iversen, P.L. Resistance to chemotherapeutic drugs overcome by c-Myc inhibition in a Lewis lung carcinoma murine model. Anticancer Drugs 2003, 14, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.P.; Liu, J.D.; Chow, J.M.; Liu, C.R.; Liu, H.E. Small-molecule c-Myc inhibitor, 10058-F4, inhibits proliferation, downregulates human telomerase reverse transcriptase and enhances chemosensitivity in human hepatocellular carcinoma cells. Anticancer Drugs 2007, 18, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Reyes-González, J.M.; Armaiz-Peña, G.N.; Mangala, L.S.; Valiyeva, F.; Ivan, C.; Pradeep, S.; Echevarría-Vargas, I.M.; Rivera-Reyes, A.; Sood, A.K.; Vivas-Mejía, P.E. Targeting c-MYC in Platinum-Resistant Ovarian Cancer. Mol. Cancer. Ther. 2015, 14, 2260–2269. [Google Scholar] [CrossRef] [PubMed]
- Gravina, G.L.; Festuccia, C.; Popov, V.M.; Di Rocco, A.; Colapietro, A.; Sanità, P.; Delle Monache, S.; Musio, D.; De Felice, F.; Di Cesare, E.; et al. c-Myc Sustains Transformed Phenotype and Promotes Radioresistance of Embryonal Rhabdomyosarcoma Cell Lines. Radiat. Res. 2016, 185, 411–422. [Google Scholar] [CrossRef] [PubMed]
- Leonetti, C.; Biroccio, A.; Candiloro, A.; Citro, G.; Fornari, C.; Mottolese, M.; Del Bufalo, D.; Zupi, G. Increase of cisplatin sensitivity by c-myc antisense oligodeoxynucleotides in a human metastatic melanoma inherently resistant to cisplatin. Clin. Cancer Res. 1999, 5, 2588–2595. [Google Scholar] [PubMed]
- Huang, M.J.; Cheng, Y.C.; Liu, C.R.; Lin, S.; Liu, H.E. A small-molecule c-Myc inhibitor, 10058-F4, induces cell-cycle arrest, apoptosis, and myeloid differentiation of human acute myeloid leukemia. Exp. Hematol. 2006, 34, 1480–1489. [Google Scholar] [CrossRef] [PubMed]
- Weinberg, R.A. E2F and cell proliferation: A world turned upside down. Cell 1996, 85, 457–459. [Google Scholar] [CrossRef]
- Maser, R.S.; Mirzoeva, O.K.; Wells, J.; Olivares, H.; Williams, B.R.; Zinkel, R.A.; Farnham, P.J.; Petrini, J.H. Mre11 complex and DNA replication: Linkage to E2F and sites of DNA synthesis. Mol. Cell. Biol. 2001, 21, 6006–6016. [Google Scholar] [CrossRef] [PubMed]
- Frame, F.M.; Rogoff, H.A.; Pickering, M.T.; Cress, W.D.; Kowalik, T.F. E2F1 induces MRN foci formation and a cell cycle checkpoint response in human fibroblasts. Oncogene 2006, 25, 3258–3266. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zhu, F.; Weaks, R.L.; Biswas, A.K.; Guo, R.; Li, Y.; Johnson, D.G. E2F1 promotes recruitment of DNA repair factors to sites of DNA double-strand breaks. Cell Cycle 2011, 10, 1287–1294. [Google Scholar] [CrossRef] [PubMed]
- Thalmeier, K.; Synovzik, H.; Mertz, R.; Winnacker, E.L.; Lipp, M. Nuclear factor E2F mediates basic transcription and trans-activation of E1a of the human MYC promoter. Genes Dev. 1989, 3, 527–536. [Google Scholar] [CrossRef] [PubMed]
- Mudryj, M.; Hiebert, S.W.; Nevins, J.R. A role for the adenovirus inducible E2F transcription factor in a proliferation dependent signal transduction pathway. EMBO J. 1990, 9, 2179–2184. [Google Scholar] [PubMed]
- Chiang, Y.C.; Teng, S.C.; Su, Y.N.; Hsieh, F.J.; Wu, K.J. c-Myc directly regulates the transcription of the NBS1 gene involved in DNA double-strand break repair. J. Biol. Chem. 2003, 278, 19286–19291. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, P.C.; Frank, S.R.; Wang, L.; Schroeder, M.; Liu, S.; Greene, J.; Cocito, A.; Amati, B. Genomic targets of the human c-Myc protein. Genes Dev. 2003, 17, 1115–1129. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Van Calcar, S.; Qu, C.; Cavenee, W.K.; Zhang, M.Q.; Ren, B. A global transcriptional regulatory role for c-Myc in Burkitt’s lymphoma cells. Proc. Natl. Acad. Sci. USA 2003, 100, 8164–8169. [Google Scholar] [CrossRef] [PubMed]
