1. Introduction
As with most animal production systems, the cost of feed in chicken meat production is a high proportion of the total farming expense, being nearly 70% of the total cost of poultry production [
1]. The selection of more efficient animals reduces production costs, decreases the area of land required for feed production and reduces environmental impact of nitrogen and phosphorus excretion resulting from the digestion/metabolic processes.
Residual feed intake (RFI), defined as the difference between the actual feed intake (FI) and predicted requirements based on growth and maintenance, was first proposed as an alternate measure of feed efficiency (FE) by Koch et al. [
2]. RFI is a sensitive and accurate indicator that is increasingly accepted as an alternative measure of FE of livestock. RFI has a moderate heritability (0.21 to 0.49) in broilers and responds to selection [
3]. Elucidation of the genomic variants for RFI could potentially be important for marker- or gene-assisted selection [
4].
Current whole genomic sequencing methodologies are important tools to unravel the mechanisms of complex traits, facilitate new understanding of the genetic regulation of phenotype and allow the identification of potential biomarkers for early or more accurate genetic prediction [
5]. In recent years, some progress has been gained in attempts to identify the molecular mechanisms underlying RFI. Transcriptome analysis of mRNA and microRNA in skeletal muscle in pigs showed that genes involved in mitochondrial energy metabolism were downregulated, while those involved in skeletal muscle differentiation and proliferation were upregulated in low RFI (LRFI), as compared to high RFI (HRFI) animals [
6]. The duodenal transcriptome architecture of extreme RFI phenotypes of six brown-egg dwarf hens showed that significant differentially-expressed genes were involved in several specific biological functions related to the processes of digestibility, metabolism, biosynthesis and energy homeostasis [
7]. Combination analysis of genome-wide association and transcriptome sequencing in chickens showed that the effective single-nucleotide polymorphisms (SNPs) related to energy utilization were located in a 1-Mb region (16.3–17.3 Mb) of chicken chromosome 12, and liver transcriptomes may contribute to RFI, which is directly influenced by appetite, cell activities and fat metabolism [
8]. Genome-level analyses of genetic markers and candidate genes for RFI in pigs by genome-wide associations and systems genetic analyses revealed that the SNPs of
MAP3K5,
PEX7 and
DSCAM might be interesting markers for RFI. Functional annotation of genes indicated that regulation of protein and lipid metabolism, gap junction formation, inositol phosphate metabolism and the insulin signaling pathway were significant biological processes and pathways associated with RFI [
