Overexpression of the OsIMP Gene Increases the Accumulation of Inositol and Confers Enhanced Cold Tolerance in Tobacco through Modulation of the Antioxidant Enzymes’ Activities
Abstract
:1. Introduction
2. Results
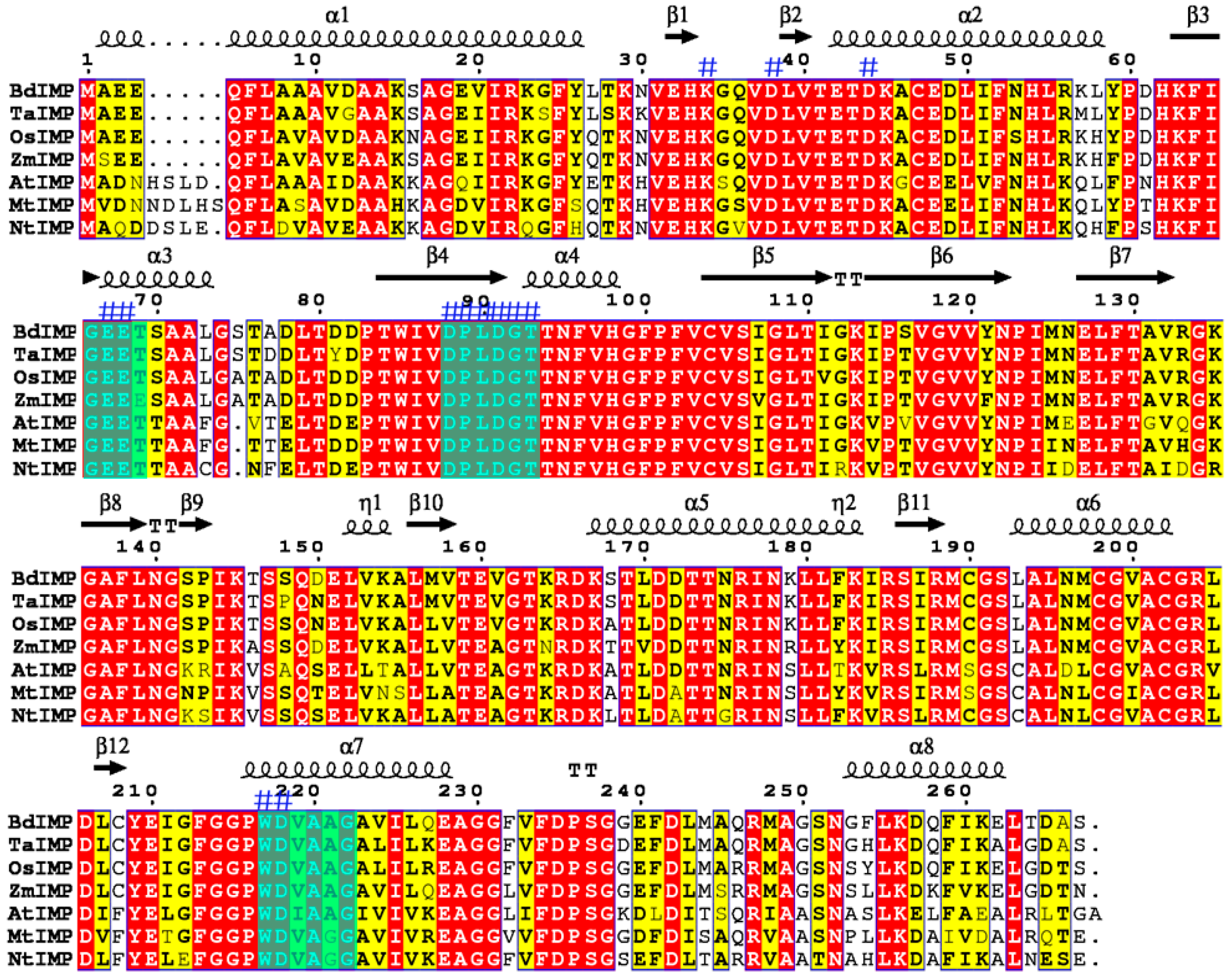
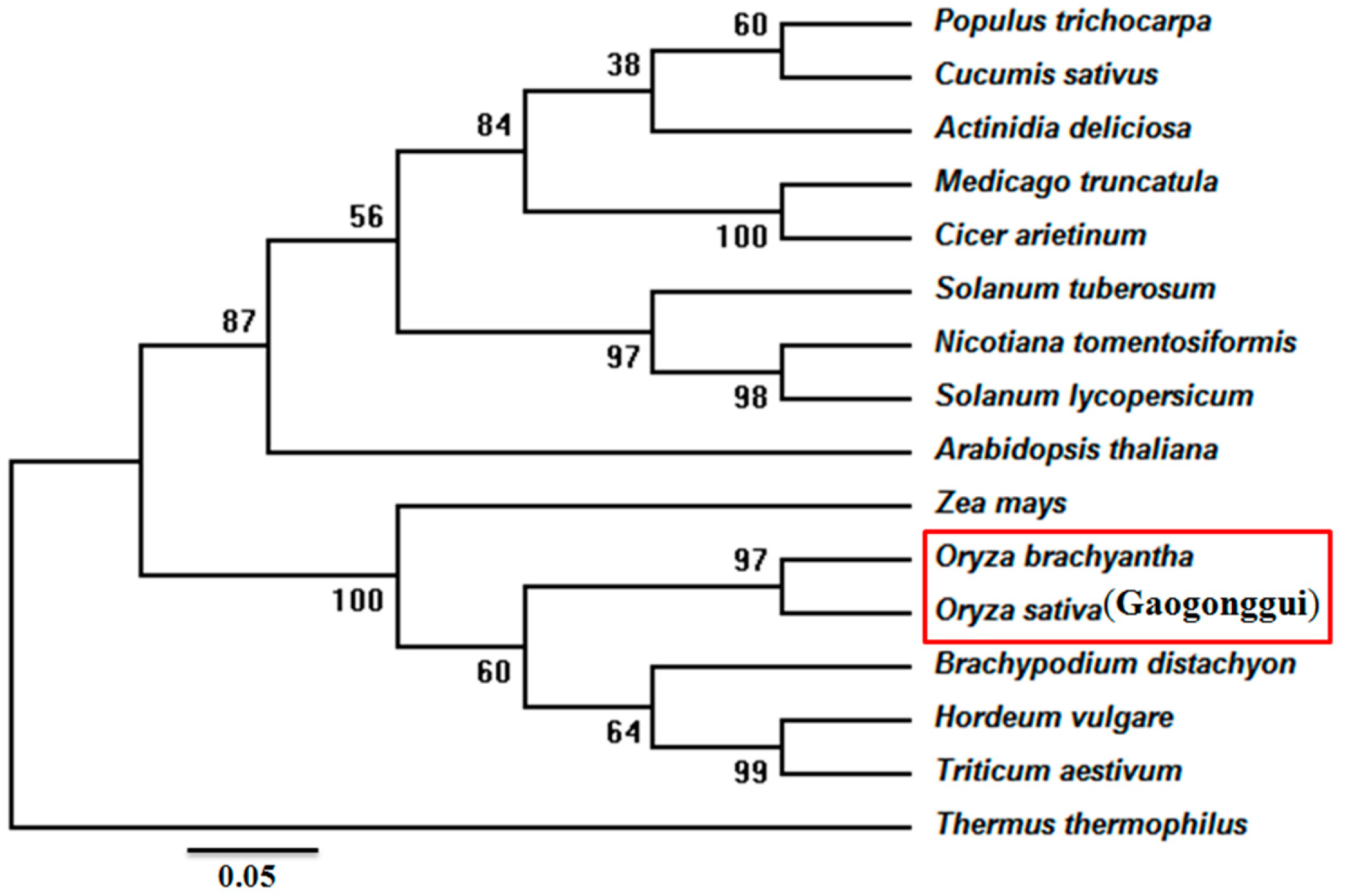
2.1. Cloning and Bioinformatic Analysis of the OsIMP Gene
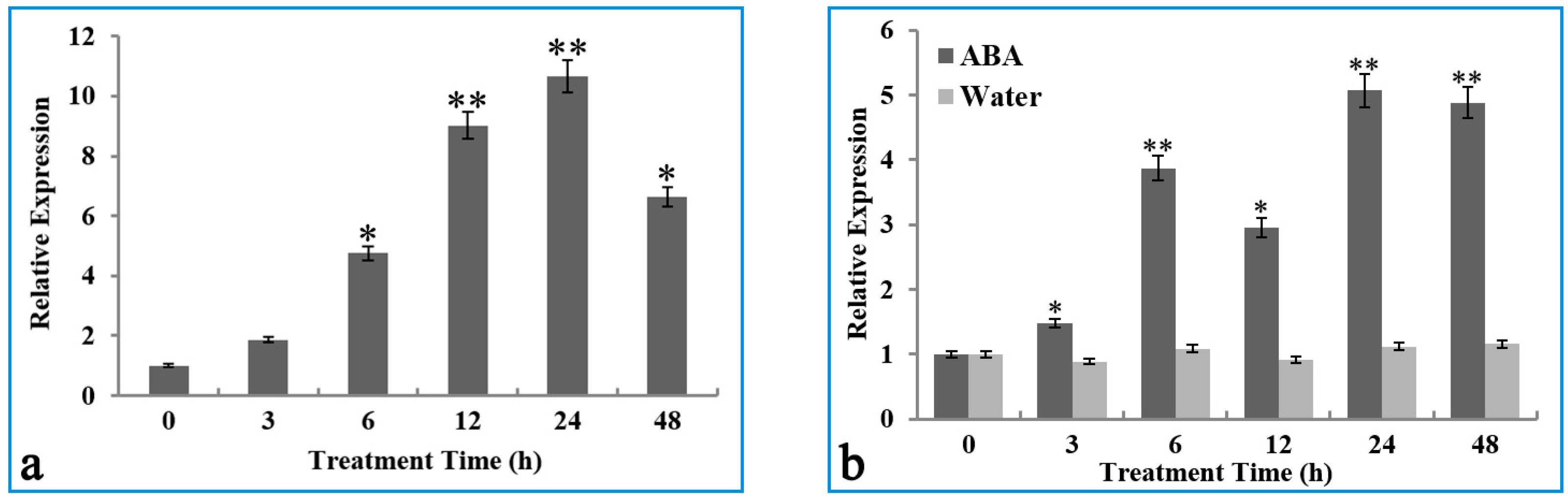
2.2. Transcription of the OsIMP Gene Was Induced by the Cold Stress and ABA Treatment
2.3. Amplification of OsIMP Gene Promoter and Cis-Acting Elements Prediction
2.4. Plasmid Construction, Generation and Confirmation of the OsIMP Transgenic Tobacco Plants
2.5. Overexpression of the OsIMP Gene Increased Inositol Content in Tobacco Plants
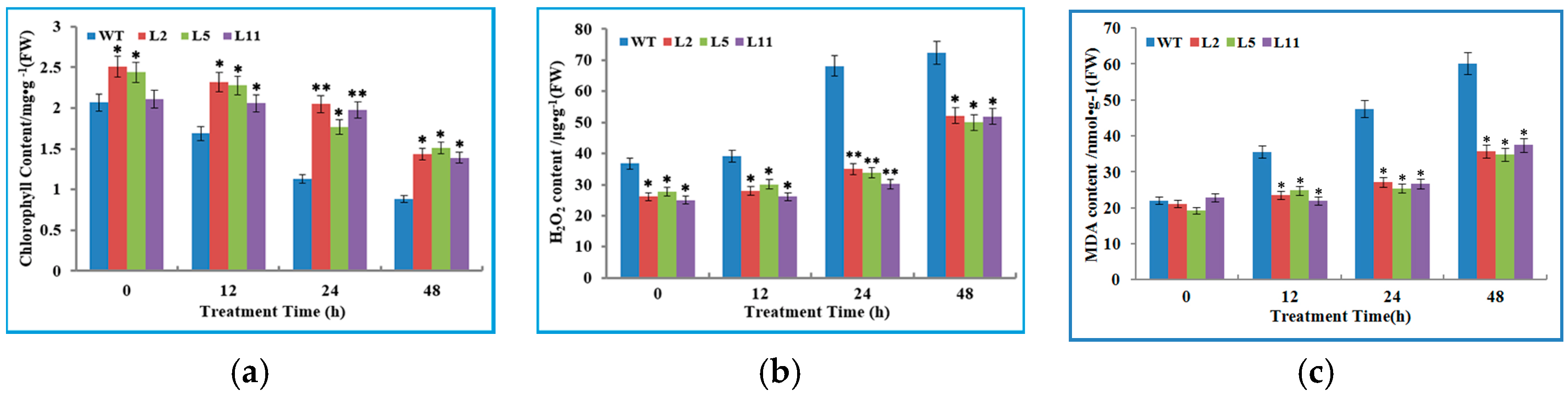
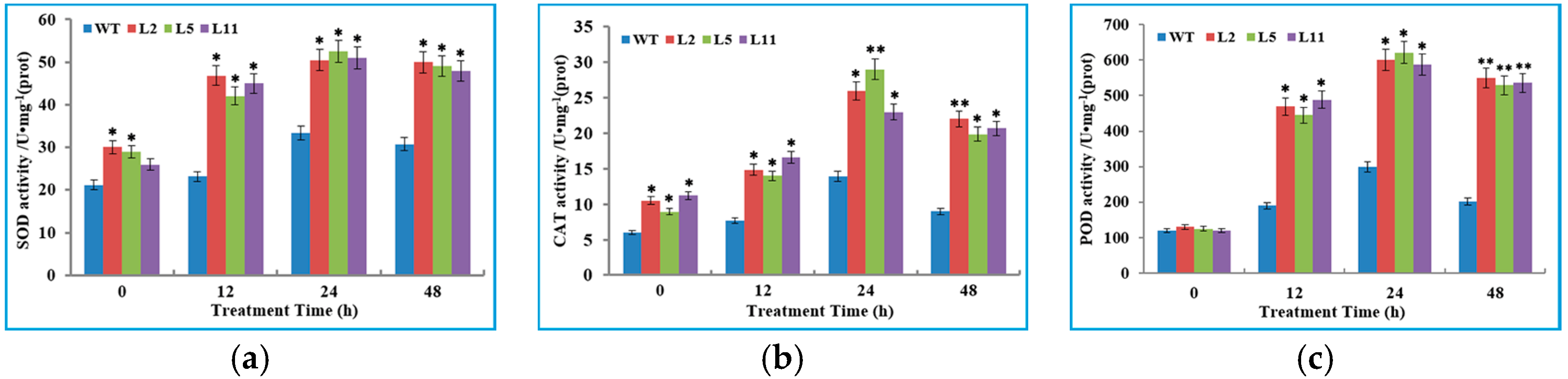
2.6. Transgenic Tobacco Plants Exhibited Improved Tolerance to Cold Stress
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Methods
4.2.1. Cloning and Bioinformatic Analysis of the OsIMP Gene
4.2.2. Expression Profile Analysis of the OsIMP Gene by Real-Time Quantitative PCR
4.2.3. Amplification of the OsIMP Gene Promoter and Cis-Acting Element Prediction
4.2.4. Plasmid Construction, the Generation of OsIMP Transgenic Tobacco Plants and Molecular Comfirmation
4.2.5. Measurement of Inositol Content in WT and T1 Transgenic Tobacco Plants
4.2.6. Cold Stress Treatment and Determination of Stress-Associated Physiological Indicators
5. Statistical Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| IMPase | l-myo-inositol monophosphatase |
| LTR | low-temperature responsiveness |
| ABA | abscisic acid |
| MDA | malondialdehyde |
| SOD | superoxide dismutase |
| CAT | catalase |
| POD | peroxidase |
| MIPS | l-myo-inositol 1-phosphate synthase |
| RFOs | raffinose family of oligosaccharides |
References
- Pandey, S.; Byerlee, D.; Dawe, D.; Dobermann, A.; Mohanty, S.; Rozelle, S.; Hardy, B. (Eds.) Rice in the Global Economy: Strategic Research and Policy Issues for Food Security; International Rice Research Institute: Los Baños, Philippines, 2010; 477p. [Google Scholar]
