Transcriptomic Studies of the Effect of nod Gene-Inducing Molecules in Rhizobia: Different Weapons, One Purpose
Abstract
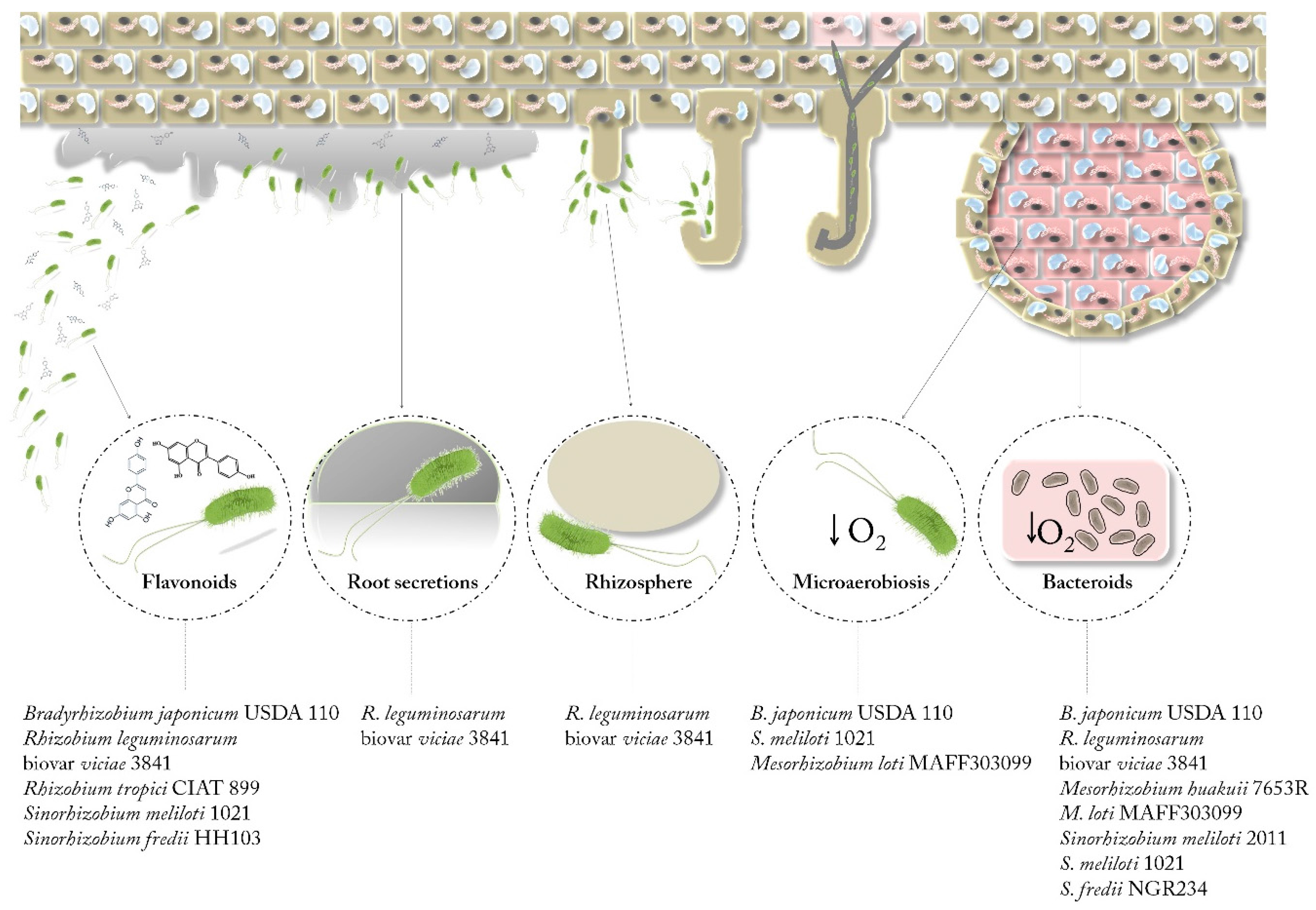
:1. Introduction
2. Rhizobial Transcriptomic Studies Related with Symbiosis
2.1. Bacteroid and Microaerobiosis Conditions
2.2. Rhizosphere and Exudates Conditions
3. Gene Expression Changes under nod Gene-Inducing Molecules: The Arsenal of Weapons
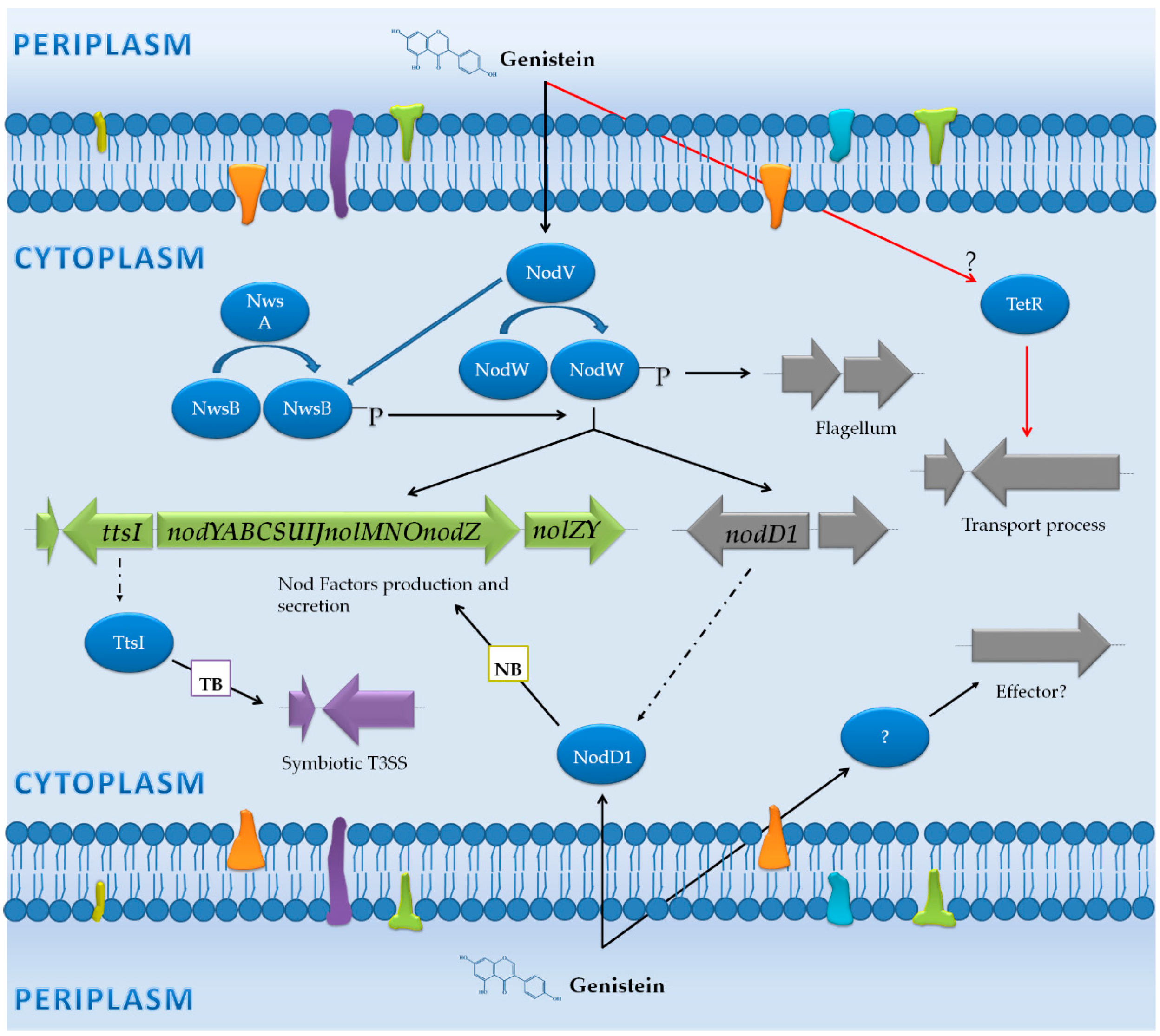
3.1. Bradyrhizobium Japonicum USDA 110: Diversification in the Activation Pathways for NF Production, Assembly of the T3SS and the Flagellum
3.1.1. Genes Controlled by NB
3.1.2. Genes Controlled by TB
3.1.3. Genes Not Preceded by a NB or a TB
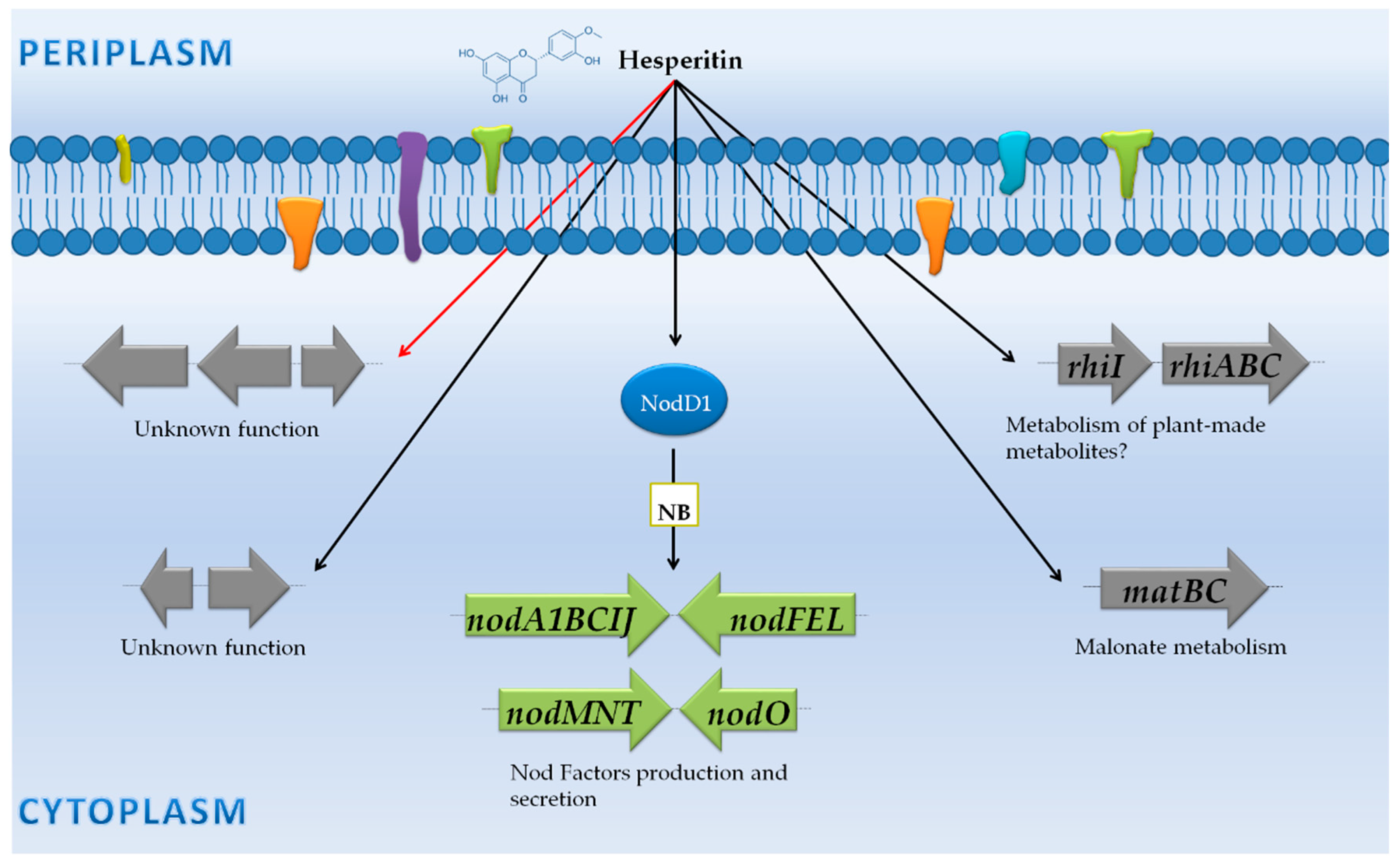
3.2. Rhizobium Leguminosarum Biovar Viciae 3841: NF Synthesis and Adaptation to the Rhizosphere Environment
3.2.1. Genes Controlled by NB
3.2.2. Genes Not Preceded by a NB
3.3. Rhizobium Tropici CIAT 899: NF Synthesis Also under Stressing Conditions, Production of a Large Variety of NF and Phytohormones