4]. Genome-wide association analysis of FI and RFI in Nellore cattle identified several markers located on chromosomes 4, 8, 14 and 21 in regions near genes regulating appetite and ion transport [
9].
There are more than one hundred domestic chicken breeds available across China, and collectively, they have roughly the same market share in China as do introduced commercial lines. Beijing-You chickens, one of the representative domestic chickens, are known for their peculiar characteristics including higher intramuscular fat content and chewy texture [
10], which are appealing palatability factors for Chinese consumers. The Cobb line has been intensely selected for high feed efficiency and growth rate and is an important commercial breed world-wide. To gain insights into the molecular alterations underlying differences between HRFI and LRFI chickens, whole genome sequencing (WGS) was conducted using HRFI and LRFI birds of local and commercial breeds.
4. Discussion
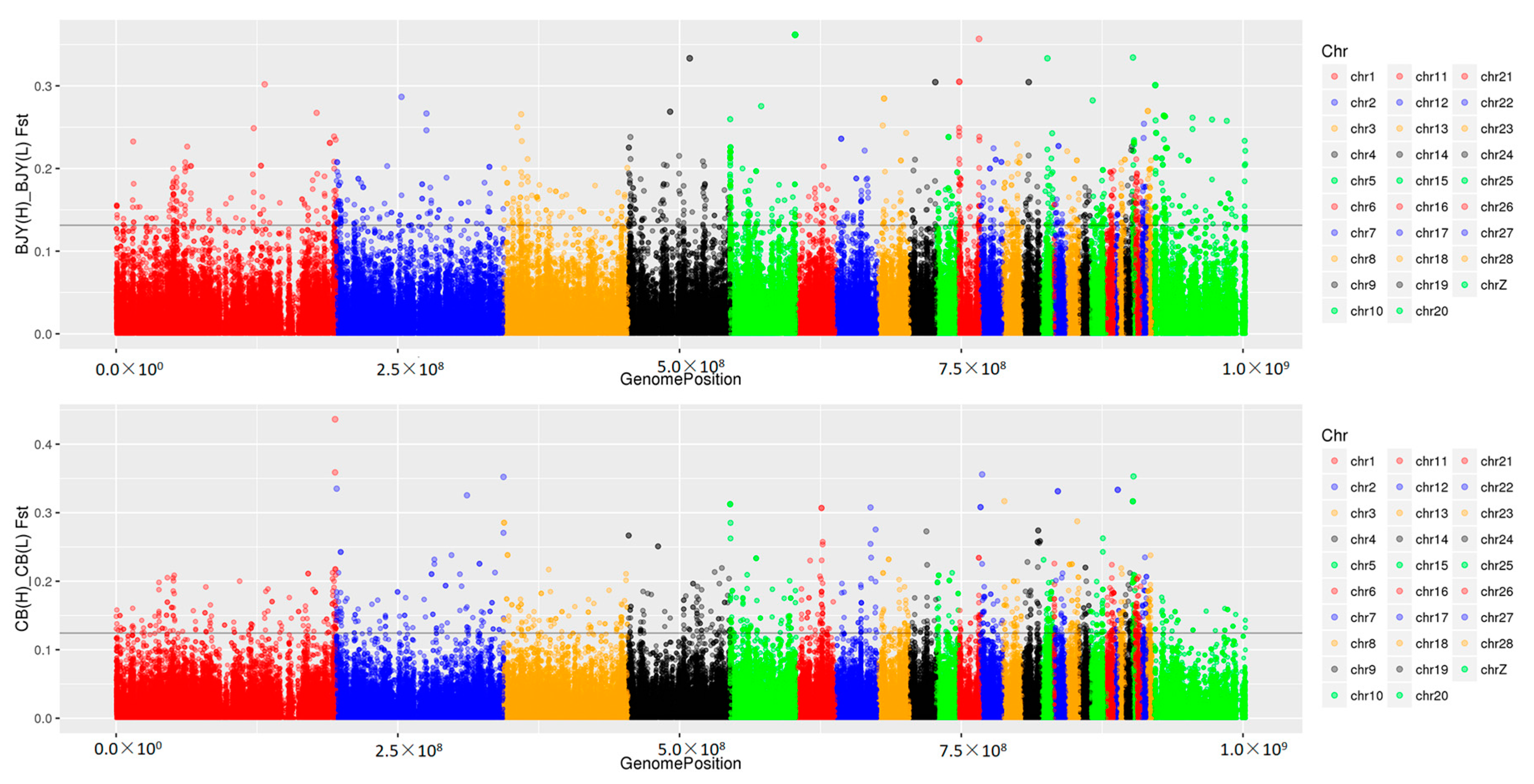
Genetic improvement of economically-important traits, such as growth rate and feed efficiency, has occurred for about 100 years in the Cobb chickens. There has been no systematic genetic selection in the conservation population of the local Beijing-You chickens. Consistent with the current findings, more genomic variants exist in the Beijing-You than Cobb chickens, because the high intensity of genetic selection in the latter would be expected to result in genomic regions with increased