Viral Infection Identifies Micropeptides Differentially Regulated in smORF-Containing lncRNAs
Abstract
:1. Introduction
2. Materials and Methods
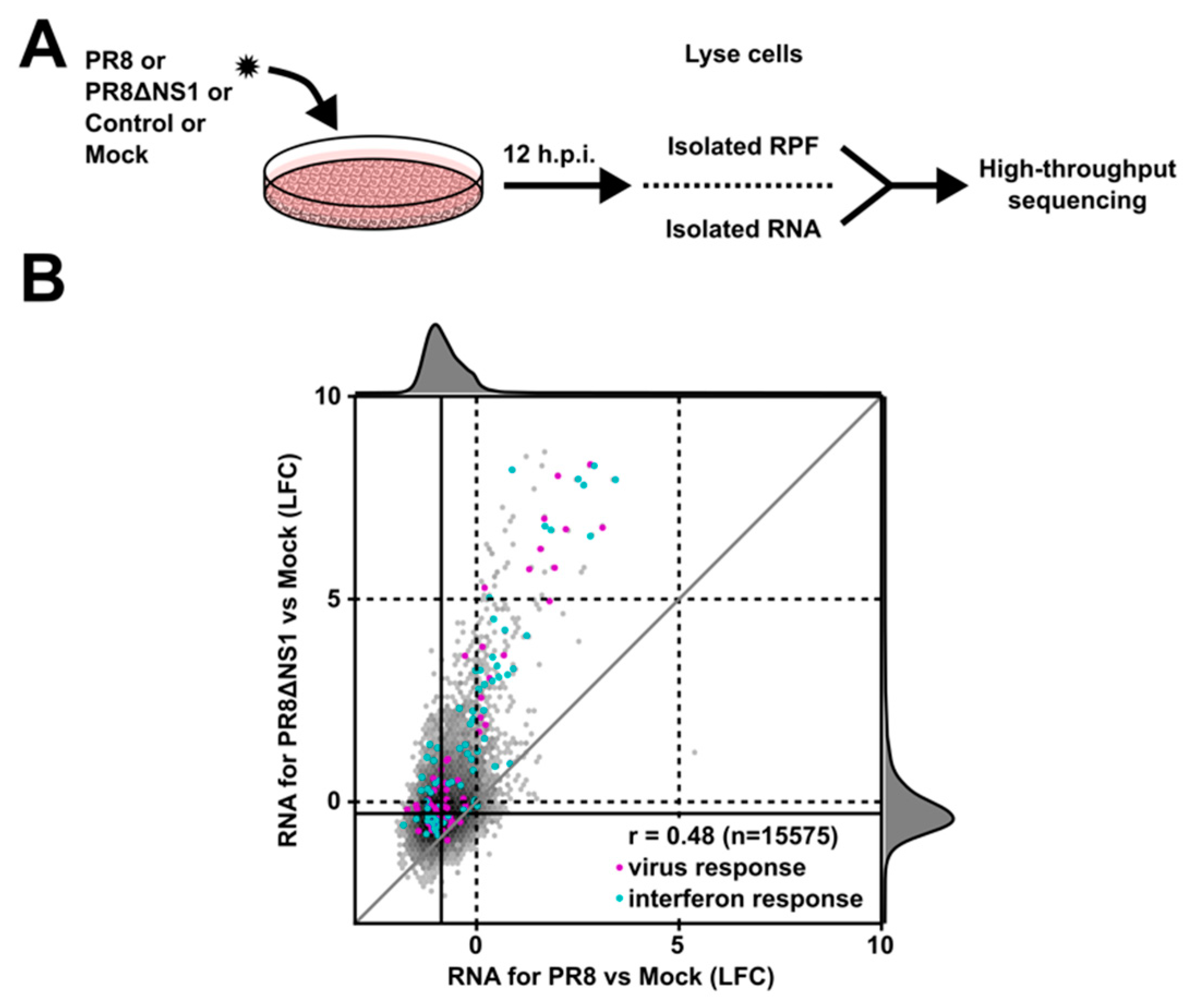
2.1. Ribosome Profiling and RNA-seq Library Preparation
2.2. High-Throughput Sequencing Analysis
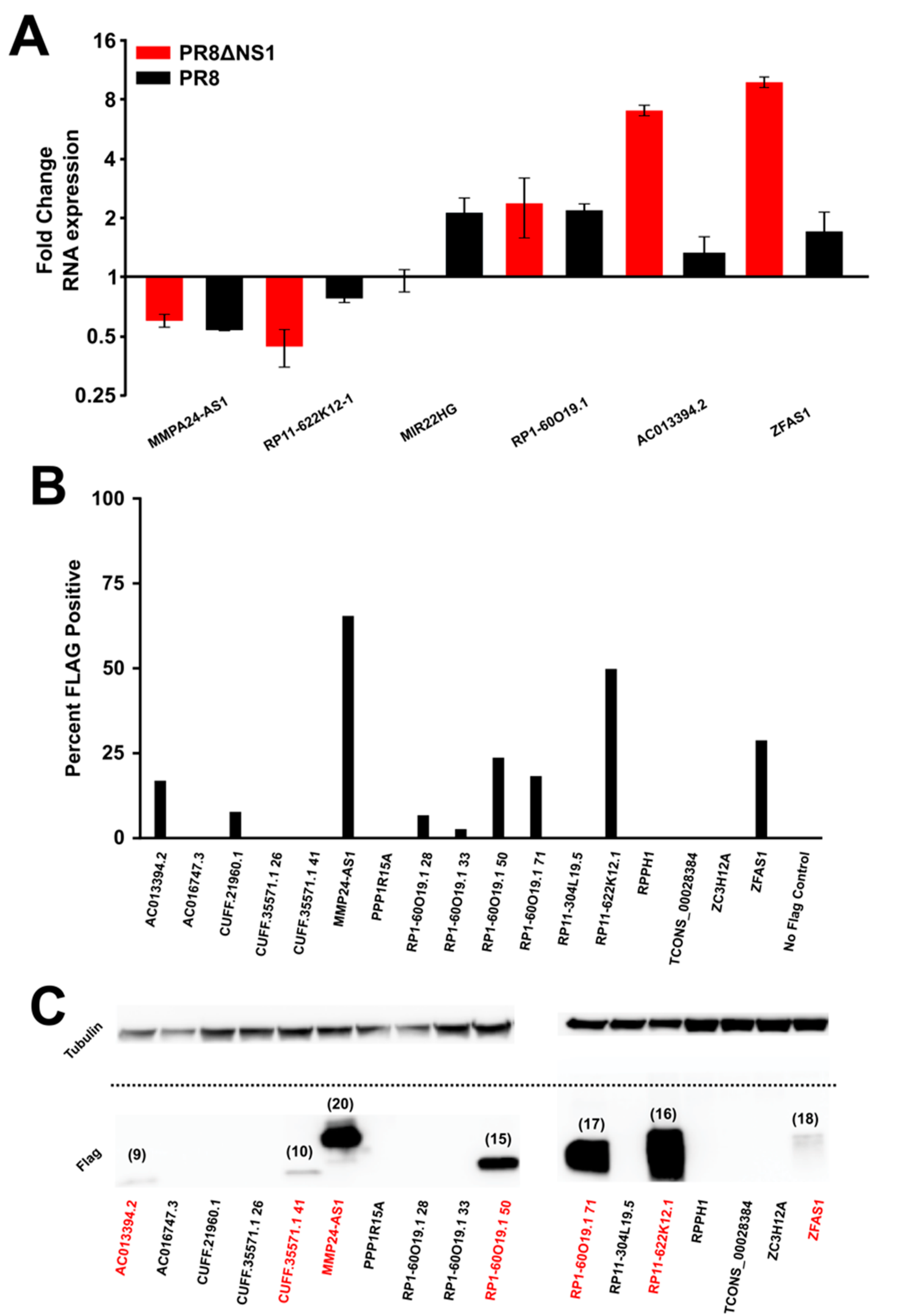
2.3. Quantitative PCR Methods
2.4. Cell and Viral Culture
2.5. Cloning
2.6. Flow Cytometry
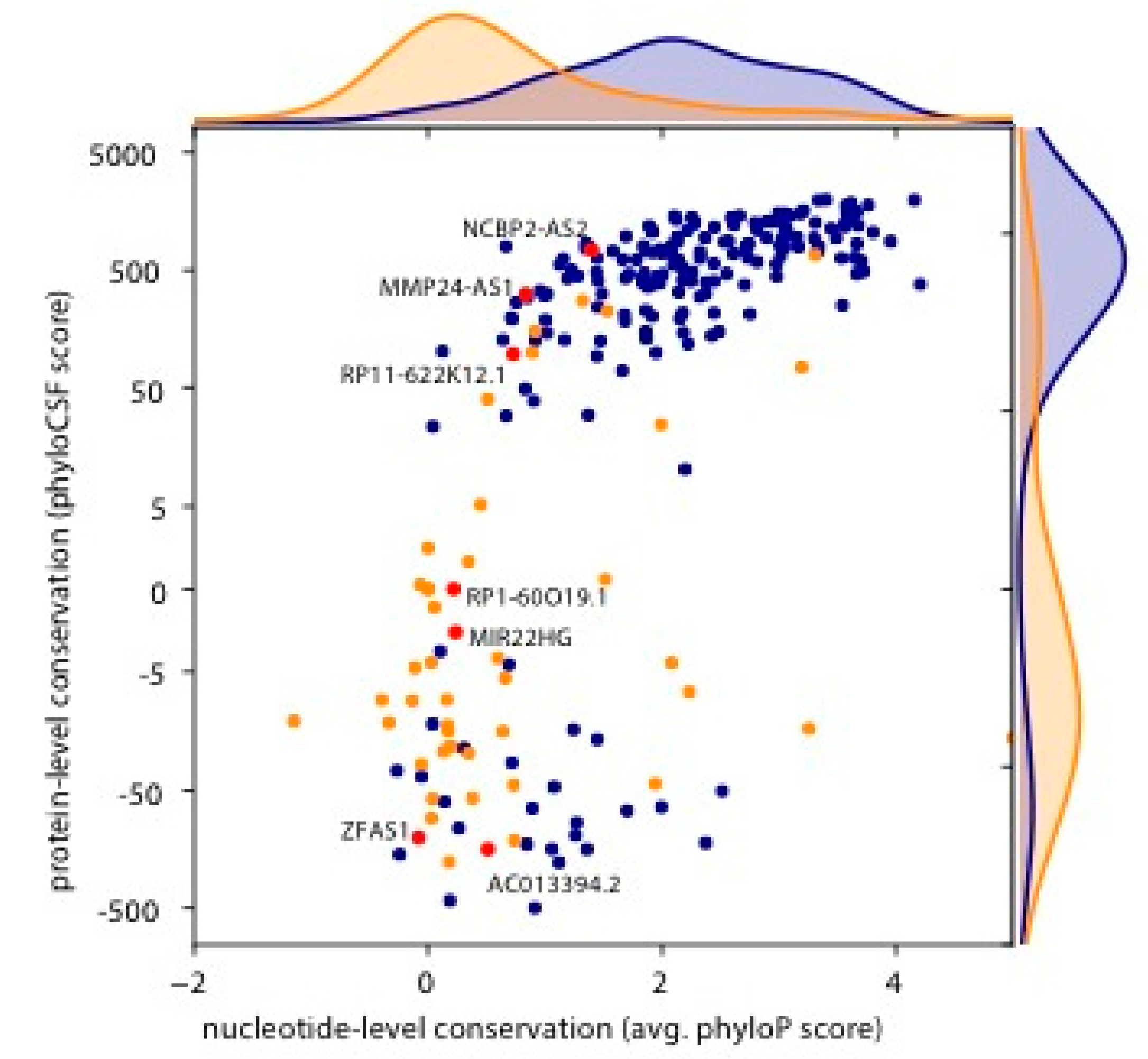
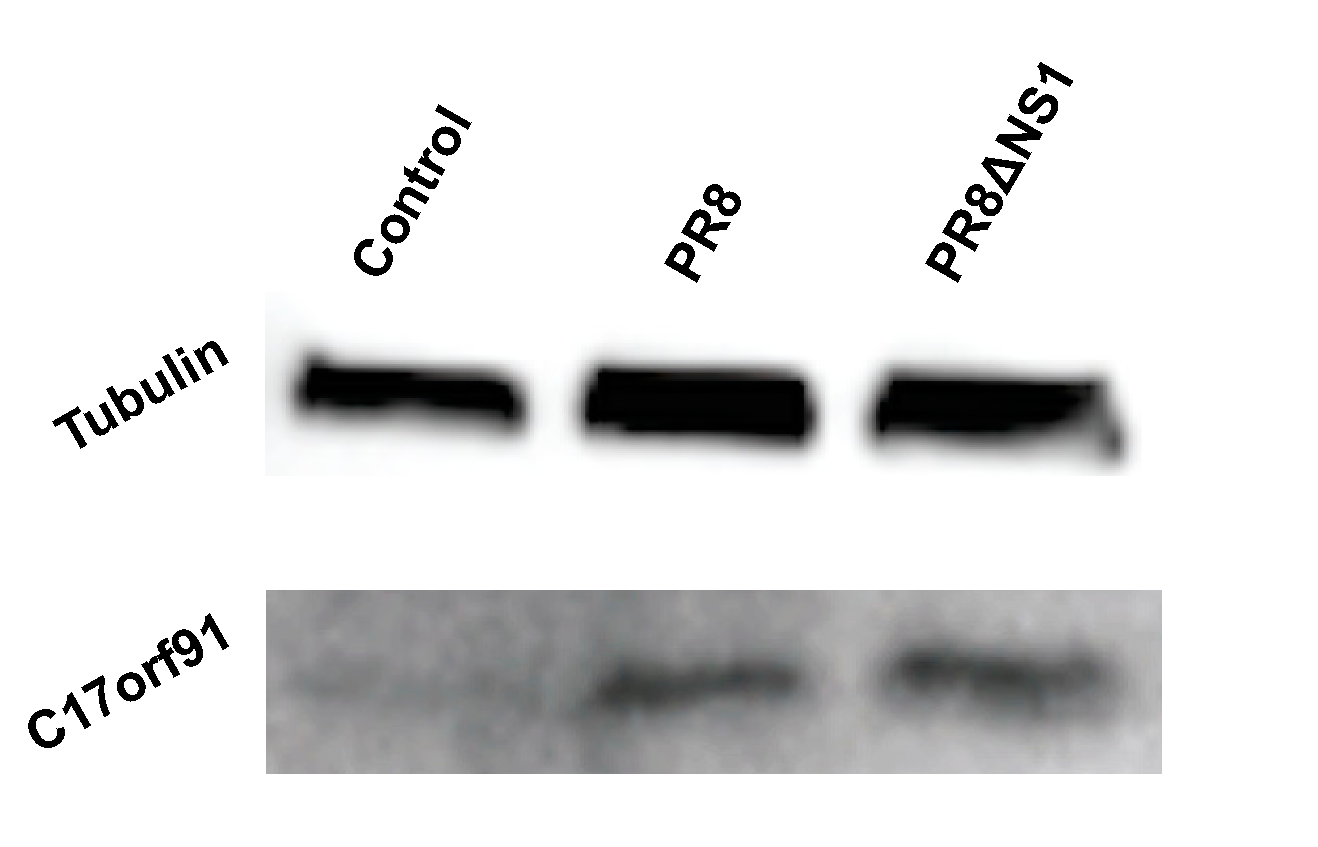
3. Results
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Van Valen, L. A new evolutionary law. Evolut. Theory 1973, 1, 1–30. [Google Scholar]
- Bieniasz, P.D. Intrinsic immunity: A front-line defense against viral attack. Nat. Immunol. 2004, 5, 1109–1115. [Google Scholar] [CrossRef] [PubMed]
- Hoebe, K.; Janssen, E.; Beutler, B. The interface between innate and adaptive immunity. Nat. Immunol. 2004, 5, 971–974. [Google Scholar] [CrossRef] [PubMed]
- Janeway, C.A. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb. Symp. Quant. Biol. 1989, 54 Pt 1, 1–13. [Google Scholar] [CrossRef]
- Satpathy, A.T.; Chang, H.Y. Long noncoding RNA in hematopoiesis and immunity. Immunity 2015, 42, 792–804. [Google Scholar] [CrossRef] [PubMed]