3.3.1. Genes Controlled by NB
3.3.2. Genes Not Preceded by a NB
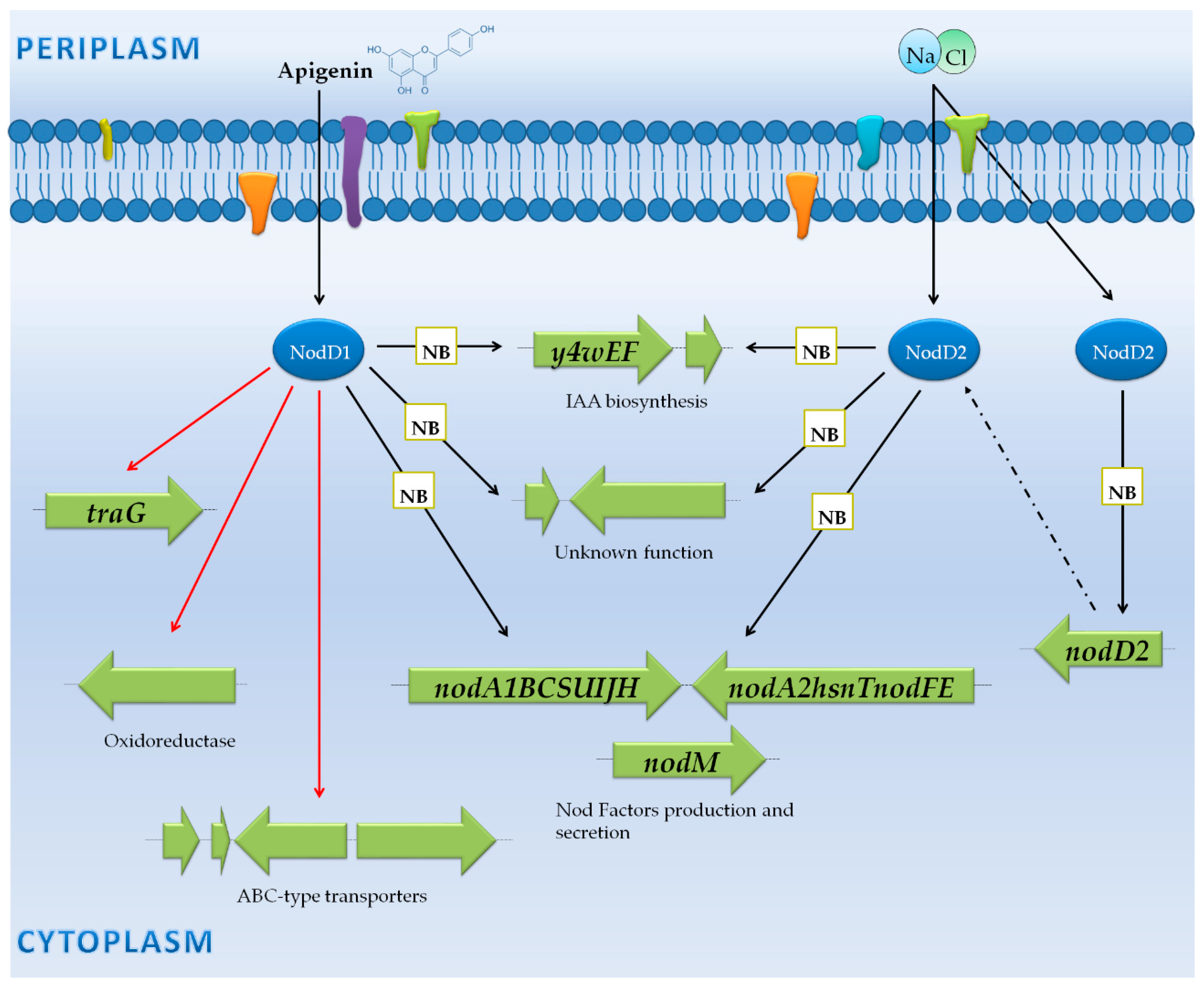
3.4. Sinorhizobium Meliloti 1021: Only Production and Export of NF?
3.4.1. Genes Controlled by NB via NodD1
3.4.2. Genes Not Preceded by a NB
3.5. Sinorhizobium Fredii HH103: A Hierarchical Regulatory Cascade Controls NF Production and Export, Biosynthesis of Phytohormones, and Assembly of the T3SS
3.5.1. Genes Controlled by NB via NodD1
3.5.2. Genes Controlled by TB via TtsI
3.5.3. Genes Not Preceded by a NB or a TB
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Ekins, R.; Chu, F.W. Microarrays: Their origins and applications. Trends Biotechnol. 1999, 17, 217–218. [Google Scholar] [CrossRef]
- Marguerat, S.; Bahler, J. RNA-seq: From technology to biology. Cell. Mol. Life Sci. 2010, 67, 569–579. [Google Scholar] [CrossRef] [PubMed]
- Ramsay, G. DNA chips: State-of-the art. Nat. Biotechnol. 1998, 16, 40–44. [Google Scholar] [CrossRef] [PubMed]
- Hegde, P.; Qi, R.; Abernathy, K.; Gay, C.; Dharap, S.; Gaspard, R.; Hughes, J.E.; Snesrud, E.; Lee, N.; Quackenbush, J. A concise guide to cDNA microarray analysis. Biotechniques 2000, 29, 548–556. [Google Scholar] [PubMed]
- Schena, M.; Shalon, D.; Davis, R.W.; Brown, P.O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 1995, 270, 467–470. [Google Scholar] [CrossRef] [PubMed]
- Xiang, C.C.; Chen, Y. cDNA microarray technology and its applications. Biotechnol. Adv. 2000, 18, 35–46. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Schlüter, J.P.; Reinkensmeier, J.; Daschkey, S.; Evguenieva-Hackenberg, E.; Janssen, S.; Jänicke, S.; Becker, J.D.; Giegerich, R.; Becker, A. A genome-wide survey of sRNAs in the symbiotic nitrogen-fixing alpha-proteobacterium Sinorhizobium meliloti. BMC Genom. 2010, 11, 245. [Google Scholar] [CrossRef] [PubMed]
- Sallet, E.; Roux, B.; Sauviac, L.; Jardinaud, M.F.; Carrère, S.; Faraut, T.; de Carvalho-Niebel, F.; Gouzy, J.; Gamas, P.; Capela, D.; et al. Next-generation annotation of prokaryotic genomes with EuGene-P: Application to Sinorhizobium meliloti 2011. DNA Res. 2013, 20, 339–354. [Google Scholar] [CrossRef] [PubMed]
- Kogenaru, S.; Qing, Y.; Guo, Y.; Wang, N. RNA-seq and microarray complement each other in transcriptome profiling. BMC Genom. 2012, 15, 629. [Google Scholar] [CrossRef] [PubMed]
- Oke, V.; Long, S.R. Bacterial genes induced within the nodule during the Rhizobium-legume symbiosis. Mol. Microbiol. 1999, 32, 837–850. [Google Scholar] [CrossRef] [PubMed]
- Kondorosi, E.; Mergaert, P.; Kereszt, A. A paradigm for endosymbiotic life: Cell differentiation of Rhizobium bacteria provoked by host plant factors. Annu. Rev. Microbiol. 2013, 67, 611–628. [Google Scholar] [CrossRef] [PubMed]
- Long, S.R. Genes and signals in the Rhizobium-legume symbiosis. Plant Physiol. 2001, 125, 69–72. [Google Scholar] [CrossRef] [PubMed]
- Prell, J.; Poole, P. Metabolic changes of rhizobia in legume nodules. Trends Microbiol. 2006, 14, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Suzaki, T.; Kawaguchi, M. Root nodulation: A developmental program involving cell fate conversion triggered by symbiotic bacterial infection. Curr. Opin. Plant Biol. 2014, 21, 16–22. [Google Scholar] [CrossRef] [PubMed]
- Soupène, E.; Foussard, M.; Boistard, P.; Truchet, G.; Batut, J. Oxygen as a key developmental regulator of Rhizobium meliloti N2-fixation gene expression within the alfalfa root nodule. Proc. Natl. Acad. Sci. USA 1995, 92, 3759–3763. [Google Scholar] [CrossRef] [PubMed]
- Becker, A.; Bergès, H.; Krol, E.; Bruand, C.; Rüberg, S.; Capela, D.; Lauber, E.; Meilhoc, E.; Ampe, F.; de Bruijn, F.J.; et al. Global changes in gene expression in Sinorhizobium meliloti 1021 under microoxic and symbiotic conditions. Mol. Plant Microbe Interact. 2004, 17, 292–303. [Google Scholar] [CrossRef] [PubMed]
- Pessi, G.; Ahrens, C.H.; Rehrauer, H.; Lindemann, A.; Hauser, F.; Fischer, H.S.; Hennecke, H. Genome-wide transcript analysis of Bradyrhizobium japonicum bacteroids in soybean root nodules. Mol. Plant Microbe Interact. 2007, 20, 1353–1363. [Google Scholar] [CrossRef] [PubMed]
- Uchiumi, T.; Ohwada, T.; Itakura, M.; Mitsui, H.; Nukui, N.; Dawadi, P.; Kaneko, T.; Tabata, S.; Yokoyama, T.; Tejima, K.; et al. Expression islands clustered on the symbiosis island of the Mesorhizobium loti genome. J. Bacteriol. 2004, 186, 2439–2448. [Google Scholar] [CrossRef] [PubMed]
- Cooper, J.E. Multiple responses of rhizobia to flavonoids during legume root infection. Adv. Bot. Res. 2004, 41, 1–62. [Google Scholar]
- Gough, C. Rhizobium symbiosis: Insight into Nod factor receptors. Curr. Biol. 2003, 13, R973–R975. [Google Scholar] [CrossRef] [PubMed]
- D’Haeze, W.; Holsters, M. Nod factor structures, responses, and perception during initiation of nodule development. Glycobiology 2002, 12, 79R–105R. [Google Scholar] [CrossRef] [PubMed]
- Broughton, W.J.; Jabbouri, S.; Perret, X. Keys to symbiotic harmony. J. Bacteriol. 2000, 182, 5641–5652. [Google Scholar] [CrossRef] [PubMed]
- Oldroyd, G.E. Speak, friend, and enter: Signaling systems that promote beneficial symbiotic associations in plants. Nat. Rev. Microbiol. 2013, 11, 252–263. [Google Scholar] [CrossRef] [PubMed]
- López-Baena, F.J.; Vinardell, J.M.; Pérez-Montaño, F.; Crespo-Rivas, J.C.; Bellogín, R.A.; Espuny, M.R.; Ollero, F.J. Regulation and symbiotic significance of nodulation outer proteins secretion in Sinorhizobium fredii HH103. Microbiology 2008, 154, 1825–1836. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pérez-Montaño, F.; Guasch-Vidal, B.; González-Barroso, S.; López-Baena, F.J.; Cubo, T.; Ollero, F.J.; Gil-Serrano, A.M.; Bellogín, R.A.; Espuny, M.R. Nodulation-gene-inducing flavonoids increase overall production of autoinducers and expression of N-acyl homoserine lactone synthesis genes in rhizobia. Res. Microbiol. 2011, 162, 715–723. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Montaño, F.; Jiménez-Guerrero, I.; Del Cerro, P.; Baena-Ropero, I.; López-Baena, F.J.; Ollero, F.J.; Bellogín, R.A.; Lloret, J.; Espuny, M.R. The Symbiotic biofilm of Sinorhizobium fredii SMH12, necessary for successful colonization and symbiosis of Glycine max cv Osumi, is regulated by quorum sensing systems and inducing flavonoids via NodD1. PLoS ONE 2014, 9, e105901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Theunis, M.; Kobayashi, H.; Broughton, W.J.; Prinsen, E. Flavonoids, NodD1, NodD2, and nod-box NB15 modulate expression of the y4wEFG locus that is required for indole-3-acetic acid synthesis in Rhizobium sp. strain NGR234. Mol. Plant Microbe Interact. 2004, 17, 1153–1161. [Google Scholar] [CrossRef] [PubMed]
- Vinardell, J.M.; Ollero, F.J.; Hidalgo, A.; López-Baena, F.J.; Medina, C.; Ivanov-Vangelov, K.; Parada, M.; Madinabeitia, N.; Espuny, M.R.; Bellogín, R.A.; et al. NolR regulates diverse symbiotic signals of Sinorhizobium fredii HH103. Mol. Plant Microbe Interact. 2004, 17, 676–685. [Google Scholar] [CrossRef] [PubMed]