homozygosity. There were numerous SNPs associated with RFI in the Beijing-You and a larger proportional difference in FCR (HRFI/LRFI = 1.2), suggesting a high potential for genetically-increasing feed efficiency in Beijing-You chickens. Obviously, FCR is already much lower in Cobb chickens, and there was less difference (HRFI/LRFI = 1.08) between the phenotypic extremes.
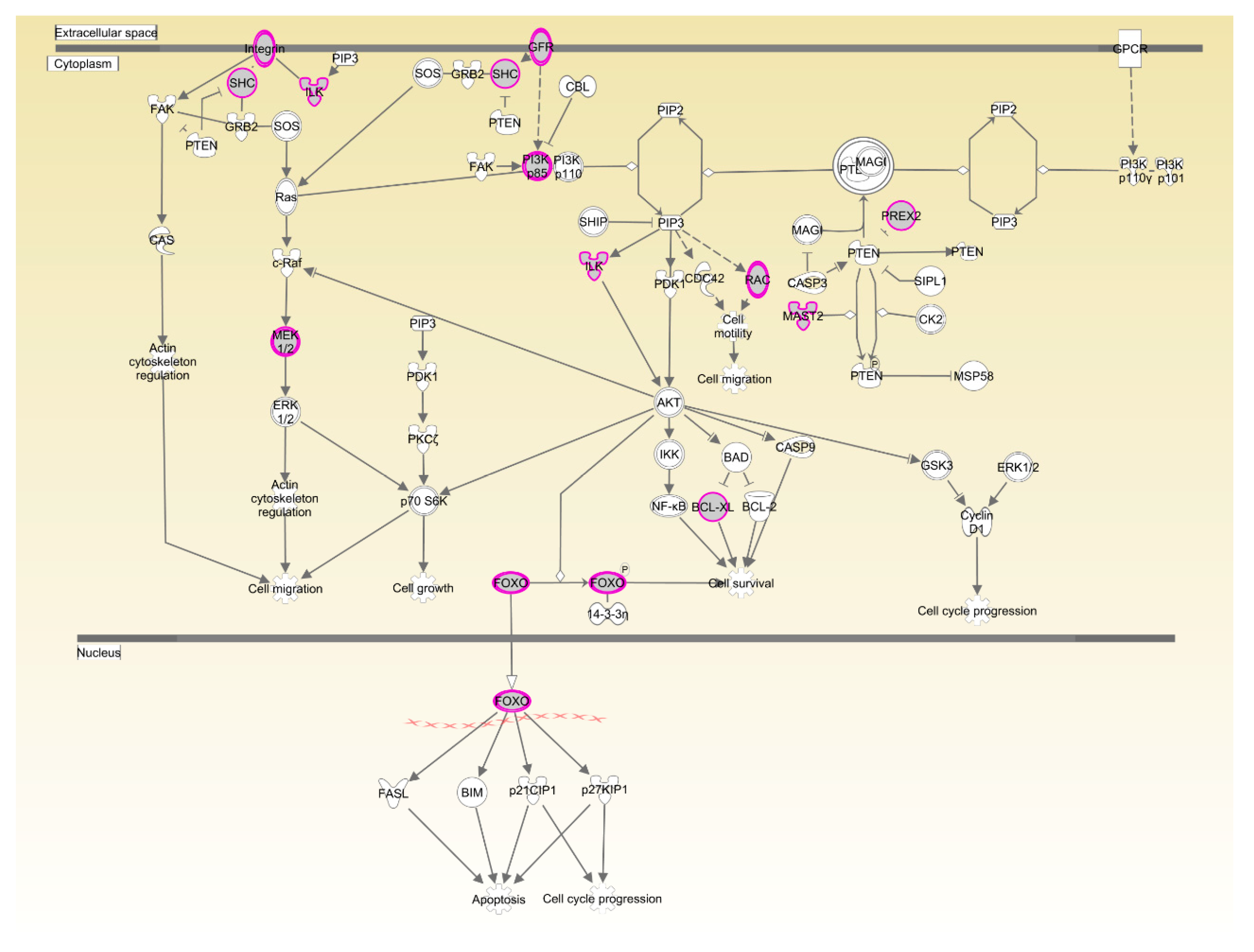
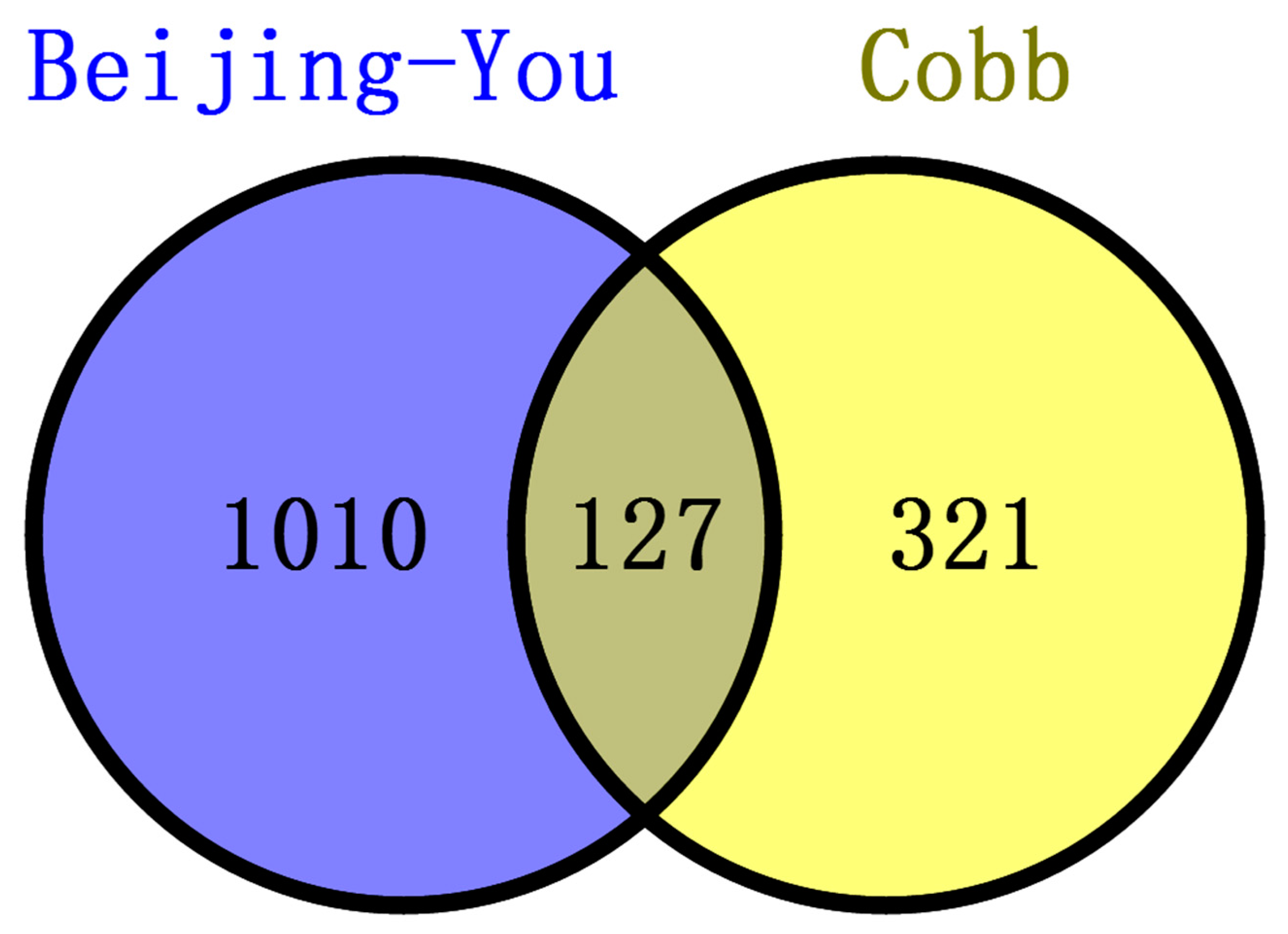
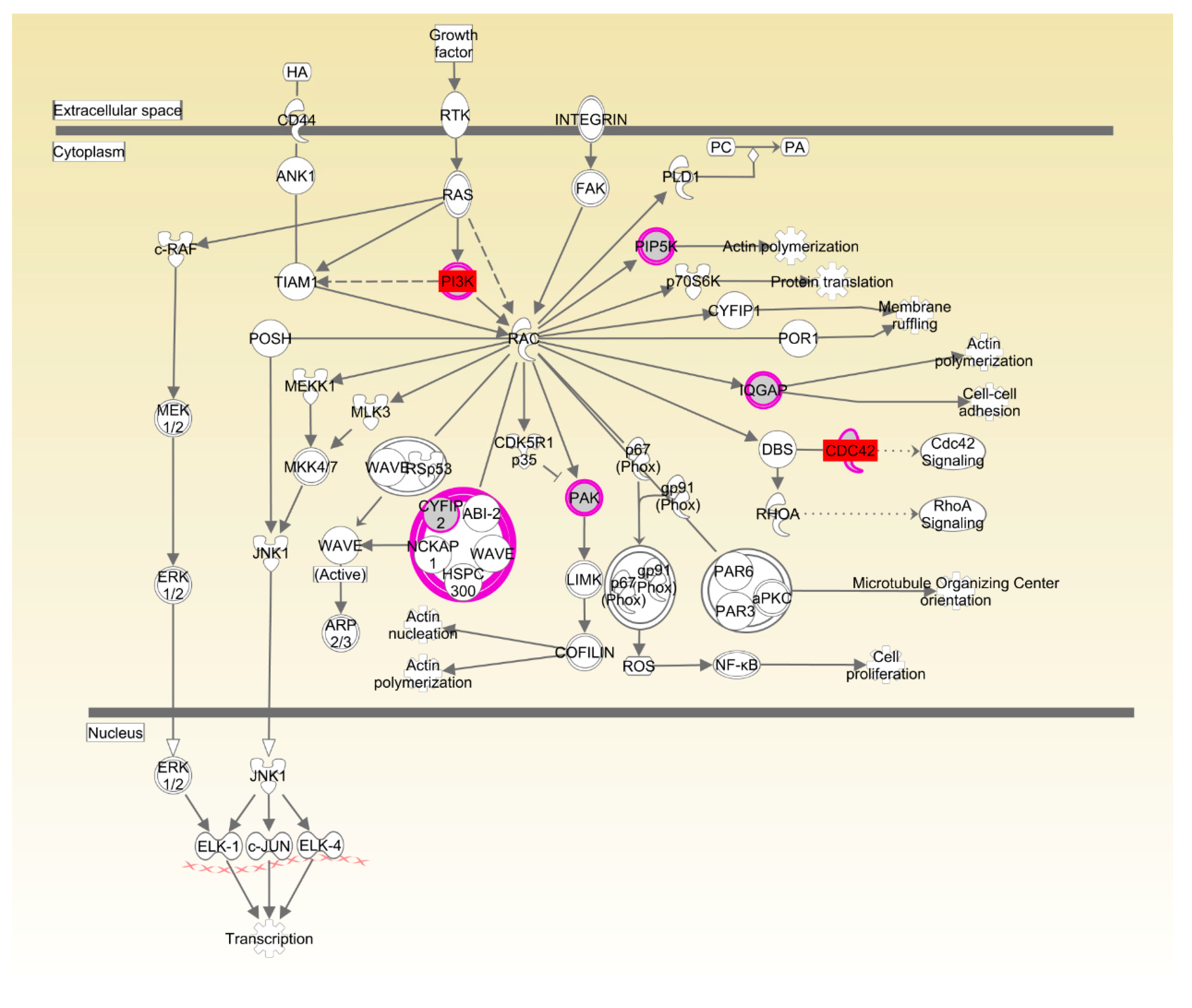
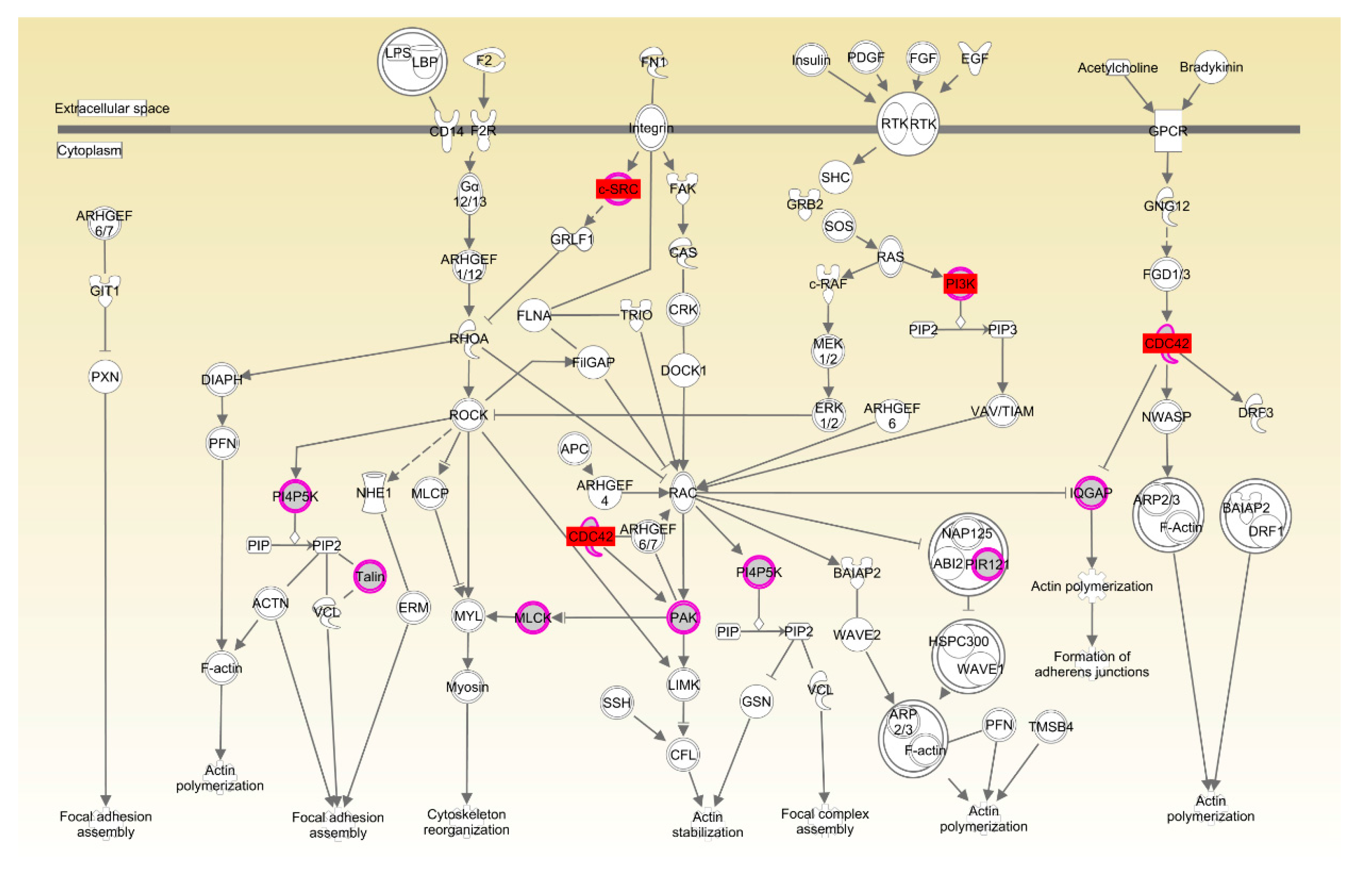
The two breeds examined here showed some unique characteristics, and only 127 genes associated with RFI were identified in both breeds. These represented a small proportion of the total (127/1137) for Beijing-You, but much higher (127/448) for Cobb chickens. PTEN signaling, as well as lipid and carbohydrate metabolism were only enriched in Beijing-You chickens. The PTEN/PI3K/AKT signaling pathway has been shown to regulate cell metabolism, growth and survival [
16,
17]. PTEN/PI3K/AKT signaling can mediate the insulin-stimulated activation of many major pathways of hepatic lipid metabolism, including lipogenesis, fatty acid β-oxidation, fatty acid esterification to triglycerides and very low density lipoprotein-triglyceride (VLDL-TG) secretion [
18,
19]. PTEN, a negative regulator, dephosphorylates PIP3 to inhibit the activity of PI3K/AKT signaling [