- Sanghera, G.S.; Wani, S.H.; Hussain, W.; Singh, N.B. Engineering cold stress tolerance in crop plants. Curr. Genom. 2011, 12, 30–43. [Google Scholar] [CrossRef] [PubMed]
- Yang, A.; Dai, X.Y.; Zhang, W.H. A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J. Exp. Bot. 2012, 63, 2541–2556. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.W.; Wu, F.Q.; Wu, W.X.; Wang, H.J.; Zheng, X.M.; Zhang, Y.H.; Chen, X.L.; Zhou, K.N.; Jin, M.N.; Cheng, Z.J.; et al. Rice LTG1 is involved in adaptive growth and fitness under low ambient temperature. Plant J. 2014, 78, 468–480. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Dai, X.Y.; Xu, Y.Y.; Luo, W.; Zheng, X.M.; Zeng, D.L.; Pan, Y.J.; Lin, X.L.; Liu, H.H.; Zhang, D.J.; et al. COLD1 confers chilling tolerance in rice. Cell 2015, 160, 1209–1221. [Google Scholar] [CrossRef] [PubMed]
- Yoon, D.H.; Lee, S.S.; Park, H.J.; Lyu, J.I.; Chong, W.S.; Liu, J.R.; Kim, B.G.; Ahn, J.C.; Cho, H.S. Overexpression of OsCYP19-4 increases tolerance to cold stress and enhances grain yield in rice (Oryza sativa). J. Exp. Bot. 2016, 67, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Loewus, F.A.; Murthy, P.P.N. Myo-inositol metabolism in plants. Plant Sci. 2000, 150, 1–19. [Google Scholar] [CrossRef]
- Bohnert, H.J.; Nelson, D.E.; Jensen, R.G. Adaptations to environmental stresses. Plant Cell 1995, 7, 1099–1111. [Google Scholar] [CrossRef] [PubMed]
- Valluru, R.; Ende, W.V.D. Myo-inositol and beyond—Emerging networks under stress. Plant Sci. 2011, 181, 387–400. [Google Scholar] [CrossRef] [PubMed]
- Chaouch, S.; Noctor, G. Myo-inositol abolishes salicylic acid-dependent cell death and pathogen defence responses triggered by peroxisomal hydrogen peroxide. New Phytol. 2010, 188, 711–718. [Google Scholar] [CrossRef] [PubMed]
- Bruggeman, Q.; Garmier, M.; Bont, L.D.; Soubigou-Taconnat, L.; Mazubert, C.; Benhamed, M.; Raynaud, C.; Bergounioux, C.; Delarue, M. The polyadenylation factor subunit CLEAVAGE AND POLYADENYLATION SPECIFICITY FACTOR30: A key factor of programmed cell death and a regulator of immunity in Arabidopsis. Plant Physiol. 2014, 165, 732–746. [Google Scholar] [CrossRef] [PubMed]