- Acosta-Jurado, S.; Navarro-Gómez, P.; Murdoch, P.S.; Crespo-Rivas, J.C.; Jie, S.; Cuesta-Berrio, L.; Ruiz-Sainz, J.E.; Rodríguez-Carvajal, M.Á.; Vinardell, J.M. Exopolysaccharide production by Sinorhizobium fredii HH103 is repressed by genistein in a NodD1-dependent manner. PLoS ONE 2016, 11, e0160499. [Google Scholar] [CrossRef] [PubMed]
- Ardissone, S.; Noel, K.D.; Klement, M.; Broughton, W.J.; Deakin, W.J. Synthesis of the flavonoid-induced lipopolysaccharide of Rhizobium sp. strain NGR234 requires rhamnosyl transferases encoded by genes rgpF and wbgA. Mol. Plant Microbe Interact. 2011, 24, 1513–1521. [Google Scholar] [CrossRef] [PubMed]
- Chang, W.S.; Franck, W.L.; Cytryn, E.; Jeong, S.; Joshi, T.; Emerich, D.W.; Sadowsky, M.J.; Xu, D.; Stacey, G. An oligonucleotide microarray resource for transcriptional profiling of Bradyrhizobium japonicum. Mol. Plant Microbe Interact. 2007, 20, 1298–1307. [Google Scholar] [CrossRef] [PubMed]
- Karunakaran, R.; Ramachandran, V.K.; Seaman, J.C.; East, A.K.; Mouhsine, B.; Mauchline, T.H.; Prell, J.; Skeffington, A.; Poole, P.S. Transcriptomic analysis of Rhizobium leguminosarum biovar viciae in symbiosis with host plants Pisum sativum and Vicia cracca. J. Bacteriol. 2009, 191, 4002–4014. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Hao, B.; Liu, L.; Wang, S.; Ma, B.; Yang, Y.; Xie, F.; Li, Y. RNA-Seq and microarrays analyses reveal global differential transcriptomes of Mesorhizobium huakuii 7653R between bacteroids and free-living cells. PLoS ONE 2014, 9, e93626. [Google Scholar] [CrossRef] [PubMed]
- Ampe, F.; Kiss, E.; Sabourdy, F.; Batut, J. Transcriptome analysis of Sinorhizobium meliloti during symbiosis. Genome Biol. 2003, 4, R15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Capela, D.; Filipe, C.; Bobik, C.; Batut, J.; Bruand, C. Sinorhizobium meliloti differentiation during symbiosis with alfalfa: A transcriptomic dissection. Mol. Plant Microbe Interact. 2007, 19, 363–372. [Google Scholar] [CrossRef] [PubMed]
- Barnett, M.J.; Toman, C.J.; Fisher, R.F.; Long, S.R. A dual-genome Symbiosis Chip for coordinate study of signal exchange and development in a prokaryote-host interaction. Proc. Natl. Acad. Sci. USA 2004, 101, 16636–16641. [Google Scholar] [CrossRef] [PubMed]
- Bobik, C.; Meilhoc, E.; Batut, J. FixJ: A major regulator of the oxygen limitation response and late symbiotic functions of Sinorhizobium meliloti. J. Bacteriol. 2006, 188, 4890–4902. [Google Scholar] [CrossRef] [PubMed]
- Roux, B.; Rodde, N.; Jardinaud, M.F.; Timmers, T.; Sauviac, L.; Cottret, L.; Carrère, S.; Sallet, E.; Courcelle, E.; Moreau, S.; et al. An integrated analysis of plant and bacterial gene expression in symbiotic root nodules using laser-capture microdissection coupled to RNA sequencing. Plant J. 2014, 77, 817–837. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Tian, C.F.; Chen, W.F.; Wang, L.; Sui, X.H.; Chen, W.X. High-resolution transcriptomic analyses of Sinorhizobium sp. NGR234 bacteroids in determinate nodules of Vigna unguiculata and indeterminate nodules of Leucaena leucocephala. PLoS ONE 2013, 8, e70531. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, V.K.; East, A.K.; Karunakaran, R.; Downie, J.A.; Poole, P.S. Adaptation of Rhizobium leguminosarum to pea, alfalfa and sugar beet rhizospheres investigated by comparative transcriptomics. Genome Biol. 2011, 12, R106. [Google Scholar] [CrossRef] [PubMed]
- Lang, K.; Lindemann, A.; Hauser, F.; Göttfert, M. The genistein stimulon of Bradyrhizobium japonicum. Mol. Genet. Genom. 2008, 279, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Montaño, F.; del Cerro, P.; Jiménez-Guerrero, I.; López-Baena, F.J.; Cubo, M.T.; Hungria, M.; Ollero, F.J. RNA-seq analysis of the Rhizobium tropici CIAT 899 transcriptome shows similarities in the activation patterns of symbiotic genes in the presence of apigenin and salt. BMC Genom. 2016, 17, 198. [Google Scholar] [CrossRef] [PubMed]
- Del Cerro, P.; Pérez-Montaño, F.; Gil-Serrano, A.; López-Baena, F.J.; Megías, M.; Hungria, M.; Ollero, F.J. The Rhizobium tropici CIAT 899 NodD2 protein regulates the production of Nod factors under salt stress in a flavonoid-independent manner. Sci. Rep. 2017, 7, 46712. [Google Scholar] [CrossRef] [PubMed]
- Capela, D.; Carrere, S.; Batut, J. Transcriptome-based identification of the Sinorhizobium meliloti NodD1 regulon. Appl. Environ. Microbiol. 2005, 71, 4910–4913. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Montaño, F.; Jiménez-Guerrero, I.; Acosta-Jurado, S.; Navarro-Gómez, P.; Ollero, F.J.; Ruiz-Sainz, J.E.; López-Baena, F.J.; Vinardell, J.M. A transcriptomic analysis of the effect of genistein on Sinorhizobium fredii HH103 reveals novel rhizobial genes putatively involved in symbiosis. Sci. Rep. 2016, 7, 31592. [Google Scholar] [CrossRef] [PubMed]
- Fischer, H.M. Genetic regulation of nitrogen fixation in rhizobia. Microbiol. Rev. 1994, 58, 352–386. [Google Scholar] [PubMed]
- Oláh, B.; Kiss, E.; Györgypál, Z.; Borzi, J.; Cinege, G.; Csanádi, G.; Batut, J.; Kondorosi, A.; Dusha, I. Mutation in the ntrR gene, a member of the vap gene family, increases the symbiotic efficiency of Sinorhizobium meliloti. Mol. Plant Microbe Interact. 2001, 14, 887–894. [Google Scholar] [CrossRef] [PubMed]
- Mitsui, H.; Sato, T.; Sato, Y.; Ito, N.; Minamisawa, K. Sinorhizobium meliloti RpoH1 is required for effective nitrogen-fixing symbiosis with alfalfa. Mol. Genet. Genom. 2004, 271, 416–425. [Google Scholar] [CrossRef] [PubMed]
- Hassan, S.; Mathesius, U. The role of flavonoids in root-rhizosphere signalling: Opportunities and challenges for improving plant-microbe interactions. J. Exp. Bot. 2012, 63, 3429–3444. [Google Scholar] [CrossRef] [PubMed]
- Makoi, J.H.J.R.; Ndakidemi, P.A. Biological, ecological and agronomic significance of plant phenolic compounds in rhizosphere of the symbiotic legumes. Afr. J. Biotechnol. 2007, 6, 1358–1368. [Google Scholar]
- Cooper, J.E. Early interactions between legumes and rhizobia: Disclosing complexity in a molecular dialogue. J. Appl. Microbiol. 2007, 103, 1355–1365. [Google Scholar] [CrossRef] [PubMed]
- Tolin, S.; Arrigoni, G.; Moscatiello, R.; Masi, A.; Navazio, L.; Sablok, G.; Squartini, A. Quantitative analysis of the naringenin-inducible proteome in Rhizobium leguminosarum by isobaric tagging and mass spectrometry. Proteomics 2013, 13, 1961–1972. [Google Scholar] [CrossRef] [PubMed]
- Meneses, N.; Taboada, H.; Dunn, M.F.; Vargas, M.D.C.; Buchs, N.; Heller, M.; Encarnación, S. The naringenin-induced exoproteome of Rhizobium etli CE3. Arch. Microbiol. 2017, 199, 737–755. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Pasayo, R.; Martinez-Romero, E. Multiresistance genes of Rhizobium etli CFN42. Mol. Plant Microbe Interact. 2000, 13, 572–577. [Google Scholar] [CrossRef] [PubMed]
