Structural and Evolutionary Relationships in the Giant Sex Chromosomes of Three Microtus Species
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chromosome Preparations
2.2. Chromosome Painting
2.3. Fluorescent In-Situ Hybridization (FISH)
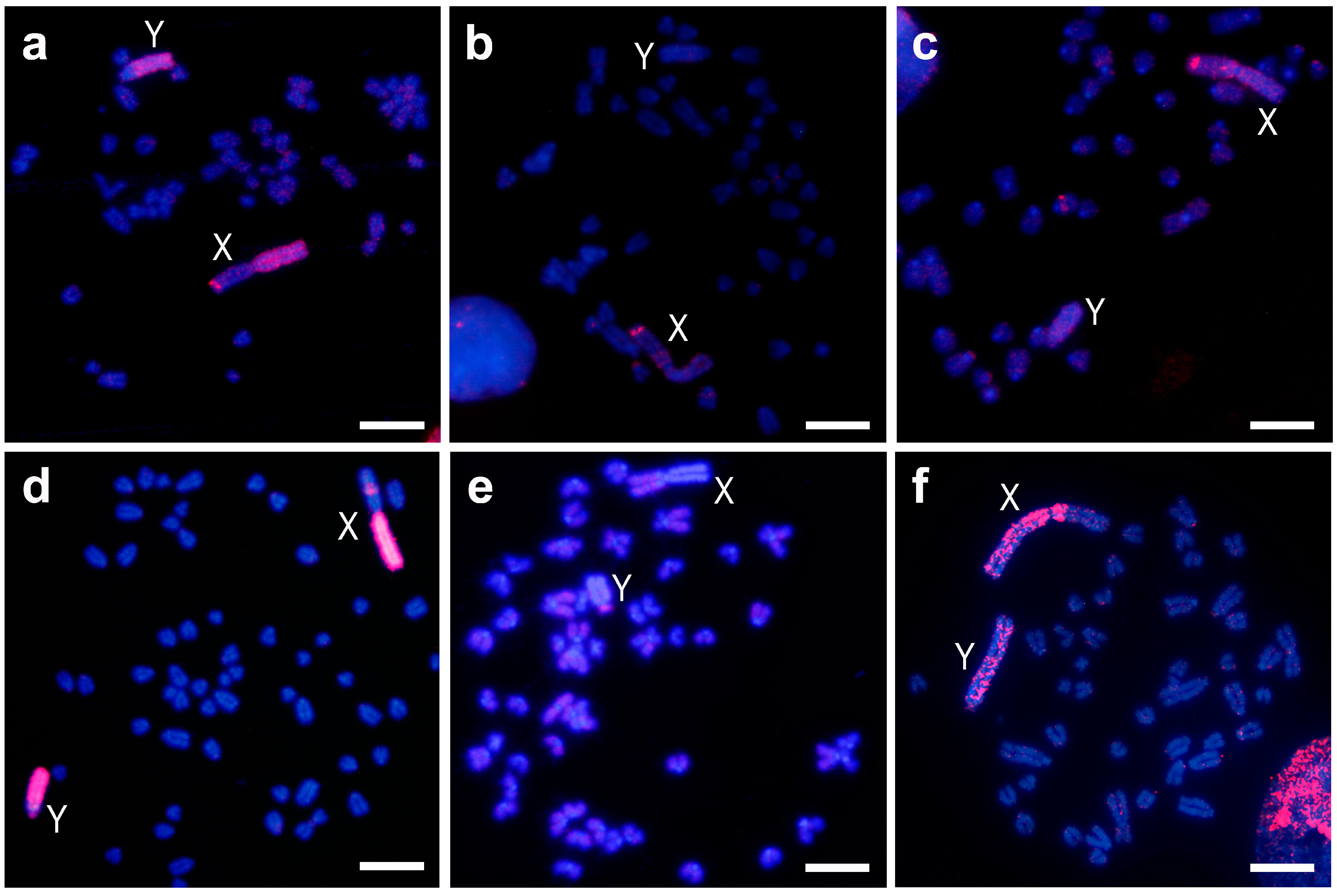
3. Results
3.1. Sex Chromosomes Heterochromatin Painting
3.2. Repetitive Sequences FISH
3.2.1. pMAHAE2
3.2.2. pMAECO14
3.2.3. Telomeric Repeat
3.2.4. Msat-160
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Fink, S.; Fischer, M.C.; Excoffier, L.; Heckel, G. Genomic scans support repetitive continental colonization events during the rapid radiation of voles (Rodentia: Microtus): The utility of AFLPs versus mitochondrial and nuclear sequence markers. Syst. Biol. 2010, 59, 548–572. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, T.; Imai, H.T. Evolutionary rate of the mammalian karyotype. J. Theor. Biol. 1981, 90, 111–121. [Google Scholar] [CrossRef]
- Libbus, B.L.; Johnson, L.A. The creeping vole, Microtus oregoni: Karyotype and sex-chromosome differences between two geographical populations. Cytogenet. Cell Genet. 1988, 47, 181–184. [Google Scholar] [CrossRef] [PubMed]
- Gornung, E.; Castiglia, R.; Rovatsos, M.; Marchal, J.A.; Díaz de la Guardia-Quiles, R.; Sanchez, A. Comparative cytogenetic study of two sister species of Iberian ground voles, Microtus (Terricola) duodecimcostatus and M. (T.) lusitanicus (Rodentia, Cricetidae). Cytogenet. Genome Res. 2011, 132, 144–150. [Google Scholar] [CrossRef] [PubMed]
- Matthey, R. Les chromosomes sexuels géants de Microtus agrestis L. Experientia 1950, 53, 163–184. [Google Scholar] [CrossRef]
- Hansen-Melander, E. The relation of sex chromosomes to chromocenters in somatic cells of Microtus agrestis (L.). Hereditas 1965, 52, 357–366. [Google Scholar] [CrossRef] [PubMed]
- Kalscheuer, V.; Singh, A.P.; Nanda, I.; Sperling, K.; Neitzel, H. Evolution of the gonosomal heterochromatin of Microtus agrestis: Rapid amplification of a large, multimeric, repeat unit containing a 3.0-kb (GATA)11-positive, middle repetitive element. Cytogenet. Cell Genet. 1996, 73, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Meylan, A. Karyotype and giant sex chromosomes of Microtus chrotorrhinus (Miller) (Mammalia: Rodentia). Can. J. Genet. Cytol. 1967, 9, 700–703. [Google Scholar] [CrossRef] [PubMed]
- Díaz de la Guardia, R.; Pascual, L.; Orozco, J.C. The karyotype of Microtus cabrerae Thomas, another species with giant chromosomes. Experientia 1979, 35, 741–742. [Google Scholar] [CrossRef]
- Modi, W.S. Phylogenetic analyses of chromosomal banding patterns among the nearctic Arvicolidae (Mammalia: Rodentia). Syst. Zool. 1987, 36, 109–136. [Google Scholar] [CrossRef]