- Majee, M.; Maitra, S.; Dastidar, K.G.; Pattnaik, S.; Chatterjee, A.; Hait, N.C.; Das, K.P.; Majumder, A.L. A novel salt-tolerant L-myo-inositol-1-phosphate synthase from Porteresia coarctata (Roxb.) Tateoka, a halophytic wild rice: Molecular cloning, bacterial overexpression, characterization, and functional introgression into tobacco conferring salt tolerance phenotype. J. Biol. Chem. 2004, 279, 28539–28552. [Google Scholar] [PubMed]
- Tan, J.L.; Wang, C.Y.; Xiang, B.; Han, R.; Guo, Z.F. Hydrogen peroxide and nitric oxide mediated cold- and dehydration-induced myo-inositol phosphate synthase that confers multiple resistances to abiotic stresses. Plant Cell Environ. 2013, 36, 288–299. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, S.; Patra, B.; Ray, S.; Majumder, A.L. Inositol methyl tranferase from a halophytic wild rice, Porteresia coarctata Roxb. (Tateoka): Regulation of pinitol synthesis under abiotic stress. Plant Cell Environ. 2008, 31, 1442–1459. [Google Scholar] [CrossRef] [PubMed]
- Nishizawa, A.; Yabuta, Y.; Shigeoka, S. Galactinol and raffinose constitute a novel function to protect plants from oxidative damage. Plant Physiol. 2008, 147, 1251–1263. [Google Scholar] [CrossRef] [PubMed]
- Peters, S.; Keller, F. Frost tolerance in excised leaves of the common bugle (Ajuga reptans L.) correlates positively with the concentrations of raffinose family oligosaccharides (RFOs). Plant Cell Environ. 2009, 32, 1099–1107. [Google Scholar] [CrossRef] [PubMed]
- Donahue, J.L.; Alford, S.R.; Torabinejad, J.; Kerwin, R.E.; Nourbakhsh, A.; Ray, W.K.; Hernick, M.; Huang, X.Y.; Lyons, B.M.; Hein, P.P.; et al. The Arabidopsis thaliana myo-inositol 1-phosphate synthase1 gene is required for myo-inositol synthesis and suppression of cell death. Plant Cell 2010, 22, 888–903. [Google Scholar] [CrossRef] [PubMed]
- Patra, B.; Ray, S.; Richter, A.; Majumder, A.L. Enhanced salt tolerance of transgenic tobacco plants by co-expression of PcINO1 and McIMT1 is accompanied by increased level of myo-inositol and methylated inositol. Protoplasma 2010, 245, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Qin, G.J.; Zhang, J.; Liang, Y.; Song, Y.Q.; Zhao, M.P.; Tsuge, T.; Aoyama, T.; Liu, J.J.; Gu, H.Y.; et al. D-myo-Inositol-3-Phosphate affects phosphatidylinositol-mediated endomembrane function in Arabidopsis and is essential for auxin-regulated embryogenesis. Plant Cell 2011, 23, 1352–1372. [Google Scholar] [CrossRef] [PubMed]