20]. When the PTEN was overexpressed, insulin signaling effects were inhibited, resulting in decreased RAC-alpha serine/threonine-protein kinase (AKT) activity and solute carrier family 2, facilitated glucose transporter member 4 (GLUT4) translocation to the cell membrane, which consequently contributed to insulin resistance and progression of non-alcoholic fatty liver disease [
21,
22,
23]. In addition to controlling the activity of signaling intermediates at the cell membrane, PI3Ks can drive gene expression programs in the nucleus. The forkhead box protein O1(FOXO) transcription factors are coupled to PI3K activity through AKT-dependent phosphorylation [
24], which are considered effectors of the pathway regulating hepatic glucose production. Matsumoto et al. [
25] showed that delivery of FOXO1 to mouse liver resulted in steatosis arising from increased triglyceride accumulation and decreased fatty acid oxidation. In the present study, the genes (
PHKG1,
PIP5K1B and
NEU3) that participate in carbohydrate and lipid metabolism [
26,
27,
28] were upregulated in the livers of HRFI Beijing-You chickens. Previous studies [
5,
29] have demonstrated that HRFI animals required more energy to maintain biological activities, such as protein turnover, stress response and muscle activity. Beijing-You chickens are characterized by high fat deposition in muscle and abdominal adipose [
30]. Therefore, lipid and carbohydrate metabolism may play crucial roles in RFI through the PTEN/PI3K/AKT signaling pathway in Beijing-You chickens.