- Luoto, K.R.; Meng, A.X.; Wasylishen, A.R.; Zhao, H.; Coackley, C.L.; Penn, L.Z.; Bristow, R.G. Tumor cell kill by c-MYC depletion: Role of MYC-regulated genes that control DNA double-strand break repair. Cancer Res. 2010, 70, 8748–8759. [Google Scholar] [CrossRef] [PubMed]
- Cui, F.; Fan, R.; Chen, Q.; He, Y.; Song, M.; Shang, Z.; Zhang, S.; Zhu, W.; Cao, J.; Guan, H.; et al. The involvement of c-Myc in the DNA double-strand break repair via regulating radiation-induced phosphorylation of ATM and DNA-PKcs activity. Mol. Cell. Biochem. 2015, 406, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Eyler, C.E.; Rich, J.N. Survival of the Fittest: Cancer Stem Cells in Therapeutic Resistance and Angiogenesis. J. Clin. Oncol. 2008, 26, 2839–2845. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Joo, K.M.; Jin, J.; Nam, D. Cancer Stem Cells and Their Mechanism of Chemo-Radiation Resistance. Int. J. Stem Cells. 2009, 2, 109–114. [Google Scholar] [CrossRef] [PubMed]
- Vinogradov, S.; Wei, X. Cancer stem cells and drug resistance: The potential of nanomedicine. Nanomedicine 2012, 7, 597–615. [Google Scholar] [CrossRef] [PubMed]
- Wei, X. DNA damage responses in cancer stem cells: Implications for cancer therapeutic strategies. World J. Biol. Chem. 2015, 6, 57–64. [Google Scholar]
- Skvortsov, S.; Debbage, P.; Lukas, P.; Skvortsova, I. Crosstalk between DNA repair and cancer stem cell (CSC) associated intracellular pathways. Semin. Cancer Biol. 2015, 31, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Eilers, M.; Eisenman, R.N. Myc’s broad reach. Genes Dev. 2008, 22, 2755–2766. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, H.; Li, Z.; Wu, Q.; Lathia, J.D.; McLendon, R.E.; Hjelmeland, A.B.; Rich, J.N. c-Myc Is Required for Maintenance of Glioma Cancer Stem Cells. PLoS ONE 2008, 3, e3769. [Google Scholar] [CrossRef] [PubMed]
- Konecny, G.E.; Kristeleit, R.S. PARP inhibitors for BRCA1/2-mutated and sporadic ovarian cancer: A current practice and future directions. Br. J. Cancer 2016, 115, 1157–1173. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.S.; Kaye, S.B.; Yap, T.A. PARP inhibitors: The race is on. Br. J. Cancer 2016, 114, 713–715. [Google Scholar] [CrossRef] [PubMed]
- Davidson, D.; Amrein, L.; Panasci, L.; Aloyz, R. Small molecules, inhibitors of DNA-PK, Targeting DNA repair, and beyond. Front Pharmacol. 2013, 4, 5. [Google Scholar] [CrossRef] [PubMed]
- Beck, C.; Boehler, C.; Guirouilh Barbat, J.; Bonnet, M.E.; Illuzi, G.; Ronde, P.; Gauthier, L.R.; Magroun, N.; Rajendran, A.; Lopez, B.S.; et al. PARP3 affects the relative contribution of homologous recombination and nonhomologous end-joining pathways. Nucleic Acids Res. 2014, 42, 5616–5632. [Google Scholar] [CrossRef] [PubMed]
- Weber, A.M.; Ryan, A.J. ATM and ATR as therapeutic targets in cancer. Pharmacol. Ther. 2015, 149, 124–138. [Google Scholar] [CrossRef] [PubMed]
- Benada, J.; Macurek, L. Targeting the checkpoint to kill cancer cells. Biomolecules 2015, 5, 1912–1937. [Google Scholar] [CrossRef] [PubMed]