- Gillaspy, G.E.; Keddie, J.S.; Oda, K.; Gruissem, W. Plant inositol monophosphatase is a lithium-sensitive enzyme encoded by a multigene family. Plant Cell 1995, 7, 2175–2185. [Google Scholar] [CrossRef] [PubMed]
- Gillaspy, G.E. Signaling and the polyphosphoinositide phosphatases from plants. In Lipid Signaling in Plants; Munnik, T., Ed.; Springer: Berlin/Heidelberg, Germany, 2009; Volume 16, pp. 117–130. [Google Scholar]
- Conklin, P.L.; Gatzek, S.; Wheeler, G.L.; Dowdle, J.; Raymond, M.J.; Rolinski, S.; Isupov, M.; Littlechild, J.A.; Smirnoff, N. Arabidopsis thaliana VTC4 encodes l-galactose-1-P phosphatase, a plant ascorbic acid biosynthetic enzyme. J. Biol. Chem. 2006, 281, 15662–15670. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Peterson, K.; Guttieri, M.; Souza, E.; Raboy, V. Barley (Hordeum vulgare L.) inositol monophosphatase: Gene structure and enzyme characteristics. Plant Mol. Biol. 2008, 67, 629–642. [Google Scholar] [CrossRef] [PubMed]
- Styer, J.C.; Keddie, J.; Spence, J.; Gillaspy, G.E. Genomic organization and regulation of the LeIMP-1 and LeIMP-2 genes encoding myo-inositol monophosphatase in tomato. Gene 2004, 326, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Torabinejad, J.; Donahue, J.L.; Gunesekera, B.N.; Allen-Daniels, M.J.; Gillaspy, G.E. VTC4 is a bifunctional enzyme that affects myo-inositol and ascorbate biosynthesis in plants. Plant Physiol. 2009, 150, 951–961. [Google Scholar] [CrossRef] [PubMed]
- Saxena, S.C.; Salvi, P.; Kaur, H.; Verma, P.; Petla, B.P.; Rao, V.; Kamble, N.; Majee, M. Differentially expressed myo-inositol monophosphatase gene (CaIMP) in chickpea (Cicer arietinum L.) encodes a lithium-sensitive phosphatase enzyme with broad substrate specificity and improves seed germination and seedling growth under abiotic stresses. J. Exp. Bot. 2013, 64, 5623–5639. [Google Scholar] [CrossRef] [PubMed]