- Yu, A.D.; Wang, Z.; Morris, K.V. Long noncoding RNAs: A potent source of regulation in immunity and disease. Immunol. Cell Biol. 2015, 93, 277–283. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, S.; Aiello, D.; Atianand, M.K.; Ricci, E.P.; Gandhi, P.; Hall, L.L.; Byron, M.; Monks, B.; Henry-Bezy, M.; Lawrence, J.B.; et al. A long noncoding RNA mediates both activation and repression of immune response genes. Science 2013, 341, 789–792. [Google Scholar] [CrossRef] [PubMed]
- Heward, J.A.; Lindsay, M.A. Long non-coding RNAs in the regulation of the immune response. Trends Immunol. 2014, 35, 408–419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, X.; Gralinski, L.; Armour, C.D.; Ferris, M.T.; Thomas, M.J.; Proll, S.; Bradel-Tretheway, B.G.; Korth, M.J.; Castle, J.C.; Biery, M.C.; et al. Unique signatures of long noncoding RNA expression in response to virus infection and altered innate immune signaling. mBio 2010, 1, e00206:10–e00206:18. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Zhang, Y.; Li, T.; Ma, Z.; Jia, H.; Chen, Q.; Zhao, Y.; Zhai, L.; Zhong, R.; Li, C.; et al. Long non-coding RNA Linc-RAM enhances myogenic differentiation by interacting with MyoD. Nat. Commun. 2017, 8, 14016. [Google Scholar] [CrossRef] [PubMed]
- Ulitsky, I.; Bartel, D.P. lincRNAs: Genomics, evolution, and mechanisms. Cell 2013, 154, 26–46. [Google Scholar] [CrossRef] [PubMed]
- Aspden, J.L.; Eyre-Walker, Y.C.; Phillips, R.J.; Amin, U.; Mumtaz, M.A.S.; Brocard, M.; Couso, J.-P. Extensive translation of small Open Reading Frames revealed by Poly-Ribo-Seq. Elife 2014, 3, e03528. [Google Scholar] [CrossRef] [PubMed]
