Translational Advances of Hydrofection by Hydrodynamic Injection
Abstract
:1. Introduction
2. Hydrodynamic Model
2.1. Hydrodynamic Injection of Naked Nucleic Acids
- (a)
- The DNA reached the muscle and was expressed [6].
- (b)
- The mouse liver could be transfected in vivo by the pressurized injection of a hyperosmotic solution containing naked human growth hormone (hGH) DNA through the portal vein [7], achieving plasma protein levels (65 ng/mL) 50-fold higher than the normal basal values.
- (c)
- High expression levels of tracer genes were achieved in the hind limb muscle of rats by pressurized DNA injection through the iliac artery [8].
2.2. Delivery Mechanism
- (1)
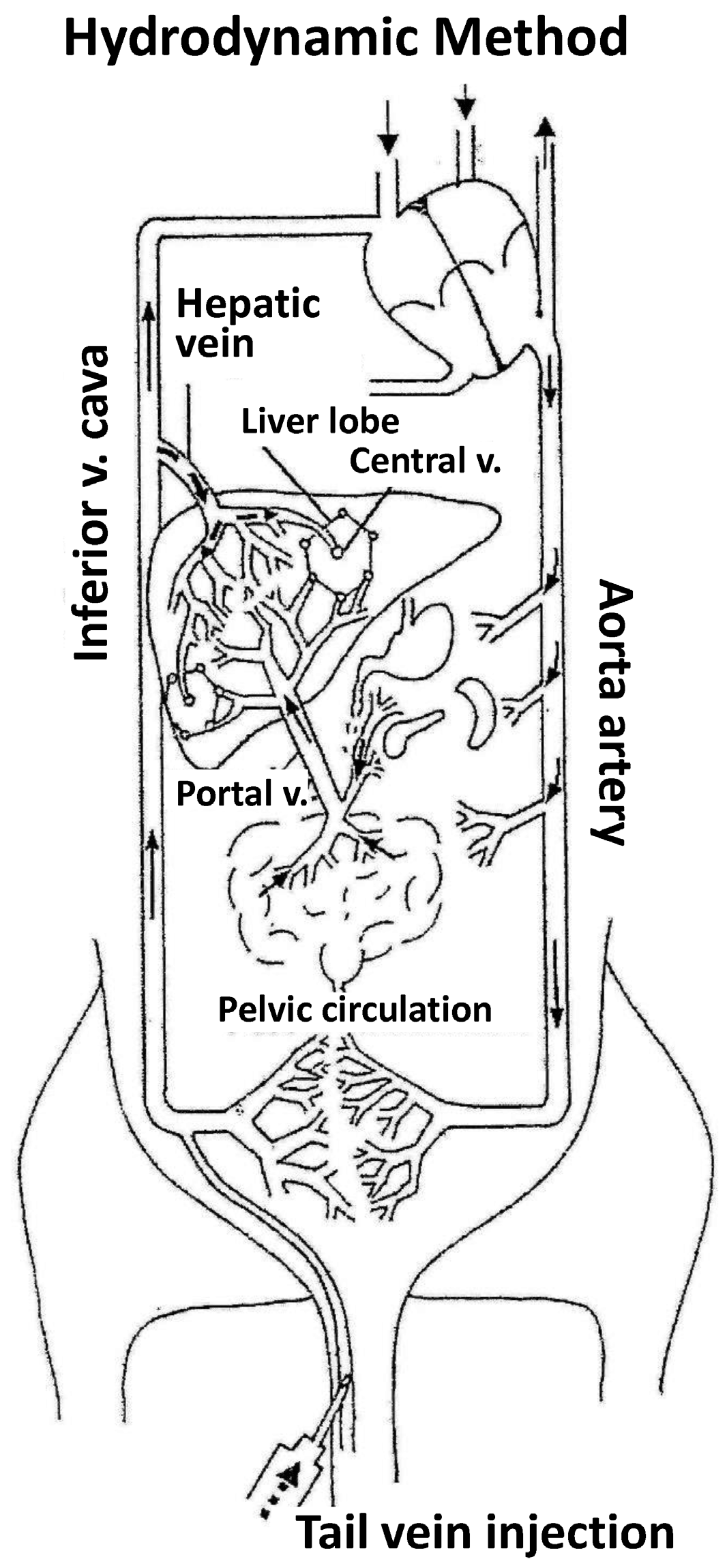
- The first experiments employing tracer genes and immunohistochemical methods to identify the liver expression of exogenous genes showed expression to be mainly located in central vein areas, thereby supporting that the inversion of flow sense mediates retrograde backflow to the liver and this could be involved in the mechanism of action (Figure 1).