- Nourbakhsh, A.; Collakova, E.; Gillaspy, G.E. Characterization of the inositol monophosphatase gene family in Arabidopsis. Front. Plant Sci. 2015, 5, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Derbyshire, M.K.; Gonzales, N.R.; Lu, S.; Chitsaz, F.; Geer, L.Y.; Geer, R.C.; He, J.; Gwadz, M.; Hurwitz, D.I.; et al. CDD: NCBI’s conserved domain database. Nucl. Acids Res. 2015, 43, 222–226. [Google Scholar] [CrossRef] [PubMed]
- Nigou, J.; Dover, L.G.; Besra, G.S. Purification and biochemical characterization of Mycobacterium tuberculosis SuhB, an inositol monophosphatase involved in inositol biosynthesis. Biochemistry 2002, 41, 4392–4398. [Google Scholar] [CrossRef] [PubMed]
- Pollack, S.J.; Knowles, M.R.; Atack, J.R.; Broughton, H.B.; Ragan, C.I.; Osborne, S.; McAllister, G. Probing the role of metal ions in the mechanism of inositol monophosphatase by site-directed mutagenesis. Eur. J. Biochem. 1993, 217, 281–287. [Google Scholar] [CrossRef] [PubMed]
- Arai, R.; Ito, K.; Ohnishi, T.; Ohba, H.; Akasaka, R.; Bessho, Y.; Hanawa-Suetsugu, K.; Yoshikawa, T.; Shirouzu, M.; Yokoyama, S. Crystal structure of human myo-inositol monophosphatase 2, the product of the putative susceptibility gene for bipolar disorder, schizophrenia, and febrile seizures. Proteins 2007, 67, 732–742. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.K.; Meng, G.Y.; Ghadbane, H.; Scott, D.J.; Dover, L.G.; Nigou, J.; Besra, G.S.; Fütterer, K. Dimerization of inositol monophosphatase Mycobacterium tuberculosis SuhB is not constitutive, but induced by binding of the activator Mg2+. BMC Struct. Biol. 2007, 7, 149–162. [Google Scholar] [CrossRef] [PubMed]
- Matsui, A.; Ishida, J.; Morosawa, T.; Mochizuki, Y.; Kaminuma, E.; Endo, T.A.; Okamoto, M.; Nambara, E.; Nakajima, M.; Kawashima, M.; et al. Arabidopsis transcriptome analysis under drought, cold, high-salinity and ABA treatment conditions using a tiling array. Plant Cell Physiol. 2008, 49, 1135–1149. [Google Scholar] [CrossRef] [PubMed]
- Mega, R.; Meguro-Maoka, A.; Endo, A.; Shimosaka, E.; Murayama, S.; Nambara, E.; Seo, M.; Kanno, Y.; Abrams, S.R.; Sato, Y. Sustained low abscisic acid levels increase seedling vigor under cold stress in rice (Oryza sativa L.). Sci. Rep. 2015, 5, 13819–13831. [Google Scholar] [CrossRef] [PubMed]