- Bazzini, A.A.; Johnstone, T.G.; Christiano, R.; Mackowiak, S.D.; Obermayer, B.; Fleming, E.S.; Vejnar, C.E.; Lee, M.T.; Rajewsky, N.; Walther, T.C.; et al. Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J. 2014, 33, 981–993. [Google Scholar] [CrossRef] [PubMed]
- Chew, G.-L.; Pauli, A.; Rinn, J.L.; Regev, A.; Schier, A.F.; Valen, E. Ribosome profiling reveals resemblance between long non-coding RNAs and 5’ leaders of coding RNAs. Development 2013, 140, 2828–2834. [Google Scholar] [CrossRef] [PubMed]
- Olexiouk, V.; Crappé, J.; Verbruggen, S.; Verhegen, K.; Martens, L.; Menschaert, G. sORFs.org: A repository of small ORFs identified by ribosome profiling. Nucleic Acids Res. 2016, 44, D324–D329. [Google Scholar] [PubMed]
- Ingolia, N.T.; Ghaemmaghami, S.; Newman, J.R.S.; Weissman, J.S. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 2009, 324, 218–223. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Diedrich, J.K.; Jungreis, I.; Donaldson, C.; Vaughan, J.; Kellis, M.; Yates, J.R.; Saghatelian, A. Improved Identification and Analysis of Small Open Reading Frame Encoded Polypeptides. Anal. Chem. 2016, 88, 3967–3975. [Google Scholar] [CrossRef] [PubMed]
- Magny, E.G.; Pueyo, J.I.; Pearl, F.M.G.; Cespedes, M.A.; Niven, J.E.; Bishop, S.A.; Couso, J.-P. Conserved regulation of cardiac calcium uptake by peptides encoded in small open reading frames. Science 2013, 341, 1116–1120. [Google Scholar] [CrossRef] [PubMed]