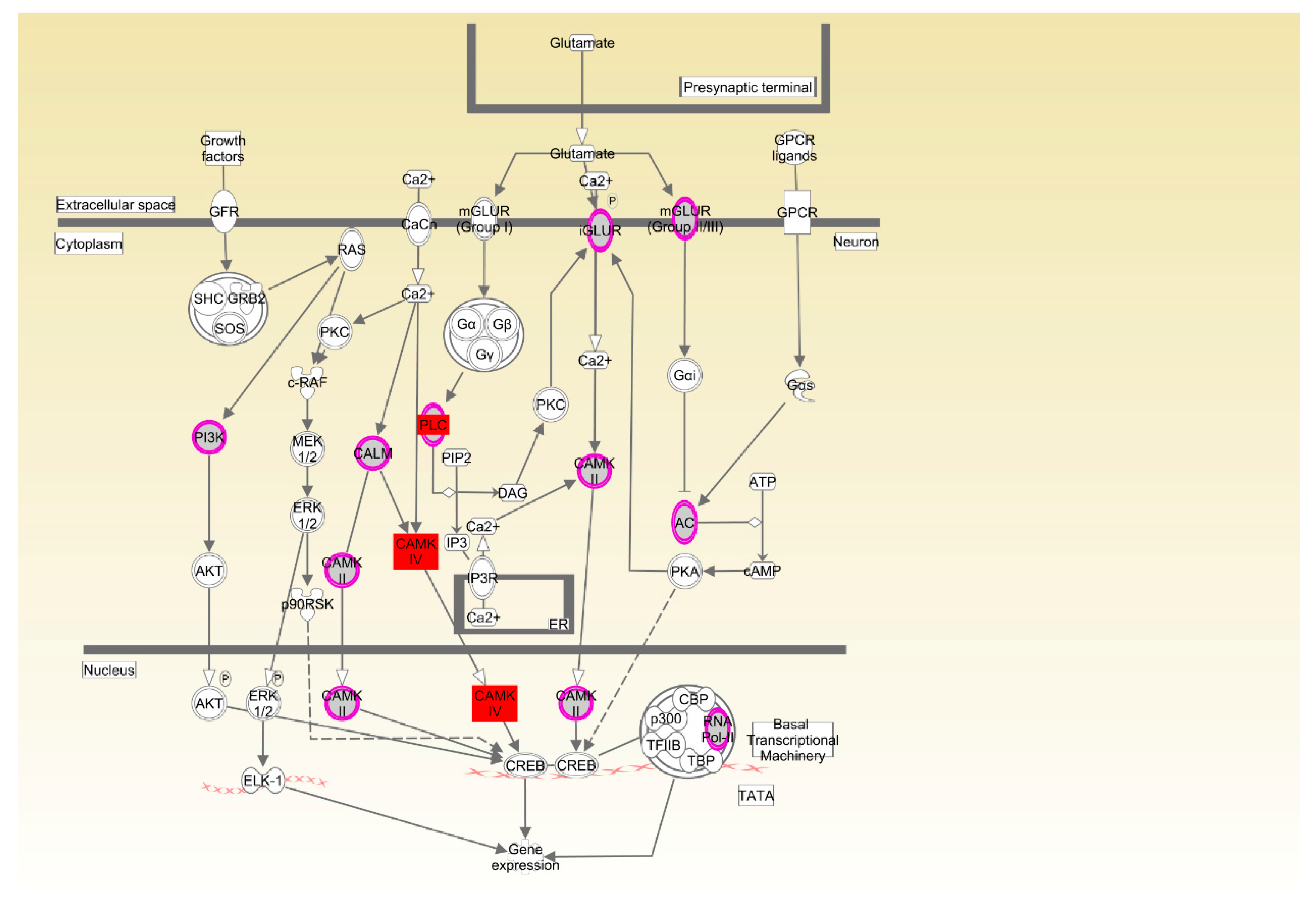
For Cobb chickens, the results showed that feed behavior and the related pathway (CREB signaling in neurons) were enriched. Feed behavior plays a key role in feed intake, and variation in feed intake is associated with variation in RFI. The CREB transcription factor has been widely investigated as a key factor in the control of appetite and FI by the hypothalamus [
31]. A previous report showed that an increase and phosphorylation of CREB in the hypothalamus, as a result of injection of NPY into the rat hypothalamus or fasting for 48 h, suggested that CREB phosphorylation might be involved in the regulation of feeding behavior [
32]. Calcium/calmodulin-dependent protein kinase (CAMK) plays a critical role in activation of CREB via Ser-133 phosphorylation [
33].