- Dunn, M.A.; White, A.J.; Vural, S.; Hughes, M.A. Identification of promoter elements in a low-temperature-responsive gene (blt4.9) from barley (Hordeum vulgare L.). Plant Mol. Biol. 1998, 38, 551–564. [Google Scholar] [CrossRef] [PubMed]
- White, T.C.; Simmonds, D.; Donaldson, P.; Singh, J. Regulation of BN115, a low-temperature-responsive gene from winter Brassica napus. Plant Physiol. 1994, 106, 917–928. [Google Scholar] [CrossRef] [PubMed]
- Yabuta, Y.; Motoki, T.; Yoshimura, K.; Takeda, T.; Ishikawa, T.; Shigeoka, S. Thylakoid membrane-bound ascorbate peroxidase is a limiting factor of antioxidative systems under photo-oxidative stress. Plant J. 2002, 32, 915–925. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Sharma, N.D. The molecular biology of the low temperature response in plants. BioEssays 2005, 27, 1048–1059. [Google Scholar] [CrossRef] [PubMed]
- Kanneganti, V.; Gupta, A.K. Overexpression of OsiSAP8, a member of stress associated protein (SAP) gene family of rice confers tolerance to salt, drought and cold stress in transgenic tobacco and rice. Plant Mol. Biol. 2008, 66, 445–462. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Zhang, H.W.; Pan, X.W.; Chen, X.L.; Zhang, Z.J.; Lu, X.Y.; Huang, R.F. Overexpression of ethylene response factor TERF2 confers cold tolerance in rice seedlings. Transgenic Res. 2011, 20, 857–866. [Google Scholar] [CrossRef] [PubMed]
- Xie, G.; Kato, H.; Sasaki, K.; Imai, R. A cold-induced thioredoxin h of rice, OsTrx23, negatively regulates kinase activities of OsMPK3 and OsMPK6 in vitro. FEBS Lett. 2009, 583, 2734–2738. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Guo, Z. Responses of antioxidative system to chilling stress in two rice cultivars differing in sensitivity. Biol. Plant. 2005, 49, 81–84. [Google Scholar] [CrossRef]
- Kang, H.; Saltveit, M.E. Reduced chilling tolerance in elongating cucumber seedling radicles is related to their reduced antioxidant enzyme and DPPH-radical scavenging activity. Physiol. Plant. 2002, 115, 244–250. [Google Scholar] [CrossRef] [PubMed]