- Nelson, B.R.; Makarewich, C.A.; Anderson, D.M.; Winders, B.R.; Troupes, C.D.; Wu, F.; Reese, A.L.; McAnally, J.R.; Chen, X.; Kavalali, E.T.; et al. A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science 2016, 351, 271–275. [Google Scholar] [CrossRef] [PubMed]
- Anderson, D.M.; Anderson, K.M.; Chang, C.-L.; Makarewich, C.A.; Nelson, B.R.; McAnally, J.R.; Kasaragod, P.; Shelton, J.M.; Liou, J.; Bassel-Duby, R.; et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell 2015, 160, 595–606. [Google Scholar] [CrossRef] [PubMed]
- Crappé, J.; Van Criekinge, W.; Trooskens, G.; Hayakawa, E.; Luyten, W.; Baggerman, G.; Menschaert, G. Combining in silico prediction and ribosome profiling in a genome-wide search for novel putatively coding sORFs. BMC Genom. 2013, 14, 648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsumoto, A.; Pasut, A.; Matsumoto, M.; Yamashita, R.; Fung, J.; Monteleone, E.; Saghatelian, A.; Nakayama, K.I.; Clohessy, J.G.; Pandolfi, P.P. mTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide. Nature 2017, 541, 228–232. [Google Scholar] [CrossRef] [PubMed]
- Talon, J.; Horvath, C.M.; Polley, R.; Basler, C.F.; Muster, T.; Palese, P.; García-Sastre, A. Activation of interferon regulatory factor 3 is inhibited by the influenza A virus NS1 protein. J. Virol. 2000, 74, 7989–7996. [Google Scholar] [CrossRef] [PubMed]