- Modi, W.S. C-banding analysis and the evolution of heterochromatin among arvicolid rodent. J. Mammal. 1987, 68, 704–714. [Google Scholar] [CrossRef]
- Singh, A.; Henschel, S.; Sperling, K.; Kalscheuer, V.; Neitzel, H. Differences in the meiotic pairing behavior of gonosomal heterochromatin between female and male Microtus agrestis: Implications for the mechanism of heterochromatin amplification on the X and Y. Cytogenet. Cell Genet. 2000, 91, 253–260. [Google Scholar] [CrossRef] [PubMed]
- Marchal, J.A.; Acosta, M.J.; Bullejos, M.; Díaz de la Guardia, R.; Sánchez, A. Sex chromosomes, sex determination, and sex-linked sequences in Microtidae. Cytogenet. Genome Res. 2003, 101, 266–273. [Google Scholar] [CrossRef] [PubMed]
- Modi, W.S. Nucleotide sequence and genomic organization of a tandem satellite array from the rock vole Microtus chrotorrhinus (Rodentia). Mamm. Genome 1992, 3, 226–232. [Google Scholar] [CrossRef] [PubMed]
- Burgos, M.; Jiménez, R.; Olmos, D.M.; Díaz de la Guardia, R. Heterogeneous heterochromatin and size variation in the sex chromosomes of Microtus cabrerae. Cytogenet. Cell Genet. 1988, 47, 75–79. [Google Scholar] [CrossRef] [PubMed]
- Burgos, M.; Jiménez, R.; Díaz de la Guardia, R. Comparative study of G- and C-banded chromosomes of five species of Microtidae. Genetica 1989, 78, 3–12. [Google Scholar] [CrossRef]
- Fernández, R.; Barragán, M.J.; Bullejos, M.; Marchal, J.A.; Martínez, S.; Díaz de la Guardia, R.; Sánchez, A. Molecular and cytogenetic characterization of highly repeated DNA sequences in the vole Microtus cabrerae. Heredity 2001, 87, 637–646. [Google Scholar] [CrossRef] [PubMed]
- Fernández, R.; Barragán, M.J.; Bullejos, M.; Marchal, J.A.; Martínez, S.; Díaz de la Guardia, R.; Sánchez, A. Mapping the SRY gene in Microtus cabrerae: A vole species with multiple SRY copies in males and females. Genome 2002, 45, 600–603. [Google Scholar] [CrossRef] [PubMed]
- Natarajan, A.T.; Sharma, R.P.; Ahnström, G. Fluorochromes and heterochromatin. A study of the chromosomes of Microtus agrestis L. Hereditas 1971, 69, 217–222. [Google Scholar] [CrossRef] [PubMed]
- Nanda, I.; Neitzel, H.; Sperling, K.; Studer, R.; Epplen, J.T. Simple GATCA repeats characterize the X chromosome heterochromatin in Microtus agrestis, European field vole (Rodentia, Cricetidae). Chromosoma 1988, 96, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Neitzel, H.; Kalscheuer, V.; Henschel, S.; Digweed, M.; Sperling, K. Beta-heterochromatin in mammals: Evidence from studies in Microtus agrestis based on the extensive accumulation of L1 and non-L1 retroposons in the heterochromatin. Cytogenet. Cell Genet. 1998, 80, 165–172. [Google Scholar] [CrossRef] [PubMed]