- Bonnecarrèrea, V.; Borsanib, O.; Diazb, P.; Capdeviellea, F.; Blancoc, P.; Monzab, J. Response to photoxidative stress induced by cold in japonica rice is genotype dependent. Plant Sci. 2011, 180, 726–732. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Sun, S.J.; Xu, D.Q.; Yang, X.; Bao, Y.M.; Wang, Z.F.; Tang, H.J.; Zhang, H. Increased tolerance of rice to cold, drought and oxidative stresses mediated by the overexpression of a gene that encodes the zinc finger protein ZFP245. Biochem. Biophys. Res. Commun. 2009, 389, 556–561. [Google Scholar] [CrossRef] [PubMed]
- Smirnoff, N.; Cumbes, Q.J. Hydroxyl radical scavenging activity of compatible solutes. Phytochemistry 1989, 28, 1057–1060. [Google Scholar] [CrossRef]
- Shen, B.; Jensen, R.G.; Bohnert, H.J. Increased resistance to oxidative stress in transgenic plants by targeting mannitol biosynthesis to chloroplasts. Plant Physiol. 1997, 113, 1177–1183. [Google Scholar] [CrossRef] [PubMed]
- Adams, P.; Thomas, J.C.; Vernon, D.M.; Bohnert, H.J.; Jensen, R.G. Distinct cellular and organismic responses to salt stress. Plant Cell Physiol. 1992, 33, 1215–1223. [Google Scholar]
- Ishitani, M.; Majumder, A.L.; Bornhouser, A.; Michalowski, C.B.; Jensen, R.G.; Bohnert, H.J. Coordinate transcriptional induction of myo-inositol metabolism during environmental stress. Plant J. 1996, 9, 537–548. [Google Scholar] [CrossRef] [PubMed]
- Cook, D.; Fowler, S.; Fiehn, O.; homashow, M.F. A prominent role for the CBF cold response pathway in configuring the low temperature metabolome of Arabidopsis. Proc. Natl. Acad. Sci. USA 2004, 101, 15243–15248. [Google Scholar] [CrossRef] [PubMed]
- Usadel, B.; Blasing, O.E.; Gibon, Y.; Poree, F.; Hohne, M.; Gunter, M.; Trethewey, R.; Kamlage, B.; Poorter, H.; Stitt, M. Multilevel genomic analysis of the response of transcripts, enzyme activities and metabolites in Arabidopsis rosettes to a progressive decrease of temperature in the non-freezing range. Plant Cell Environ. 2008, 31, 518–547. [Google Scholar] [CrossRef] [PubMed]
- Hincha, D.K. Effects of calcium-induced aggregation on the physical stability of liposomes containing plant glycolipids. Biochim. Biophys. Acta 2003, 1611, 180–186. [Google Scholar] [CrossRef]
- Primer Quest Tool. Available online: http://www.idtdna.com/primerquest/Home/Index (accessed on 16 March 2017).