- Bryant, H.E.; Helleday, T. Inhibition of poly (ADP-ribose) polymerase activates ATM which is required for subsequent homologous recombination repair. Nucleic Acids Res. 2006, 34, 1685–1691. [Google Scholar] [CrossRef] [PubMed]
- Wiggans, A.J.; Cass, G.K.S.; Bryant, A.; Lawrie, T.A.; Morrison, J. Poly(ADP-ribose) polymerase (PARP) inhibitors for the treatment of ovarian cancer. Cochrane Database Syst. Rev. 2015. [Google Scholar] [CrossRef]
- Kumari, A.; Iwasaki, T.; Pyndiah, S.; Cassimere, E.K.; Palani, C.D.; Sakamuro, D. Regulation of E2F1-induced apoptosis by poly(ADP-ribosyl)ation. Cell Death Differ. 2015, 22, 311–322. [Google Scholar] [CrossRef] [PubMed]
- Denekamp, J.; Dasu, A.; Waites, A. Vasculature and microenvironmental gradients: The missing links in novel approaches to cancer therapy? Adv. Enzyme Regul. 1998, 38, 281–299. [Google Scholar] [CrossRef]
- Harrison, L.; Blackwell, K. Hypoxia and anemia: Factors in decreased sensitivity to radiation therapy and chemotherapy? Oncologist 2004, 9, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Wrann, S.; Kaufmann, M.R.; Wirthner, R.; Stiehl, D.P.; Wenger, R.H. HIF mediated and DNA damage independent histone H2AX phosphorylation in chronic hypoxia. Biol. Chem. 2013, 394, 519–528. [Google Scholar] [CrossRef] [PubMed]
- Tu, W.Z.; Li, B.; Huang, B.; Wang, Y.; Liu, X.D.; Guan, H.; Zhang, S.M.; Tang, Y.; Rang, W.Q.; Zhou, P.K. γH2AX foci formation in the absence of DNA damage: Mitotic H2AX phosphorylation is mediated by the DNA-PKcs/CHK2 pathway. FEBS Lett. 2013, 587, 3437–3443. [Google Scholar] [CrossRef] [PubMed]
- Turinetto, V.; Giachino, C. Multiple facets of histone variant H2AX: A DNA double-strand-break marker with several biological functions. Nucleic Acids Res. 2015, 43, 2489–2498. [Google Scholar] [CrossRef] [PubMed]
- Cremona, C.A.; Behrens, A. ATM signalling and cancer. Oncogene 2014, 33, 3351–3360. [Google Scholar] [CrossRef] [PubMed]
- Olcina, M.M.; Grand, R.J.; Hammond, E.M. ATM activation in hypoxia—Causes and consequences. Mol. Cell. Oncol. 2014, 1, e29903. [Google Scholar] [CrossRef] [PubMed]
- Guerra, L.; Albihn, A.; Tronnersjö, S.; Yan, Q.; Guidi, R.; Stenerlöw, B.; Sterzenbach, T.; Josenhans, C.; Fox, J.G.; Schauer, D.B.; et al. Myc is required for activation of the ATM-Dependent checkpoints in response to DNA damage. PLoS ONE 2010, 5, e8924. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Reinhardt, H.C.; Bartkova, J.; Tommiska, J.; Blomqvist, C.; Nevanlinna, H.; Bartek, J.; Yaffe, M.B.; Hemann, M.T. The combined status of ATM and p53 link tumor development with therapeutic response. Genes Dev. 2009, 23, 1895–1909. [Google Scholar] [CrossRef] [PubMed]
- Godwin, A.K.; Meister, A.; O’Dwyer, P.J.; Huang, C.S.; Hamilton, T.C.; Anderson, M.E. High resistance to cisplatin in human ovarian cancer cell lines is associated with marked increase of glutathione synthesis. Proc. Natl. Acad. Sci. USA 1992, 89, 3070–3074. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumari, A.; Folk, W.P.; Sakamuro, D. The Dual Roles of MYC in Genomic Instability and Cancer Chemoresistance. Genes 2017, 8, 158. https://doi.org/10.3390/genes8060158
Kumari A, Folk WP, Sakamuro D. The Dual Roles of MYC in Genomic Instability and Cancer Chemoresistance. Genes. 2017; 8(6):158. https://doi.org/10.3390/genes8060158
Chicago/Turabian StyleKumari, Alpana, Watson P. Folk, and Daitoku Sakamuro. 2017. "The Dual Roles of MYC in Genomic Instability and Cancer Chemoresistance" Genes 8, no. 6: 158. https://doi.org/10.3390/genes8060158