- Modi, W.S. Rapid, localized amplification of a unique satellite DNA family in the rodent Microtus chrotorrhinus. Chromosoma 1993, 102, 484–490. [Google Scholar] [CrossRef] [PubMed]
- Modi, W.S. Heterogeneity in the concerted evolution process of a tandem satellite array in meadow mice (Microtus). J. Mol. Evol. 1993, 37, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Modi, W.S. Comparative analyses of heterochromatin in Microtus: Sequence heterogeneity and localized expansion and contraction of satellite DNA arrays. Cytogenet. Cell Genet. 1993, 62, 142–148. [Google Scholar] [CrossRef] [PubMed]
- Modi, W.S.; Serdyukova, N.A.; Vorobieva, N.V.; Graphodatsky, A.S. Chromosomal localization of six repeated DNA sequences among species of Microtus (Rodentia). Chromosome Res. 2003, 11, 705–713. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, S.V.; Modi, W.S. Molecular characterization of the complex sex-chromosome heterochromatin in the rodent Microtus chrotorrhinus. Cytogenet. Cell Genet. 1996, 75, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Khrapov, E.A.; Elisafenko, E.A.; Rogozin, I.B.; Pavlova, S.V.; Vorob’eva, N.V.; Serdyukova, N.A.; Sablina, O.V.; Grafodatskii, A.S.; Zakiyan, S.M. Novel family of STR47 tandem repeats in the Microtus rossiaemeridionalis genome. Mol. Biol. 1998, 32, 830–834. [Google Scholar]
- Acosta, M.J.; Marchal, J.A.; Mitsainas, G.P.; Rovatsos, M.T.; Fernández-Espartero, C.H.; Giagia-Athanasopoulou, E.B.; Sánchez, A. A new pericentromeric repeated DNA sequence in Microtus thomasi. Cytogenet. Genome Res. 2009, 124, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Acosta, M.J.; Marchal, J.A.; Fernández-Espartero, C.; Romero-Fernández, I.; Rovatsos, M.T.; Giagia-Athanasopoulou, E.B.; Gornung, E.; Castiglia, R.; Sánchez, A. Characterization of the satellite DNA Msat-160 from species of Terricola (Microtus) and Arvicola (Rodentia, Arvicolinae). Genetica 2010, 138, 1085–1098. [Google Scholar] [CrossRef] [PubMed]
- Mayorov, V.I.; Adkison, L.R.; Vorobyeva, N.V.; Khrapov, E.A.; Kholodhov, N.G.; Rogozin, I.B.; Nesterova, T.B.; Protopopov, A.I.; Sablina, O.V.; Graphodatsky, A.S.; et al. Organization and chromosomal localization of a B1-like containing repeat of Microtus subarvalis. Mamm. Genome 1996, 7, 593–597. [Google Scholar] [CrossRef] [PubMed]
- Elisaphenko, E.A.; Nesterova, T.B.; Duthie, S.M.; Ruldugina, O.V.; Rogozin, I.B.; Brockdorff, N.; Zakian, S.M. Repetitive DNA sequences in the common vole: Cloning, characterization and chromosome localization of two novel complex repeats MS3 and MS4 from the genome of the East European vole Microtus rossiaemeridionalis. Chromosome Res. 1998, 6, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Acosta, M.J.; Marchal, J.A.; Fernández-Espartero, C.; Bullejos, M.; Sánchez, A. Retroelements (LINES and SINES) in vole genomes: Differential distribution in the constitutive heterochromatin. Chromosome Res. 2008, 16, 949–959. [Google Scholar] [CrossRef] [PubMed]