- Dylan, T.; Helinski, D.R.; Ditta, G.S. Hypoosmotic adaptation in Rhizobium meliloti requires β-1,2-glucan. J. Bacteriol. 1990, 172, 1400–1408. [Google Scholar] [CrossRef] [PubMed]
- Regensburger, B.; Hennecke, H. RNA polymerase from Rhizobium japonicum. Arch. Microbiol. 1983, 135, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Göttfert, M.; Grob, P.; Hennecke, H. Proposed regulatory pathway encoded by the nodV and nodW genes, determinants of host specificity in Bradyrhizobium japonicum. Proc. Natl. Acad. Sci. USA 1990, 87, 2680–2684. [Google Scholar] [CrossRef] [PubMed]
- Loh, J.; Garcia, M.; Stacey, G. NodV and NodW, a second flavonoid recognition system regulating nod gene expression in Bradyrhizobium japonicum. J. Bacteriol. 1997, 179, 3013–3020. [Google Scholar] [CrossRef] [PubMed]
- Sanjuan, J.; Carlson, R.W.; Spaink, H.P.; Bhat, U.R.; Barbour, W.M.; Glushka, J.; Stacey, G. A 2-O-methylfucose moiety is present in the lipo-oligosaccharide nodulation signal of Bradyrhizobium japonicum. Proc. Natl. Acad. Sci. USA 1992, 89, 8789–8793. [Google Scholar] [CrossRef] [PubMed]
- Banfalvi, Z.; Nieuwkoop, A.; Schell, M.; Besl, L.; Stacey, G. Regulation of nod gene expression in Bradyrhizobium japonicum. Mol. Gen. Genet. 1988, 214, 420–424. [Google Scholar] [CrossRef] [PubMed]
- Göttfert, M.; Holzhäuser, D.; Bäni, D.; Hennecke, H. Structural and functional analysis of two different nodD genes in Bradyrhizobium japonicum USDA 110. Mol. Plant Microbe Interact. 1992, 5, 257–265. [Google Scholar] [CrossRef] [PubMed]
- Grob, P.; Hennecke, H.; Göttfert, M. Cross-talk between the two component regulatory systems NodVW and NwsAB of Bradyrhizobium japonicum. FEMS Microbiol. Lett. 1994, 120, 349–353. [Google Scholar] [CrossRef]
- Wang, S.P.; Stacey, G. Studies of the Bradyrhizobium japonicum nodD1 promoter: A repeated structure for the nod box. J. Bacteriol. 1991, 173, 3356–3365. [Google Scholar] [CrossRef] [PubMed]
- Dockendorff, T.C.; Sharma, A.J.; Stacey, G. Identification and characterization of the nolYZ genes of Bradyrhizobium japonicum. Mol. Plant Microbe Interact. 1994, 7, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Göttfert, M.; Lamb, J.W.; Gasser, R.; Semenza, J.; Hennecke, H. Mutational analysis of the Bradyrhizobium japonicum common nod genes and further nod box-linked genomic DNA regions. Mol. Gen. Genet. 1989, 215, 407–415. [Google Scholar] [CrossRef] [PubMed]
- Krause, A.; Doerfel, A.; Göttfert, M. Mutational and transcriptional analysis of the type III secretion system of Bradyrhizobium japonicum. Mol. Plant Microbe Interact. 2002, 15, 1228–1235. [Google Scholar] [CrossRef] [PubMed]
- Göttfert, M.; Hennecke, H.; Tabata, S. Facets of the Bradyrhizobium japonicum 110 genome. In Genomes and Genomics of Nitrogen-Fixing Organisms; Palacios, R., Newton, W.E., Eds.; Springer: Berlin, Germany, 2005; pp. 99–111. [Google Scholar]
- Süß, C.; Hempel, J.; Zehner, S.; Krause, A.; Patschkowski, T.; Göttfert, M. Identification of genistein-inducible and type III-secreted proteins of Bradyrhizobium japonicum. J. Biotechnol. 2006, 126, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Ellison, D.W.; Miller, V.L. Regulation of virulence by members of the MarR/SlyA family. Curr. Opin. Microbiol. 2006, 9, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, S.P.; Grove, A. Ligand-responsive transcriptional regulation by members of the MarR family of winged helix proteins. Curr. Issues Mol. Biol. 2006, 8, 51–62. [Google Scholar] [PubMed]
- Ramos, J.L.; Martínez-Bueno, M.; Molina-Henares, A.J.; Terán, W.; Watanabe, K.; Zhang, X.; Gallegos, M.T.; Brennan, R.; Tobes, R. The TetR family of transcriptional repressors. Microbiol. Mol. Biol. Rev. 2005, 69, 326–356. [Google Scholar] [CrossRef] [PubMed]
- Johnston, A.W.B.; Beringer, J.E. Identification of Rhizobium strains in pea root nodules using genetic markers. J. Gen. Microbiol. 1975, 87, 343–350. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Liu, S.; Yang, Y.; Chang, W.; Hong, G. In Rhizobium leguminosarum, NodD represses its own transcription by competing with RNA polymerase for binding sites. Nucleic Acids Res. 2000, 28, 2784–2793. [Google Scholar] [CrossRef] [PubMed]
- Rossen, L.; Shearman, C.A.; Johnston, A.W.; Downie, J.A. The nodD gene of Rhizobium leguminosarum is autoregulatory and in the presence of plant exudate induces the nodA,B,C genes. EMBO J. 1985, 4, 3369–3373. [Google Scholar] [PubMed]
- Rostas, K.; Kondorosi, E.; Horvath, B.; Simoncsits, A.; Kondorosi, A. Conservation of extended promoter regions of nodulation genes in Rhizobium. Proc. Natl. Acad. Sci. USA 1986, 83, 1757–1761. [Google Scholar] [CrossRef] [PubMed]
- Shearman, C.A.; Rossen, L.; Johnston, A.W.; Downie, J.A. The Rhizobium leguminosarum nodulation gene nodF encodes a polypeptide similar to acyl-carrier protein and is regulated by nodD plus a factor in pea root exudate. EMBO J. 1986, 5, 647–652. [Google Scholar] [PubMed]
- Spaink, H.P.; Okker, R.J.H.; Wijffelman, C.A.; Pees, E.; Lugtenberg, B.J. Promoters in the nodulation region of the Rhizobium leguminosarum Sym plasmid pRL1JI. Plant Mol. Biol. 1987, 9, 27–39. [Google Scholar] [CrossRef] [PubMed]
- Fisher, R.F.; Egelhoff, T.T.; Mulligan, J.T.; Long, S.R. Specific binding of proteins from Rhizobium meliloti cell-free extracts containing NodD to DNA sequences upstream of inducible nodulation genes. Genes Dev. 1988, 2, 282–293. [Google Scholar] [CrossRef] [PubMed]
- Surin, B.P.; Downie, J.A. Characterization of the Rhizobium leguminosarum genes nodLMN involved in efficient host-specific nodulation. Mol. Microbiol. 1988, 2, 173–183. [Google Scholar] [CrossRef] [PubMed]
- De Maagd, R.A.; Wijfjes, A.H.; Spaink, H.P.; Ruiz-Sainz, J.E.; Wijffelman, C.A.; Okker, R.J.; Lugtenberg, B.J. nodO, a new nod gene of the Rhizobium leguminosarum biovar viciae sym plasmid pRL1JI, encodes a secreted protein. J. Bacteriol. 1989, 171, 6764–6770. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.E.; Hou, B.; Zong, C.; Hong, G. Identification of a NodD repressible gene adjacent to nodM in Rhizobium leguminosarum biovar viciae. Acta Biochim. Biophys. Sin. 2012, 44, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Dibb, N.J.; Downie, J.A.; Brewin, N.J. Identification of a rhizosphere protein encoded by the symbiotic plasmid of Rhizobium leguminosarum. J. Bacteriol. 1984, 158, 621–627. [Google Scholar] [PubMed]
- Cubo, T.; Economou, A.; Murphy, G.; Johnston, A.W.B.; Downie, J.A. Molecular characterization and regulation of the rhizosphere-expressed genes rhiABCR that can influence nodulation by Rhizobium leguminosarum biovar viciae. J. Bacteriol. 1992, 174, 4026–4035. [Google Scholar] [CrossRef] [PubMed]