- Dodt, M.; Roehr, J.T.; Ahmed, R.; Dieterich, C. FLEXBAR-Flexible Barcode and Adapter Processing for Next-Generation Sequencing Platforms. Biology 2012, 1, 895–905. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016, 44, D733–D745. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Pollard, K.S.; Hubisz, M.J.; Rosenbloom, K.R.; Siepel, A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010, 20, 110–121. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.F.; Jungreis, I.; Kellis, M. PhyloCSF: A comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics 2011, 27, i275–i282. [Google Scholar] [CrossRef] [PubMed]
- Mackowiak, S.D.; Zauber, H.; Bielow, C.; Thiel, D.; Kutz, K.; Calviello, L.; Mastrobuoni, G.; Rajewsky, N.; Kempa, S.; Selbach, M.; Obermayer, B. Extensive identification and analysis of conserved small ORFs in animals. Genome Biol. 2015, 16, 179. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Tarasov, A.; Vilella, A.J.; Cuppen, E.; Nijman, I.J.; Prins, P. Sambamba: Fast processing of NGS alignment formats. Bioinformatics 2015, 31, 2032–2034. [Google Scholar] [CrossRef] [PubMed]
- Marazzi, I.; Ho, J.S.Y.; Kim, J.; Manicassamy, B.; Dewell, S.; Albrecht, R.A.; Seibert, C.W.; Schaefer, U.; Jeffrey, K.L.; Prinjha, R.K.; et al. Suppression of the antiviral response by an influenza histone mimic. Nature 2012, 483, 428–433. [Google Scholar] [CrossRef] [PubMed]