- (2)
- Hydrodynamic injection mediates a remarkable pressure increase in the cava vein, since the tail vein is its direct tributary. This is due to the fact that the administration of a large volume (2 mL in the mouse) in this area (cardiac preload) means doubling the volemia—thereby generating an important pressure increase. This process inverts the pressures at cava vein level with respect to the portal vein and causes retrograde backflow of blood to the liver. This idea is supported by experimental data. The simultaneous measurement of pressures in portal and cava vein areas shows that inversion of portal versus cava pressures exists both during injection and at least 5 min later.
- (3)
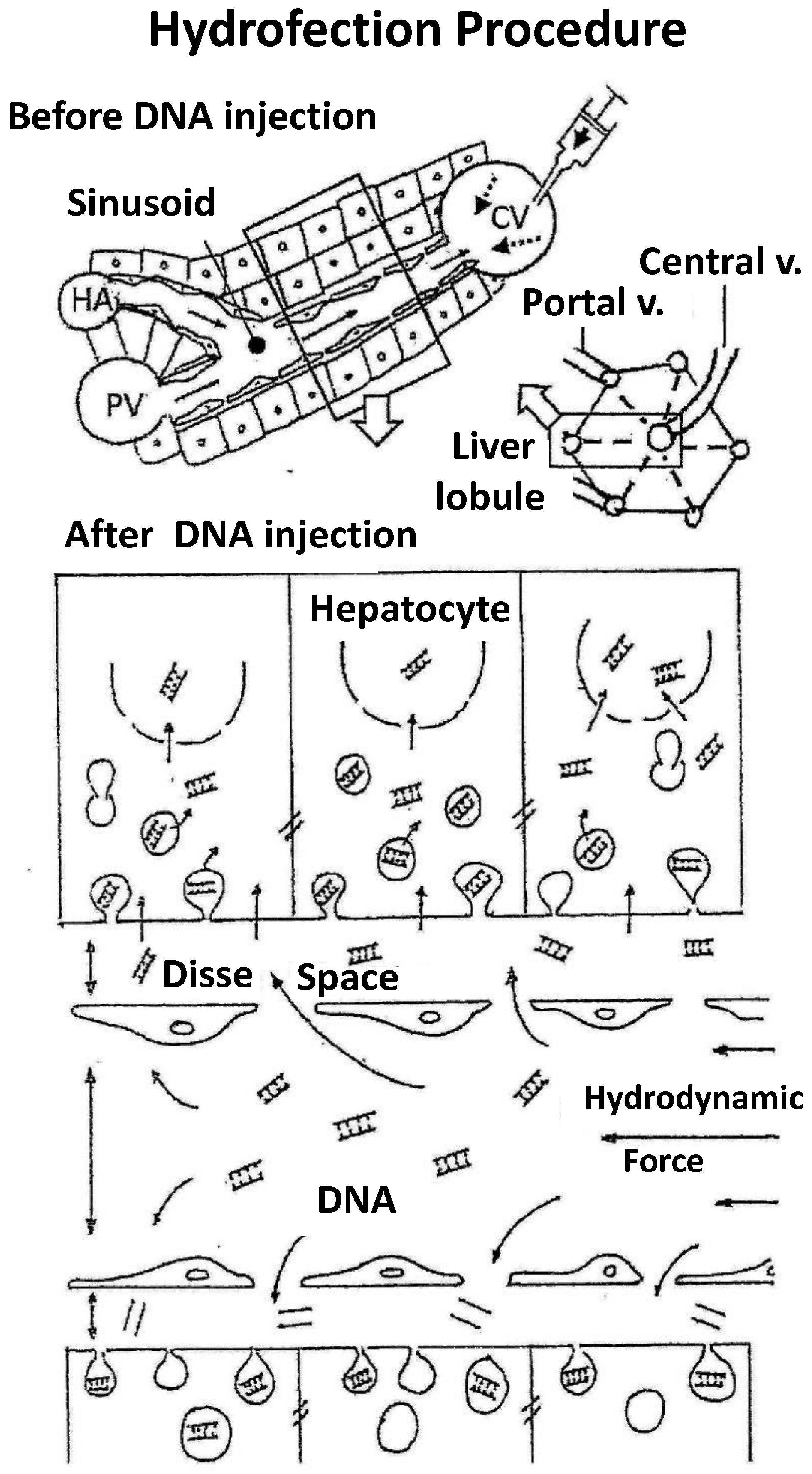
- Flow inversion has been observed by intravital microscopy [17] during hydrodynamic injection and minutes later. During rapid injection, flow is inverted and remains static but pulsatile during the first minute. Anterograde flow recovers slowly and progressively until normalization is reached within 5 or 6 min after hydrodynamic injection. It is assumed that the transfection process occurs during this short period of time.