- Rodelas, B.; Lithgow, J.K.; Wisniewski-Dye, F.; Hardman, A.; Wilkinson, A.; Economou, A.; Downie, J.A. Analysis of quorum-sensing-dependent control of rhizosphere-expressed (rhi) genes in Rhizobium leguminosarum bv. viciae. J. Bacteriol. 1999, 181, 3816–3823. [Google Scholar] [PubMed]
- An, J.H.; Lee, G.Y.; Jung, J.W.; Lee, W.; Kim, Y.S. Identification of residues essential for a two-step reaction by malonyl-CoA synthetase from Rhizobium trifolii. Biochem. J. 1999, 344, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.W.; Hong, S.Y.; Ryoo, H.D.; Rhyu, K.I.; Kim, Y.S. Kinetics of malonyl-CoA synthetase from Rhizobium trifolii and evidences for malonyl-AMP formation as a reaction intermediate. Bull. Korean Chem. Soc. 1994, 15, 394–399. [Google Scholar]
- Stumpf, D.K.; Burris, R.H. Organic acid content of soybean: Age and source of nitrogen. Plant Physiol. 1981, 68, 989–991. [Google Scholar] [CrossRef] [PubMed]
- Pini, F.; East, A.K.; Appia-Ayme, C.; Tomek, J.; Karunakaran, R.; Mendoza-Suárez, M.; Edwards, A.; Terpolilli, J.J.; Roworth, J.; Downie, J.; et al. Bacterial biosensors for in vivo spatiotemporal mapping of root secretion. Plant Physiol. 2017, 174, 1289–1306. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Romero, E.; Segovia, L.; Mercante, F.M.; Franco, A.A.; Graham, P.; Pardo, M.A. Rhizobium tropici, a novel species nodulating Phaseolus vulgaris L. Beans and Leucaena sp. Trees. Int. J. Syst. Bacteriol. 1991, 41, 417–426. [Google Scholar] [CrossRef] [PubMed]
- Del Cerro, P.; Rolla-Santos, A.A.P.; Gomes, D.F.; Marks, B.B.; Pérez-Montaño, F.; Rodríguez-Carvajal, M.A.; Nakatani, A.S.; Gil-Serrano, A.; Megías, M.; Ollero, F.J.; et al. Regulatory nodD1 and nodD2 genes of Rhizobium tropici strain CIAT 899 and their roles in the early stages of molecular signaling and host-legume nodulation. BMC Genom. 2015, 16, 251. [Google Scholar] [CrossRef] [PubMed]
- Del Cerro, P.; Rolla-Santos, A.A.P.; Gomes, D.F.; Marks, B.B.; Espuny, M.R.; Rodríguez-Carvajal, M.A.; Soria-Díaz, M.E.; Nakatani, A.S.; Hungria, M.; Ollero, F.J.; et al. Opening the “black box” of nodD3, nodD4 and nodD5 genes of Rhizobium tropici strain CIAT 899. BMC Genom. 2015, 16, 864. [Google Scholar] [CrossRef] [PubMed]
- Poupot, R.; Martínez-Romero, E.; Promé, J.C. Nodulation factors from Rhizobium tropici are sulfated or nonsulfated chitopentasaccharides containing an N-methyl-N-acylglucosaminyl terminus. Biochemistry 1993, 32, 10430–10435. [Google Scholar] [CrossRef] [PubMed]
- Garcia, M.L.J.; Dunlap, J.; Loh, J.; Stacey, G. Phenotypic characterization and regulation of the nolA gene of Bradyrhizobium japonicum. Mol. Plant Microbe Interact. 1996, 9, 625–635. [Google Scholar] [CrossRef] [PubMed]
- Estévez, J.; Soria-Díaz, M.E.; de Córdoba, F.F.; Morón, B.; Manyani, H.; Gil, A.; Thomas-Oates, J.; van Brussel, A.A.; Dardanelli, M.S.; Sousa, C.; et al. Different and new Nod factors produced by Rhizobium tropici CIAT899 following Na+ stress. FEMS Microbiol. Lett. 2009, 293, 220–231. [Google Scholar]
- Guasch-Vidal, B.; Estévez, J.; Dardanelli, M.S.; Soria-Díaz, M.E.; de Córdoba, F.F.; Balog, C.I.; Manyani, H.; Gil-Serrano, A.; Thomas-Oates, J.; Hensbergen, P.J.; et al. High NaCl concentrations induce the nod genes of Rhizobium tropici CIAT899 in the absence of flavonoid inducers. Mol. Plant Microbe Interact. 2013, 26, 451–460. [Google Scholar] [CrossRef] [PubMed]
- Ormeño-Orrillo, E.; Menna, P.; Gonzaga, L.A.; Ollero, F.J.; Nicolas, M.F.; Rodrigues, E.P.; Nakatani, S.A.; Silva Batista, J.S.; Oliveira Chueire, L.M.; Souza, R.C.; et al. Genomic basis of broad host range and environmental adaptability of Rhizobium tropici CIAT 899 and Rhizobium sp. PRF 81 which are used in inoculants for common bean (Phaseolus vulgaris L.). BMC Genom. 2012, 13, 735. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Rhijn, P.J.; Feys, B.; Verreth, C.; Vanderleyden, J. Multiple copies of nodD in Rhizobium tropici CIAT899 and BR816. J. Bacteriol. 1993, 175, 438–447. [Google Scholar] [CrossRef] [PubMed]
- Ling, J.; Wang, H.; Wu, P.; Li, T.; Tang, Y.; Naseer, N.; Zheng, H.; Masson-Boivin, C.; Zhong, Z.; Zhu, J. Plant nodulation inducers enhance horizontal gene transfer of Azorhizobium caulinodans symbiosis island. Proc. Natl. Acad. Sci. USA 2016, 113, 13875–13880. [Google Scholar] [CrossRef] [PubMed]
- Györgypal, Z.; Iyer, N.; Kondorosi, A. Three regulatory nodD alleles of diverged flavonoid-specificity are involved in host-dependent nodulation by Rhizobium meliloti. Mol. Gen. Genet. 1988, 212, 85–92. [Google Scholar] [CrossRef]
- Maillet, F.; Debelle, F.; Denarie, J. Role of the nodD and syrM genes in the activation of the regulatory gene nodD3, and of the common and host-specific nod genes of Rhizobium meliloti. Mol. Microbiol. 1990, 4, 1975–1984. [Google Scholar] [CrossRef] [PubMed]
- Galibert, F.; Finan, T.M.; Long, S.R.; Pühler, A.; Abola, P.; Ampe, F.; Barloy-Hubler, F.; Barnett, M.J.; Becker, A.; Boistard, P.; et al. The composite genome of the legume symbiont Sinorhizobium meliloti. Science 2001, 293, 668–672. [Google Scholar] [CrossRef] [PubMed]
- Pueppke, S.G.; Broughton, W.J. Rhizobium sp. strain NGR234 and R. fredii USDA257 share exceptionally broad, nested host ranges. Mol. Plant Microbe Interact. 1999, 12, 293–318. [Google Scholar] [CrossRef] [PubMed]
- Margaret, I.; Becker, A.; Blom, J.; Bonilla, I.; Goesmann, A.; Göttfert, M.; Lloret, J.; Mittard-Runte, V.; Rückert, C.; Ruiz-Sainz, J.E.; et al. Symbiotic properties and first analyses of the genomic sequence of the fast growing model strain Sinorhizobium fredii HH103 nodulating soybean. J. Biotechnol. 2011, 155, 11–19. [Google Scholar] [CrossRef] [PubMed]
- López-Baena, F.J.; Ruiz-Sainz, J.E.; Rodríguez-Carvajal, M.A.; Vinardell, J.M. Bacterial molecular signals in the Sinorhizobium fredii-soybean symbiosis. Int. J. Mol. Sci. 2016, 17, 755. [Google Scholar] [CrossRef] [PubMed]
- Schmeisser, C.; Liesegang, H.; Krysciak, D.; Bakkou, N.; Le Quéré, A.; Wollherr, A.; Heinemeyer, I.; Morgenstern, B.; Pommerening-Röser, A.; Flores, M.; et al. Rhizobium sp. strain NGR234 possesses a remarkable number of secretion systems. Appl. Environ. Microbiol. 2009, 75, 4035–4045. [Google Scholar] [CrossRef] [PubMed]
- Schuldes, J.; Rodriguez Orbegoso, M.; Schmeisser, C.; Krishnan, H.B.; Daniel, R.; Streit, W.R. Complete genome sequence of the broad-host-range strain Sinorhizobium fredii USDA257. J. Bacteriol. 2012, 194, 4483. [Google Scholar] [CrossRef] [PubMed]
- Vinardell, J.M.; Acosta-Jurado, S.; Zehner, S.; Göttfert, M.; Becker, A.; Baena, I.; Blom, J.; Crespo-Rivas, J.C.; Goesmann, A.; Jaenicke, S.; et al. The Sinorhizobium fredii HH103 genome: A comparative analysis with S. fredii strains differing in their symbiotic behavior with soybean. Mol. Plant Microbe Interact. 2015, 28, 811–824. [Google Scholar] [CrossRef] [PubMed]