- Bercovich-Kinori, A.; Tai, J.; Gelbart, I.A.; Shitrit, A.; Ben-Moshe, S.; Drori, Y.; Itzkovitz, S.; Mandelboim, M.; Stern-Ginossar, N. A systematic view on influenza induced host shutoff. Elife 2016, 5, 600. [Google Scholar] [CrossRef] [PubMed]
- Tirosh, O.; Cohen, Y.; Shitrit, A.; Shani, O.; Le-Trilling, V.T.K.; Trilling, M.; Friedlander, G.; Tanenbaum, M.; Stern-Ginossar, N. The Transcription and Translation Landscapes during Human Cytomegalovirus Infection Reveal Novel Host-Pathogen Interactions. PLoS Pathog. 2015, 11, e1005288. [Google Scholar] [CrossRef] [PubMed]
- D’Lima, N.G.; Ma, J.; Winkler, L.; Chu, Q.; Loh, K.H.; Corpuz, E.O.; Budnik, B.A.; Lykke-Andersen, J.; Saghatelian, A.; Slavoff, S.A. A human microprotein that interacts with the mRNA decapping complex. Nat. Chem. Biol. 2017, 13, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Tani, H.; Torimura, M. Identification of short-lived long non-coding RNAs as surrogate indicators for chemical stress response. Biochem. Biophys. Res. Commun. 2013, 439, 547–551. [Google Scholar] [CrossRef] [PubMed]
- Tani, H.; Onuma, Y.; Ito, Y.; Torimura, M. Long non-coding RNAs as surrogate indicators for chemical stress responses in human-induced pluripotent stem cells. PLoS ONE 2014, 9, e106282. [Google Scholar] [CrossRef] [PubMed]