- Marchal, J.A.; Acosta, M.J.; Bullejos, M.; Díaz de la Guardia, R.; Sánchez, A. A repeat DNA sequence from the Y chromosome in species of the genus Microtus. Chromosome Res. 2004, 12, 757–765. [Google Scholar] [CrossRef] [PubMed]
- Kholodilov, N.G.; Mayorov, V.I.; Mullokandov, M.R.; Cheryaukene, O.V.; Nesterova, T.B.; Rogozin, I.B.; Zakian, S.M. LINE-1 element in the vole Microtus subarvalis. Mamm. Genome 1993, 4, 624–626. [Google Scholar] [CrossRef] [PubMed]
- Marchal, J.A.; Acosta, M.J.; Bullejos, M.; Puerma, E.; Díaz de la Guardia, R.; Sánchez, A. Distribution of L-retroposons on the giant sex chromosomes of Microtus cabrerae (Arvicolidae, Rodentia): Functional and evolutionary implications. Chromosome Res. 2006, 14, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Marchal, J.A.; Acosta, M.J.; Bullejos, M.; Díaz de la Guardia, R.; Sanchez, A. Origin and spread of the SRY gene on the X and Y chromosomes of the rodent Microtus cabrerae: Role of L1 elements. Genomics 2008, 91, 142–151. [Google Scholar] [CrossRef] [PubMed]
- Rovatsos, M.T.; Marchal, J.A.; Romero-Fernández, I.; Fernández, F.J.; Giagia-Athanosopoulou, E.B.; Sánchez, A. Rapid, independent, and extensive amplification of telomeric repeats in pericentromeric regions in karyotypes of arvicoline rodents. Chromosome Res. 2011, 19, 869–882. [Google Scholar] [CrossRef] [PubMed]
- Marchal, J.A.; Acosta, M.J.; Nietzel, H.; Sperling, K.; Bullejos, M.; Díaz de la Guardia, R.; Sánchez, A. X chromosome painting in Microtus: Origin and evolution of the giant sex chromosomes. Chromosome Res. 2004, 12, 767–776. [Google Scholar] [CrossRef] [PubMed]
- Sitnikova, N.A.; Romanenko, S.A.; O’Brien, P.C.; Perelman, P.L.; Fu, B.; Rubtsova, N.V.; Serdukova, N.A.; Golenishchev, F.N.; Trifonov, V.A.; Ferguson-Smith, M.A.; et al. Chromosomal evolution of Arvicolinae (Cricetidae, Rodentia). I. The genome homology of tundra vole, field vole, mouse and golden hamster revealed by comparative chromosome painting. Chromosome Res. 2007, 15, 447–456. [Google Scholar] [CrossRef] [PubMed]
- Acosta, M.J.; Romero-Fernández, I.; Sánchez, A.; Marchal, J.A. Comparative analysis by chromosome painting of the sex chromosomes in Arvicolid rodents. Cytogenet. Genome Res. 2011, 132, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Jaarola, M.; Martinkova, N.; Gunduz, I.; Brunhoff, C.; Zima, J.; Nadachowski, A.; Amori, G.; Bulatova, N.S.; Chondropoulos, B.; Fraguedakis-Tsolis, S.; et al. Molecular phylogeny of the speciose vole genus Microtus (Arvicolinae, Rodentia) inferred from mitochondrial DNA sequences. Mol. Phylogenet. Evol. 2004, 33, 647–663. [Google Scholar] [CrossRef] [PubMed]