- Tian, C.F.; Zhou, Y.J.; Zhang, Y.M.; Li, Q.Q.; Zhang, Y.Z.; Li, D.F.; Wang, S.; Wang, J.; Gilbert, L.B.; Li, Y.R.; et al. Comparative genomics of rhizobia nodulating soybean suggests extensive recruitment of lineage-specific genes in adaptations. Proc. Natl. Acad. Sci. USA 2012, 109, 8629–8634. [Google Scholar] [CrossRef] [PubMed]
- Jiao, J.; Wu, L.J.; Zhang, B.; Hu, Y.; Li, Y.; Zhang, X.X.; Guo, H.J.; Liu, L.X.; Chen, W.X.; Zhang, Z.; et al. MucR is required for transcriptional activation of conserved ion transporters to support nitrogen fixation of Sinorhizobium fredii in soybean nodules. Mol. Plant Microbe Interact. 2016, 29, 352–361. [Google Scholar] [CrossRef] [PubMed]
- Heron, D.S.; Pueppke, S.G. Mode of infection, nodulation specificity, and indigenous plasmids of 11 fast-growing Rhizobium japonicum strains. J. Bacteriol. 1984, 160, 1061–1066. [Google Scholar] [PubMed]
- Dowdle, S.F.; Bohlool, B.B. Predominance of fast growing Rhizobium japonicum in a soybean field in the People’s Republic of China. Appl. Environ. Microbiol. 1985, 50, 1171–1176. [Google Scholar] [PubMed]
- Mimmack, M.L.; Borthakur, D.; Jones, M.A.; Downie, J.A.; Johnston, A.W. The psi operon of Rhizobium leguminosarum biovar phaseoli: Identification of two genes whose products are located at the bacterial cell surface. Microbiology 1994, 140, 1223–1229. [Google Scholar] [CrossRef] [PubMed]
- Soto, M.J.; Sanjuán, J.; Olivares, J. Rhizobia and plant-pathogenic bacteria: Common infection weapons. Microbiology 2006, 152, 3167–3174. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Guerrero, I.; Pérez-Montaño, F.; Monreal, J.A.; Preston, G.M.; Fones, H.; Vioque, B.; Ollero, F.J.; López-Baena, F.J. The Sinorhizobium (Ensifer) fredii HH103 Type 3 secretion system suppresses early defense responses to effectively nodulate soybean. Mol. Plant Microbe Interact. 2015, 28, 790–799. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Guerrero, I.; Pérez-Montaño, F.; Medina, C.; Ollero, F.J.; López-Baena, F.J. NopC Is a Rhizobium-Specific Type 3 Secretion System Effector Secreted by Sinorhizobium (Ensifer) fredii HH103. PLoS ONE 2015, 10, e0142866. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Guerrero, I.; Pérez-Montaño, F.; Medina, C.; Ollero, F.J.; López-Baena, F.J. The Sinorhizobium (Ensifer) fredii HH103 nodulation outer protein NopI is a determinant for efficient nodulation of soybean and cowpea plants. Appl. Environ. Microbiol. 2017, 83, e02770-16. [Google Scholar] [CrossRef] [PubMed]

| Comparison | Bacteria | Method | Strains | Specific Conditions | Results Related with the Symbiotic Process | References |
|---|---|---|---|---|---|---|
| Free-living cells vs. Bacteroids | Bradyrhizobium japonicum USDA 110 | Microarray (Affymetrix GeneChip) | Wild-type and RNA polymerase σ54 factor (rpoN1-2) double mutant | -Free-living cultures in either aerobic or microaerobic conditions (mid-exponential phase). -Bacteroids in soybean (10, 13, 21 and 31 days post inoculation, dpi). | -Distinction among genes expressed in early and late bacteroids. -1/3 of the genes induced in bacteroids (21 dpi) are also upregulated in microaerobiosis, including fix and nif genes. The other induced genes are regulated by factors other than O2 limitation. -Determination of genes transcribed in bacteroids in a σ54 (rpoN1)-dependent manner (several fix, nif and hup genes included). | [19] |
| B. japonicum USDA 110 | Microarray (Operon Biotechnologies) | Wild-type | -Minimal and rich media (mid-exponential phase). -Osmotic stress conditions (mid-exponential phase). -Bacteroids in soybean (28 dpi). | -15% of the genes of the genome were differentially expressed in bacteroids: N2 fixation (nif and fix) and H2 uptake (hup) genes up-regulated. Nodulation (nod) genes also up-regulated. T3SS (hrc) genes are repressed. | [33] | |
| Rhizobium leguminosarum biovar viciae 3841 | Microarray (Operon Biotechnologies) | Wild-type | -Several carbon substrates (glucose, pyruvate, succinate, inositol, acetate, and acetoacetate). -Bacteroids in vetch (28 dpi). -Bacteroids in pea (7, 15, 21 and 28 dpi). | -386 genes were differentially expressed in at least one stage of bacteroid development. -The decarboxylating arm of the tricarboxylic acid cycle and the aminobutyrate metabolism were highly induced in early nodule bacteroids (7 dpi). -Changes in the expression of regulators, exported and cell surface molecules, multidrug exporters, and heat and cold shock proteins in bacteroids of early pea nodules (7 dpi). -The fix genes were induced early but continued to increase in mature bacteroids (15 and 21 dpi), while nif genes were induced strongly in older bacteroids of pea nodules (28 dpi). | [34] | |
| Mesorhizobium huakuii 7653R | RNA-Seq Microarray (Roche NimbleGen) | Wild-type | -Bacteroids in Astragalus sinicus (32 dpi). | -Mostly, bacteroid up-regulated genes are located on plasmid while downregulated genes are chromosomal. -Free-living cells have a primary role in maintaining basal metabolism, whereas bacteroids in N2 fixation (nif, fix and suf clusters up-regulated). -Up-regulation of rpoN (σ54) and rpoH (RNA polymerase σ32 factor) indicates an essential role of both transcriptional factors in the activation of genes implied in the N2 fixation. | [35] | |
| Mesorhizobium loti MAFF303099 | Microarray (Made by Authors) | Wild-type | -Free-living cultures in either aerobic or microaerobic conditions (mid-exponential phase). -Bacteroids in Lotus japonicus (42 dpi). -C-starving conditions. | -Clusters of genes within the symbiotic islands are collectively up-regulated in bacteroids (including nif, fix, fdx, and rpoN genes) through σ54 (rpoN). -Most of the upregulated genes in bacteroids are induced in a NifA-independent manner. -Transcription outside the symbiotic island is strongly repressed in bacteroids (genes implied in cell division, cell wall and flagella synthesis). | [20] | |
| Sinorhizobium meliloti 1021 | Macroarrays (Made by authors) | Wild-type and bacA mutant | -Free-living cultures (minimal and rich media) with and without luteolin. -Free-living cultures in either aerobic or microaerobic conditions. -Bacteroids in young (8 dpi) nodules and in mature nitrogen-fixing nodules (18 dpi) in Medicago truncatula. -Bacteroids in Medicago sativa mature nodules (18 dpi). | -15 out of 214 tested genes were induced in young and mature nodules of the two strains (specific of the infection stage), including genes that code for hemolysin calcium-binding proteins, adenylate cyclases and type-IV secretion system proteins. -24 out of 214 tested genes were up-regulated in mature nodules of both plants (specific of symbiosis), including nif, fix and cya genes. -18 out of 214 tested genes were down-regulated in mature nodules of both plants (specific of symbiosis), including genes implied in cell division (ftsZ) and chaperonins. | [36] | |