- Slavoff, S.A.; Mitchell, A.J.; Schwaid, A.G.; Cabili, M.N.; Ma, J.; Levin, J.Z.; Karger, A.D.; Budnik, B.A.; Rinn, J.L.; Saghatelian, A. Peptidomic discovery of short open reading frame-encoded peptides in human cells. Nat. Chem. Biol. 2013, 9, 59–64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Q.; Vashisht, A.A.; O’Rourke, J.; Corbel, S.Y.; Moran, R.; Romero, A.; Miraglia, L.; Zhang, J.; Durrant, E.; Schmedt, C.; et al. The microprotein Minion controls cell fusion and muscle formation. Nat. Commun. 2017, 8, 15664. [Google Scholar] [CrossRef] [PubMed]
- Couso, J.-P.; Patraquim, P. Classification and function of small open reading frames. Nat. Rev. Mol. Cell Biol. 2017, 17, 669. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Razooky, B.S.; Obermayer, B.; O’May, J.B.; Tarakhovsky, A. Viral Infection Identifies Micropeptides Differentially Regulated in smORF-Containing lncRNAs. Genes 2017, 8, 206. https://doi.org/10.3390/genes8080206
Razooky BS, Obermayer B, O’May JB, Tarakhovsky A. Viral Infection Identifies Micropeptides Differentially Regulated in smORF-Containing lncRNAs. Genes. 2017; 8(8):206. https://doi.org/10.3390/genes8080206
Chicago/Turabian StyleRazooky, Brandon S., Benedikt Obermayer, Joshua Biggs O’May, and Alexander Tarakhovsky. 2017. "Viral Infection Identifies Micropeptides Differentially Regulated in smORF-Containing lncRNAs" Genes 8, no. 8: 206. https://doi.org/10.3390/genes8080206