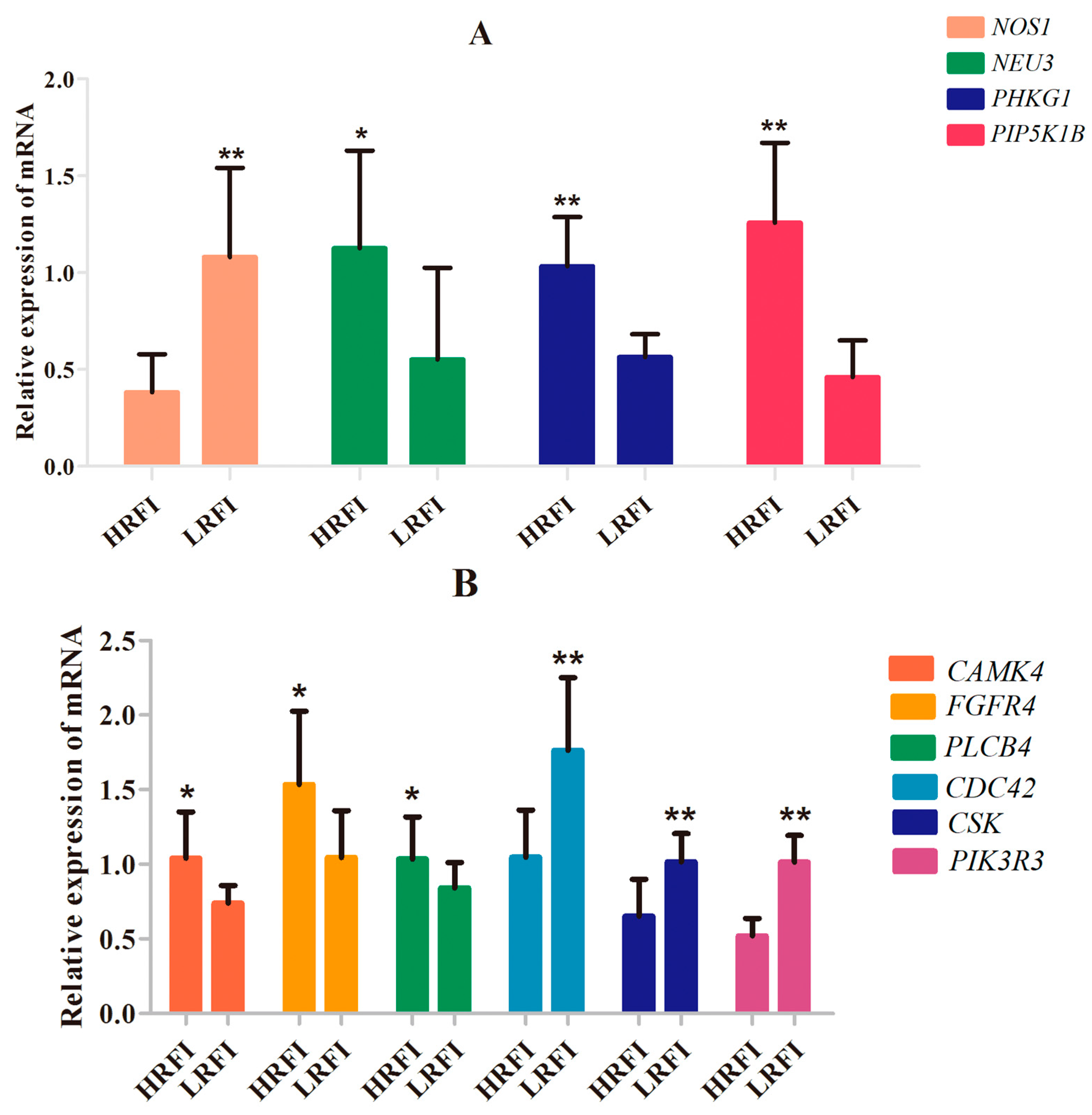
PLCB4 and
CAMK4 mRNA levels were higher in the hypothalamus of HRFI than LRFI chickens. The upregulation of
CAMK4 mRNA levels may increase FI through induction of CREB phosphorylation and activation of downstream signaling pathways, and upregulation of
PLCB4 expression may affect activation of CAMKs through changes in Ca
2+ concentrations. Therefore, the CREB signaling pathway may contribute to RFI in Cobb chickens by affecting feed behavior. Compared to the Beijing-You, the Cobb is a specialized broiler breed as a result of long-term selection for increased feed efficiency and growth rate, which may have resulted in part from changes in feed behavior and its related genetic basis.