| S. meliloti 1021 | Microarray and Macroarray (Made by authors) | Wild-type | -Free-living cultures in either aerobic or microaerobic conditions. -Bacteroids from M. sativa nodules (18 to 22 dpi). | -982 genes differentially expressed in bacteroids. -Genes downregulated were implied in cell division (ftsZ), DNA, RNA and protein metabolisms (dna and rpo cluster genes), motility and chemotaxis (che genes), phosphorus uptake and utilization (pho genes), nitrogen assimilation (gln genes) and glucolysis and aerobic respiration (nuoACEFHIKLMN operon). -Genes upregulated included mainly those related with symbiosis (several nod and noe genes), nitrogen fixation (nif and fix operons) and transport of peptides and aminoacids. | [18] | |
| S. meliloti 1021 | Microarray (Made by authors) | Wild-type and bacA mutant | -Free-living cultures in early and late exponential and stationary phase. -Bacteroids from M. sativa nodules at 5, 8, 14–18 dpi in the wild-type. -Bacteroids from M. sativa nodules at 11 dpi in the mutant. | -36 genes were specifically induced in early stages of the symbiosis (nod genes and genes implied in the synthesis of the cytochrome c). -Bacterial transcriptomic profile changes during nodule development. -The nif and fix cluster genes were up-regulated even in young nodules. | [37] | |
| S. meliloti 1021 | Microarray (Affymetrix GeneChip) | Wild-type, triple nodD1 nodD2 nodD3 mutant, rpoN and fixJ mutants, and triple nodD1 nodD2 nodD3 mutant overexpressing either nodD1 or nodD3 | -Minimal and rich media (mid-exponential phase). -Late exponential cultures diluted to 0.15–0.2 OD600 and induced for 4 h with luteolin. -Bacteroids from M. truncatula nodules at 33–35 dpi. | -RpoN controlled genes that are not differentially expressed in free-living bacteria (fdx, nif and fix operons). -Study of gene expression simultaneously in both symbiotic partners indicate that in total more than 5000 genes are differentially expressed in both organisms. -Most of the plant genes upregulated in wild-type nodules were also induced in Fix− nodules (fixJ mutant), indicating that the nitrogen fixing status does not significantly affect plant transcriptomic changes. | [38] | |
| S. meliloti 1021 | Microarray (Made by authors) | Wild-type and and fixJ, nifA, fixK, and nifH mutants | -Free-living cultures in either aerobic or microaerobic conditions. -Bacteroids from M. truncatula nodules at 14 dpi. | -122 genes were activated by FixJ via NifA and FixK (97% located in the symbiotic plasmid), including fix and nif genes. -FixJ controls the majority of genes expressed in mature bacteroids. -NifA activated genes implied in nitrogen fixation and exopolysaccharides (EPS) production and FixK up-regulated genes involved in respiration, arginine metabolism, denitrification and stress responses. | [39] | |
| S. meliloti 2011 | RNA-seq (Illumina Hiseq 2000) | Wild-type and rpoE2 mutant | -Free-living cultures in mid-exponential or early stationary phase. -Bacteroids from M. truncatula nodules at 10 dpi. | -Authors designed EuGene-P, a tool that enables the automated prediction of coding sequences, untranslated regions, transcription start sites and non-coding RNA genes. -Prediction of 6308 coding sequences, 1876 non-coding RNAs and 4077 transcription start sites using cDNA of all conditions. | [10] | |
| S. meliloti 2011 | RNA-seq (Illumina Hiseq 2000) | Wild-type | -Bacteroids from M. truncatula nodules at 10 and 15 dpi. | -Coupling bacterial and plant gene transcriptome determination to laser dissection, authors determined expression changes at the tissue level on indeterminate nodules. | [40] | |
| Sinorhizobium fredii NGR234 | RNA-seq (Illumina Hiseq 2000) | Wild-type | -Bacteroids from Vigna unguiculata nodules (21 dpi nodules) -Bacteroids from Leucaena leucocephala (35 dpi nodules). | -3143 genes differentially expressed in bacteroids of V. unguiculata and 2780 in bacteroids of L. leucocephala. -Upregulated genes in bacteroids from both hosts were implied in the synthesis of ATP-binding cassette (ABC) transporters, type 3 secretion system (T3SS), nitrogen metabolism, nitrogen fixation (nif and fix operons), fatty acid metabolism, benzoate degradation and exopolysaccharide biosynthesis (exo cluster genes). -Downregulated genes in bacteroids of both plants were involved in synthesis of DNA, RNA and protein metabolisms and flagellar assembly. | [41] | |
| Aerobiosis (Free-living cells) vs. Microaerobiosis | B. japonicum USDA 110 | Microarray (Affymetrix GeneChip) | Wild-type and rpoN1-2 double mutant | -Free-living cultures in either aerobic or microaerobic conditions. -Bacteroids in soybean (10, 13, 21 and 31 dpi). | -Microaerobiosis triggered upregulation of symbiotic relevant genes, including fix and nif clusters, mostly in a σ54-dependent manner. -1/3 of the genes induced in bacteroids (21 dpi) are also upregulated in microaerobic conditions. | [19] |
| M. loti MAFF303099 | Microarray (Made by authors) | Wild-type | -Free-living cultures in either aerobic or microaerobic conditions. -Bacteroids in L. japonicus MG20 (42 dpi). -C-starved cells. | -The genome region containing the fixNOPQ genes (outside the symbiosis island) and the fix and nif regions (in the symbiotic island) were upregulated as under both microaerobic and symbiotic conditions. | [20] | |
| S. meliloti 021 | Macroarrays (Made by authors) | Wild-type and bacA mutant | -Free-living cultures (minimal and rich media) with and without luteolin. -Free-living cultures in either aerobic or microaerobic conditions. -Bacteroids in young (8 dpi) nodules and in mature nitrogen-fixing nodules (18 dpi) in M. truncatula J6. -Bacteroids in M. sativa mature nodules (18 dpi). | -8 out of 214 tested genes were found to be induced under microoxic and bacteroids conditions, including fix and nif genes. | [36] | |
| S. meliloti 1021 | Microarray (Made by authors) | Wild-type and and fixJ, nifA, fixK, and nifH mutants | -Free-living cultures in either aerobic or microaerobic conditions (mid-exponential phase). -Bacteroids from M. truncatula nodules at 14 dpi. | -FixJ controlled 74% of the genes induced in microaerobiosis and the majority of genes expressed in mature bacteroids (including fix and nif genes). | [39] | |
| S. meliloti 1021 | Microarray and Macroarray (Made by authors) | Wild-type | -Free-living cultures in either aerobic or microaerobic conditions. -Bacteroids from M. sativa nodules (18 to 22 dpi). | -377 genes regulated by oxygen concentration (266 induced and 111 repressed in microaerobic conditions). -31 genes were induced both under microoxic and bacteroids conditions, including genes implied in N2-fixation (nif and fix operons), proline metabolism and denitrification. | [18] | |