- Chomczynski, P.; Sacchi, N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987, 162, 156–159. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using Real-Time Quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Horsch, R.B.; Fry, J.E.; Hoffmann, N.L.; Wallroth, M.; Eichholtz, D.; Rogers, S.G.; Fraley, R.T. A simple and general method for transferring genes into plants. Science 1985, 227, 1229–1231. [Google Scholar]
- Lisec, J.; Schauer, N.; Kopka, J.; Willmitzer, L.; Fernie, A.R. Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat. Protoc. 2006, 1, 387–396. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, X.Y.; Guo, J.Z.; Xia, Q.L.; Zhao, G.; Zhou, H.N.; Xi, F.W. Metabolic profiling of Chinese tobacco leaf of different geographical origins by GC-MS. J. Agric. Food Chem. 2013, 61, 2597–2605. [Google Scholar] [CrossRef] [PubMed]
- Roessner, U.; Wagner, C.; Kopka, J.; Trethewey, R.N.; Willmitzer, L. Technical advance: Simultaneous analysis of metabolites in potato tuber by gas chromatography-mass spectrometry. Plant J. 2000, 23, 131–142. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.J.; Zhao, D.; Zhao, D.G. Overexpression of NrCN improved TMV resistance in selection marker-free tobacco generated by Gene-Deletor system. Plant Mol. Biol. Rep. 2015, 33, 1619–1633. [Google Scholar] [CrossRef]
- Hodges, D.M.; Delong, J.M.; Forney, C.F.; Prange, R.K. Improving the thiobarbituric acid-reactive-substances assay for estimating lipid peroxidation in plant tissues containing anthocyanin and other interfering compounds. Planta 1999, 207, 604–611. [Google Scholar] [CrossRef]
- Jana, S.; Choudhuri, M.A. Glycolate metabolism of three submersed aquatic angiosperms during ageing. Aquat. Bot. 1982, 12, 345–354. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]

| Code | Start | Strand | Motif | Function |
|---|---|---|---|---|
| ABRE | 25 | − | TACGGTC | abscisic acid responsiveness |
| Box-W1 | 1190 | + | TTGACC | fungal elicitor responsive element |
| Box-W1 | 1838 | − | TTGACC | fungal elicitor responsive element |
| CAAT-box | 40 | + | CAAAT | promoter and enhancer regions |
| CAT-box | 1149 | + | GGAGATG | part of a light responsive element |
| G-box | 450 | − | CACATGG | light responsiveness |
| GAG-motif | 616 | − | AGAGATG | part of a light responsive element |
| GARE-motif | 273 | + | AAACAGA | gibberellin-responsive responsive element |
| GARE-motif | 1394 | + | TCTGTTG | gibberellin-responsive responsive element |
| HSE | 1085 | − | AAAAAATTTC | heat stress responsiveness |
| LTR | 2000 | + | CCGAAA | low-temperature responsiveness |
| MBS | 382 | − | TAACTG | MYB Binding Site |
| TC-rich repeats | 439 | + | ATTTTATTCA | defense and stress responsiveness |
| TC-rich repeats | 836 | + | ATTTTCTTCA | defense and stress responsiveness |
| TC-rich repeats | 1875 | + | ATTTTCTTCA | defense and stress responsiveness |
| TC-rich repeats | 1060 | + | ATTTTCTTCA | defense and stress responsiveness |
| TC-rich repeats | 2012 | + | ATTTTCTTCA | defense and stress responsiveness |
| TCA-element | 1041 | − | CCATCTTTTT | salicylic acid responsiveness |
| TCA-element | 2013 | − | GAGAAGAAAA | salicylic acid responsiveness |
| TGA-element | 35 | + | AACGAC | auxin-responsive element |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, R.-X.; Qin, L.-J.; Zhao, D.-G. Overexpression of the OsIMP Gene Increases the Accumulation of Inositol and Confers Enhanced Cold Tolerance in Tobacco through Modulation of the Antioxidant Enzymes’ Activities. Genes 2017, 8, 179. https://doi.org/10.3390/genes8070179
Zhang R-X, Qin L-J, Zhao D-G. Overexpression of the OsIMP Gene Increases the Accumulation of Inositol and Confers Enhanced Cold Tolerance in Tobacco through Modulation of the Antioxidant Enzymes’ Activities. Genes. 2017; 8(7):179. https://doi.org/10.3390/genes8070179
Chicago/Turabian StyleZhang, Rong-Xiang, Li-Jun Qin, and De-Gang Zhao. 2017. "Overexpression of the OsIMP Gene Increases the Accumulation of Inositol and Confers Enhanced Cold Tolerance in Tobacco through Modulation of the Antioxidant Enzymes’ Activities" Genes 8, no. 7: 179. https://doi.org/10.3390/genes8070179