- (4)
- Ultrastructural morphological changes [17] in liver tissue during hydrodynamic injection evidence that the hydrodynamic force exerted upon the liver sinusoids promotes vascular distension, widening sinusoidal pore diameter and the endothelial junctions and facilitating access of the DNA solution to the likewise widened virtual Disse space (Figure 2). This process allows interaction of the aqueous solution with hepatocytes in a high-pressure scenario that promotes the formation of multiple endocytic vesicles without solution of continuity of the cell membrane, as observed by electron microscopy. This excludes the possibility of DNA access to the hepatocyte through membrane disruption and the formation of large permissive pores instead of endocytic vesicles. Moreover, large pores could compromise hepatocyte viability. It must be underscored that the existence of narrow junctions (tight junctions) among hepatocytes limits free DNA diffusion through the intercellular spaces. Thus, hepatocyte hydrofection (gene transfer mediated by hydric forces) could be due to DNA saline solution entry into the hepatocyte, which is mediated by the hydrodynamic force exerted.
- (5)
- Although the possibility that part of the DNA may access the hepatocyte through a receptor-mediated process [7] cannot be discarded, this process would contribute only slightly to the efficiency of hydrodynamic delivery.
- (6)
- The early experimental data, employing molecules of different size and weight, as well as recent observations employing colloidal gold nanoparticles of known diameter and electron microscopy [18,19], support the idea that hydrofection implies a passive process without energy consumption but driven by hydrodynamic force through more permissive sites of the cell membrane, including endocytic vesicles. The dimension of the membrane sinusoids, depending on the species, can be as much as 100 nm in diameter [20]. However, only particles with diameters smaller than 10 nm can access the cell [21], whereas larger particles are virtually refractory to hepatocyte entry but can be observed within the cytoplasm of phagocytic cells (Kupffer cells). This interpretation of the delivery mechanism combines DNA access to the hepatocyte through the cell membrane with no important liver toxicity and justifies the acute plasma increase in transaminases due to marginal cell destruction, which is rapidly reverted within the first days after hydrodynamic injection. This suggests that injured cells are eliminated, whereas the efficiency of transfection remains in those cells in which DNA has gained access in a less aggressive manner. Nevertheless, further studies are needed to establish the exact mechanism of gene delivery mediated by the hydrodynamic procedure.