| Free-living cells vs. Rhizosphere | R. leguminosarum biovar viciae 3841 | Microarray (Operon Biotechnologies) | Wild-type and mutants in many different genes | -Free-living cells vs. rhizosphere (pea, alfalfa or sugar beet) attached bacteria (7 dpi). -Free-living cultures with and without root exudates (pea). -Free-living cultures with and without the flavonoid hesperetin. | -A common core of 106 genes were rhizosphere-induced genes (70 genes encode for proteins with unknown functions). -The increase of gene expression of the glyoxylate cycle only occurred in the pea rhizosphere. -Many genes on pRL8 (plasmid 8 of R. leguminosarum) were specifically up-regulated in the pea rhizosphere. | [42] |
| Non-induced (Free-living cells) vs. root secretions | R. leguminosarum biovar viciae 3841 | Microarray (Operon Biotechnologies) | Wild-type and mutants in many different genes | -Free living cells vs. rhizosphere (pea, alfalfa or sugar beet) attached bacteria (7 dpi).-Free-living cultures with and without root exudates (pea). -Free-living cultures with and without the flavonoid hesperetin. | -21 genes up-regulated including the nodulation (nod) and rhi gene clusters on pRL10 (the symbiotic plasmid). | [42] |
| Non-induced (Free-living cells) vs. flavonoids | B. japonicum USDA 110 | Microarray (Affymetrix GeneChip) | Wild-type, nodW mutant and nodW mutant overexpressing nwsB | -Free-living cultures with and without genistein (8 hpi). | -101 genes up-regulated in the presence of genistein, including nod genes, the flagellar cluster and transport genes. -NodW was essencial for induction of most of these genes. -The phenotype and the gene expression levels in the nodW mutant were partially restored by overexpression of nwsB gene. | [43] |
| R. leguminosarum biovar viciae 3841 | Microarray (Operon Biotechnologies) | Wild-type and mutants in many different genes | -Free-living cells vs. rhizosphere (pea, alfalfa or sugar beet) attached bacteria (7 dpi). -Free-living cultures with and without root exudates (pea). -Free-living cultures with and without the flavonoid hesperetin. | -27 genes up-regulated in the presence of flavonoids, including the nod and rhi gene clusters on pRL10 (the symbiotic plasmid). -6 genes down-regulated, including flaD. | [42] | |
| Rhizobium tropici CIAT 899 | RNA-seq (Illumina Hiseq 2000) | Wild-type, nodD1 mutant and nodD2 mutant | -Free-living cultures with and without the flavonoid apigenin. -Free-living cultures with and without salt. | -13 symbiotic-related genes up-regulated in the presence of apigenin, including nod genes and the IAA synthesis genes. -2 hypothetical-protein genes putatively related with symbiosis were up-regulated via NodD1 and apigenin. -NodD1 activated the expression of the 13 symbiotic-related found genes. | [44,45] | |
| S. meliloti 1021 | Macroarrays (Made by authors) | Wild-type and bacA mutant | -Free-living cultures (minimal and rich media) with and without the flavonoid luteolin. -Aerobic and microaerobic conditions. -Bacteroids in young (8 dpi) nodules and in mature nitrogen-fixing nodules (18 dpi) in M. truncatula J6. -Bacteroids in M. sativa mature nodules (18 dpi). | -7 out of the 214 tested genes were induced with luteolin, including the nod genes, traA and three genes related to iron metabolism (also in nodules). | [36] | |
| S. meliloti 1021 | Microarray (Made by authors) | Wild-type and wild-type overexpressing nodD1 | -Late exponential cultures diluted to 0.15–0.2 OD600 and induced for 4 and 24 h with luteolin. | -26 and 5 genes after 4 and 24 h, respectively, were affected in the presence of genistein, including those belonging to nod/nol/noe operons. -Other genes encoding for hyphotetical proteins were also identified (varying among replicates). | [46] | |
| S. meliloti 1021 | Microarray (Affymetrix GeneChip) | Wild-type, triple nodD1 nodD2 nodD3 mutant, rpoN and fixJ mutants, and triple nodD1 nodD2 nodD3 mutant overexpressing either nodD1 or nodD3 | -Minimal and rich media (mid-exponential phase). -Late exponential cultures diluted to 0.15–0.2 OD600 and induced for 4 h with luteolin.-Bacteroids from M. truncatula nodules at 33–35 dpi. | -Luteolin induced significant expression changes in nod/nol/noe operons (12 genes). -Other 12 genes did not show true flavonoid induction but instead vary in expression depending on replicate. | [38] | |
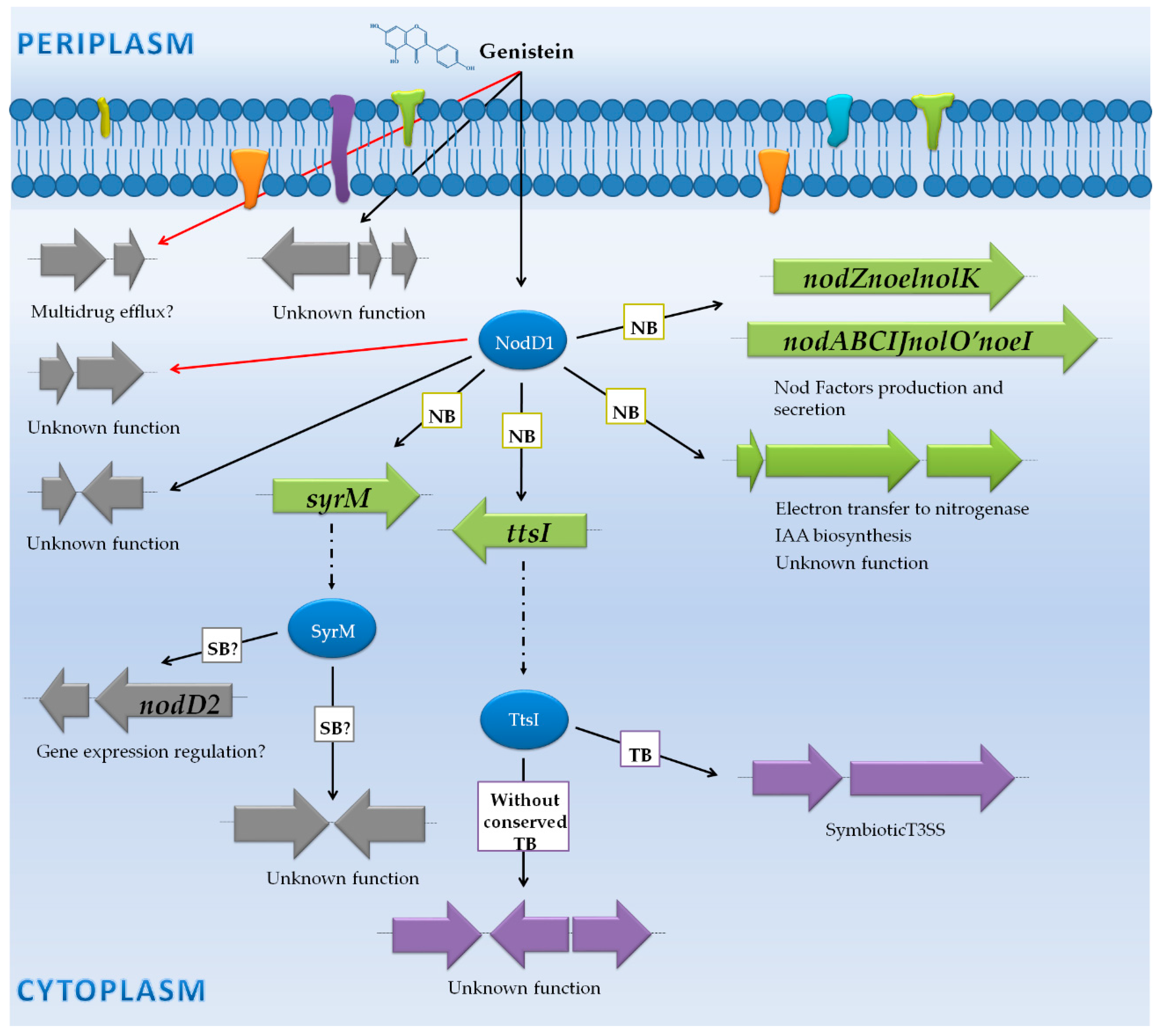
| Sinorhizobium fredii HH103 | RNA-seq (Illumina Hiseq 2000) | Wild-type, nodD1 and ttsI mutants | -Free-living cultures (early stationary phase) with and without the flavonoid genistein. | -100 genes were affected in the presence of genistein: 70 genes induced through nod boxes (nod, nol and noe genes) and tts boxes (T3SS genes) were upregulated. 30 genes not controlled by NB or TB were differentially expressed in the presence of genistein. | [47] | |
| Non-induced (Free-living cells) vs. saline stress (nod gene inducing conditions) | R. tropici CIAT 899 | RNA-seq (Illumina Hiseq 2000) | Wild-type, nodD1 mutant and nodD2 mutant | -Free-living cultures with and without the flavonoid apigenin. -Free-living cultures with and without salt. | -17 symbiotic-related genes up-regulated in the presence of salt, including nod genes and the IAA synthesis genes. -All the symbiotic-related genes upregulated in presence of apigenin are upregulated in salt conditions as well. -2 hypothetical-protein genes putatively related with symbiosis were upregulated via NodD2 and salt. -In general, higher nod gene expression was detected with salt than with apigenin. -NodD2 activated the expression of the 17 symbiotic-related found genes. -NodD1 enhanced the expression of nodD2 under salt conditions. | [44,45] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiménez-Guerrero, I.; Acosta-Jurado, S.; Del Cerro, P.; Navarro-Gómez, P.; López-Baena, F.J.; Ollero, F.J.; Vinardell, J.M.; Pérez-Montaño, F. Transcriptomic Studies of the Effect of nod Gene-Inducing Molecules in Rhizobia: Different Weapons, One Purpose. Genes 2018, 9, 1. https://doi.org/10.3390/genes9010001
Jiménez-Guerrero I, Acosta-Jurado S, Del Cerro P, Navarro-Gómez P, López-Baena FJ, Ollero FJ, Vinardell JM, Pérez-Montaño F. Transcriptomic Studies of the Effect of nod Gene-Inducing Molecules in Rhizobia: Different Weapons, One Purpose. Genes. 2018; 9(1):1. https://doi.org/10.3390/genes9010001
Chicago/Turabian StyleJiménez-Guerrero, Irene, Sebastián Acosta-Jurado, Pablo Del Cerro, Pilar Navarro-Gómez, Francisco Javier López-Baena, Francisco Javier Ollero, José María Vinardell, and Francisco Pérez-Montaño. 2018. "Transcriptomic Studies of the Effect of nod Gene-Inducing Molecules in Rhizobia: Different Weapons, One Purpose" Genes 9, no. 1: 1. https://doi.org/10.3390/genes9010001