Organismal development as the common physiological process was enriched in both breeds. The concept of RFI was initially developed to determine differences among animals in feed conversion into body tissue. A previous study in cattle showed a variation in RFI of 37%, which was explained by tissue metabolism, protein turnover and stress response [
34]. Global RNA expression in breast muscle obtained from a male broiler line phenotyped for high or low feed efficiency showed that significant differences in gene expression profiles were associated with muscle fiber development, muscle function and cytoskeletal organization [
35]. In the present study, skeletal and muscular system development and function were enriched in both Beijing-You and Cobb breeds. Phenotypic measurement showed that the percentages of breast and thigh muscle were higher in LRFI than HRFI Beijing-You chickens (
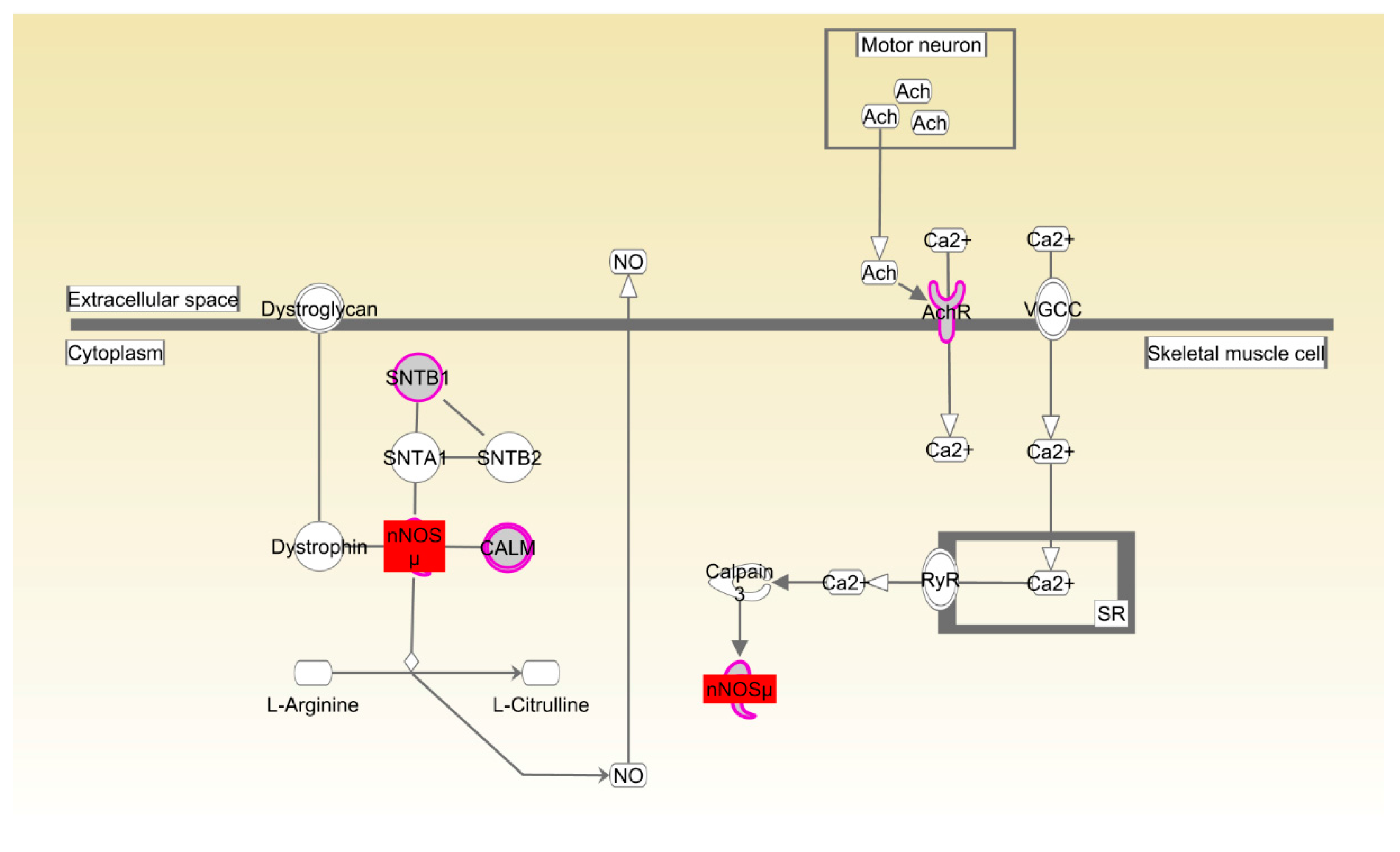
Table S8), suggesting that the development and function of the skeletal and muscular systems might control efficient conversion of dietary nutrients to lean tissue and growth. The canonical pathways that participated in the organismal development were also enriched in both Beijing-You and Cobb breeds. nNOS plays an important role in the regulation of many muscle functions, such as blood flow, contraction and metabolism [
36]. For example, nitric oxide (NO), which is physiologically synthesized in a tightly-controlled manner by NO synthases (NOSs), directly inhibits mitochondrial respiration by interfering with electron transport chain complexes, which consequently affects skeletal muscle metabolism and stimulates glucose transport by activating upstream signaling that leads to increased production of GLUT4 [
37]. Rac and actin cytoskeleton signaling has been identified in various morphogenetic events associated with tissue organization during organ development. For instance, the conserved role of paxillin in promoting signals from RAC to the actin cytoskeleton is crucial for normal tissue morphogenesis during wing and leg development of
Drosophila [
38]. Ras-related C3 botulinum toxin substrate 1 (Rac1) and Cell division control protein 42 homolog (CDC42) have also been implicated in neural development and myogenesis [
39]. CSK-1 was also essential in the organization of pharyngeal muscle filaments [
40]. In the present study, expression in muscle of
CDC42,
CSK and
PIK3R was significantly upregulated in LRFI chickens, as compared to HRFI chickens. Therefore, all of these results showed that although there were small breed differences in the participating pathways, organismal development as the common physiological process contributed to the RFI in the two breeds.
In order to further validate the reliability of the WGS results, 191 SNPs were selected for genotyping and association analysis. The results showed that there were 46 SNPs (48 genes) significantly associated with the RFI, which demonstrated that the WGS result was reliable, and these SNPs could prove to be interesting markers for RFI.