3. Gene Transfer Applications
3.1. Muscle
3.2. Liver
3.3. Other Inherited Diseases
3.4. Infectious Diseases
3.5. Cancer
3.6. Other Acquired Diseases
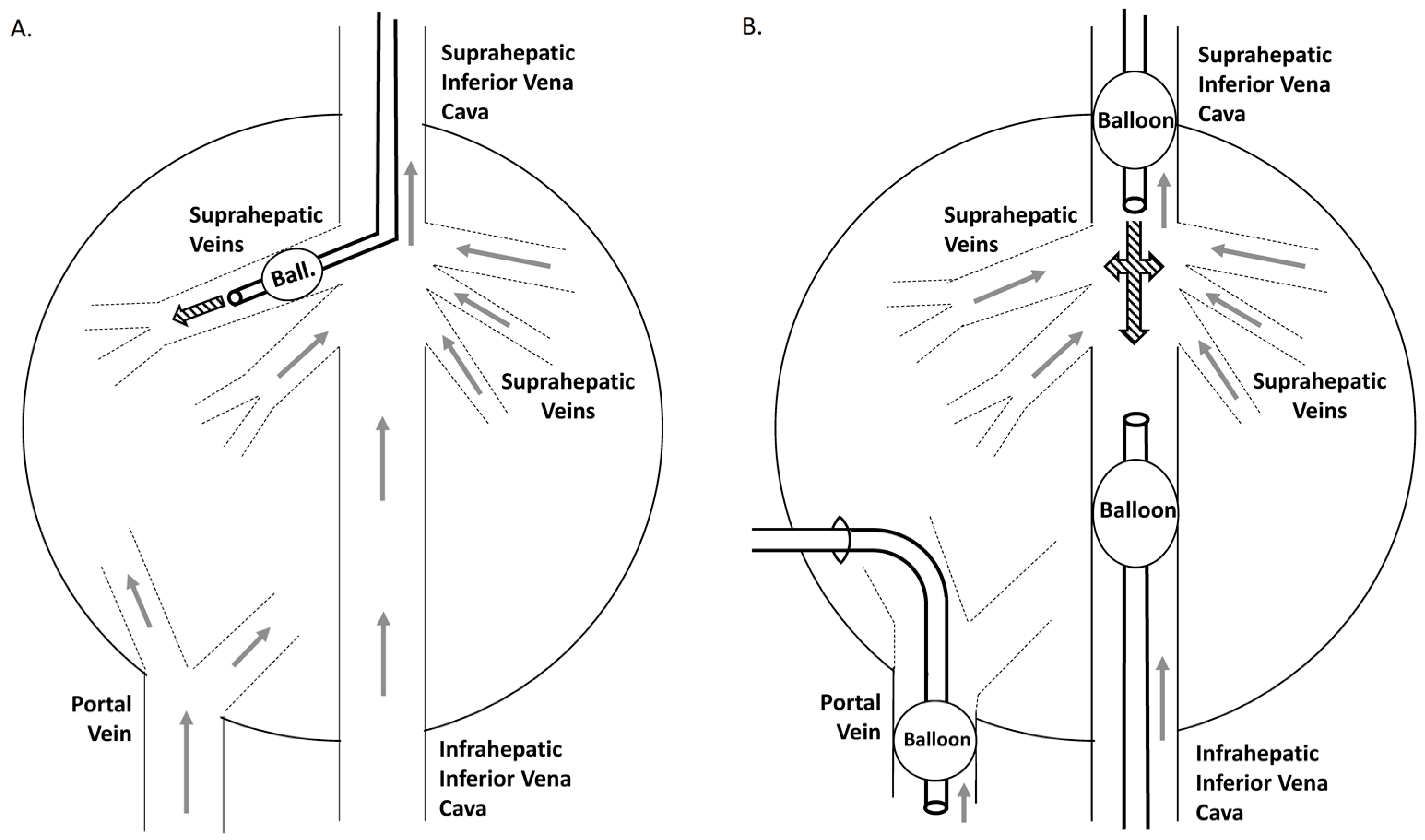
4. Translation of the Hydrodynamic Method: From Mouse to Large Animals
4.1. Rodents
4.2. Rabbit to Pig
4.3. Primates
5. Parameters of the Genes Transfer Process
- (a)
- Indexes, for the absolute number of copies of each molecular species (DNA, RNA or protein) referred to a normalized cell.
- (b)
- Intrinsic activities, representing the index ratio between consecutive steps (transcription (RNA/DNA) and translation (protein/RNA)) of the decoding process, in order to evaluate how efficient each step is, in a normalized cell.
- (c)
- Expression efficacy, defining the final efficacy of the procedure in the tissue relating the amount of protein copies per gene copy, in a normalized cell.
6. Hydrodynamic Genes Transfer to Human Organ Ex Vivo
6.1. Isolated Human Organ Segments
6.2. Future Perspectives: Isolated Organ with Continuous Vascular Perfusion and CRISPR Gene Edition
Acknowledgments
Conflicts of Interest
References
- Aliño, S.F.; Crespo, J.; Bobadilla, M.; Lejarreta, M.; Blaya, C.; Crespo, A. Expression of human alpha 1-antitrypsin in mouse after in vivo gene transfer to hepatocytes by small liposomes. Biochem. Biophys. Res. Commun. 1994, 204, 1023–1030. [Google Scholar] [CrossRef]
- Crepso, J. Long-term expression of the human alpha1-antitrypsin gene in mice employing anionic and cationic liposome vectors. Biochem. Pharmacol. 1996, 51, 1309–1314. [Google Scholar]
- Dasí, F.; Benet, M.; Crespo, J.; Crespo, A.; Aliño, S.F. Asialofetuin liposome-mediated human alpha1-antitrypsin gene transfer in vivo results in stationary long-term gene expression. J. Mol. Med. 2001, 79, 205–212. [Google Scholar] [CrossRef]
- Zhang, G.; Song, Y.K.; Liu, D. Long-term expression of human alpha1-antitrypsin gene in mouse liver achieved by intravenous administration of plasmid DNA using a hydrodynamics-based procedure. Gene Ther. 2000, 7, 1344–1349. [Google Scholar] [CrossRef]
- Alino, S.F.; Crespo, A.; Dasi, F. Long-term therapeutic levels of human alpha-1 antitrypsin in plasma after hydrodynamic injection of nonviral DNA. Gene Ther. 2003, 10, 1672–1679. [Google Scholar] [CrossRef]
- Wolff, J.A.; Malone, R.W.; Williams, P.; Chong, W.; Acsadi, G.; Jani, A.; Felgner, P.L. Direct gene transfer into mouse muscle in vivo. Science 1990, 247, 1465–1468. [Google Scholar] [CrossRef]
- Budker, V.; Zhang, G.; Knechtle, S.; Wolff, J.A. Naked DNA delivered intraportally expresses efficiently in hepatocytes. Gene Ther. 1996, 3, 593–598. [Google Scholar]