- Cuenca-Bescós, G.; López-García, J.M.; Galindo-Pellicena, M.A.; García-Perea, R.; Gisbert, J.; Rofes, J.; Ventura, J. Pleistocene history of Iberomys, an endangered endemic rodent from southwestern Europe. Integr. Zool. 2014, 9, 481–497. [Google Scholar] [CrossRef] [PubMed]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Funke, R. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef] [PubMed]
- Waterston, R.H.; Lindblad-Toh, K.; Birney, E.; Rogers, J.; Abril, J.F.; Agarwal, P.; Agarwala, R.; Ainscough, R.; Alexandersson, M.; An, P.; et al. Initial sequencing and comparative analysis of the mouse genome. Nature 2002, 420, 520–562. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, R.A.; Weinstock, G.M.; Metzker, M.L.; Muzny, D.M.; Sodergren, E.J.; Scherer, S.; Scott, G.; Steffen, D.; Worley, K.C.; Burch, P.E.; et al. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature 2004, 428, 493–521. [Google Scholar] [CrossRef] [PubMed]
- Zenzes, M.T.; Wolf, U. Pairing behaviour of the sex chromosomes during male meiosis of Microtus agrestis. Chromosoma 1971, 33, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Jiménez, R.; Carnero, A.; Burgos, M.; Sánchez, A.; Díaz de la Guardia, R. Achiasmatic giant sex chromosomes in the vole Microtus cabrerae (Rodentia, Microtidae). Cytogenet. Cell Genet. 1991, 57, 56–58. [Google Scholar] [CrossRef] [PubMed]
- Rovatsos, M.T.; Marchal, J.A.; Romero-Fernández, I.; Arroyo, M.; Athanasopoulou, E.B.; Sánchez, A. Extensive sex chromosome polymorphism of Microtus thomasi/Microtus atticus species complex associated with cryptic chromosomal rearrangements and independent accumulation of heterochromatin. Cytogenet. Genome Res. 2017, 151, 198–207. [Google Scholar] [CrossRef] [PubMed]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lamelas, L.; Arroyo, M.; Fernández, F.J.; Marchal, J.A.; Sánchez, A. Structural and Evolutionary Relationships in the Giant Sex Chromosomes of Three Microtus Species. Genes 2018, 9, 27. https://doi.org/10.3390/genes9010027
Lamelas L, Arroyo M, Fernández FJ, Marchal JA, Sánchez A. Structural and Evolutionary Relationships in the Giant Sex Chromosomes of Three Microtus Species. Genes. 2018; 9(1):27. https://doi.org/10.3390/genes9010027
Chicago/Turabian StyleLamelas, Luz, María Arroyo, Francisco Javier Fernández, Juan Alberto Marchal, and Antonio Sánchez. 2018. "Structural and Evolutionary Relationships in the Giant Sex Chromosomes of Three Microtus Species" Genes 9, no. 1: 27. https://doi.org/10.3390/genes9010027