- Budker, V.; Zhang, G.; Danko, I.; Williams, P.; Wolff, J. The efficient expression of intravascularly delivered DNA in rat muscle. Gene Ther. 1998, 5, 272–276. [Google Scholar] [CrossRef]
- Liu, F.; Song, Y.; Liu, D. Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Ther. 1999, 6, 1258–1266. [Google Scholar] [CrossRef]
- Gao, M.; Zhang, C.; Ma, Y.; Bu, L.; Yan, L.; Liu, D. Hydrodynamic delivery of mIL10 gene protects mice from high-fat diet-induced obesity and glucose intolerance. Mol. Ther. 2013, 21, 1852–1861. [Google Scholar] [CrossRef]
- Gao, M.; Ma, Y.; Cui, R.; Liu, D. Hydrodynamic delivery of FGF21 gene alleviates obesity and fatty liver in mice fed a high-fat diet. J. Control. Release 2014, 185, 1–11. [Google Scholar] [CrossRef]
- Ma, Y.; Gao, M.; Sun, H.; Liu, D. Interleukin-6 gene transfer reverses body weight gain and fatty liver in obese mice. Biochim. Biophys. Acta 2015, 1852, 1001–1011. [Google Scholar] [CrossRef]
- Hamar, P.; Song, E.; Kökény, G.; Chen, A.; Ouyang, N.; Lieberman, J. Small interfering RNA targeting Fas protects mice against renal ischemia-reperfusion injury. Proc. Natl. Acad. Sci. USA 2004, 101, 14883–14888. [Google Scholar] [CrossRef]
- Zhang, G.; Budker, V.; Williams, P.; Subbotin, V.; Wolff, J.A. Efficient expression of naked DNA delivered intraarterially to limb muscles of nonhuman primates. Hum. Gene Ther. 2001, 12, 427–438. [Google Scholar] [CrossRef]
- Zhang, G.; Ludtke, J.J.; Thioudellet, C.; Kleinpeter, P.; Antoniou, M.; Herweijer, H.; Braun, S.; Wolff, J.A. Intraarterial delivery of naked plasmid DNA expressing full-length mouse dystrophin in the mdx mouse model of duchenne muscular dystrophy. Hum. Gene Ther. 2004, 15, 770–782. [Google Scholar] [CrossRef]
- Mann, M.J.; Gibbons, G.H.; Hutchinson, H.; Poston, R.S.; Hoyt, E.G.; Robbins, R.C.; Dzau, V.J. Pressure-mediated oligonucleotide transfection of rat and human cardiovascular tissues. Proc. Natl. Acad. Sci. USA 1999, 96, 6411–6416. [Google Scholar] [CrossRef]
- Crespo, A.; Peydró, A.; Dasí, F.; Benet, M.; Calvete, J.J.; Revert, F.; Aliño, S.F. Hydrodynamic liver gene transfer mechanism involves transient sinusoidal blood stasis and massive hepatocyte endocytic vesicles. Gene Ther. 2005, 12, 927–935. [Google Scholar] [CrossRef]
- Aliño, S.F.; José Herrero, M.; Bodi, V.; Noguera, I.; Mainar, L.; Dasí, F.; Sempere, A.; Sánchez, M.; Díaz, A.; Sabater, L.; et al. Naked DNA delivery to whole pig cardiac tissue by coronary sinus retrograde injection employing non-invasive catheterization. J. Gene Med. 2010, 12, 920–926. [Google Scholar] [CrossRef]
- Sendra, L.; Pérez, D.; Miguel, A.; Herrero, M.J.; Noguera, I.; Díaz, A.; Barettino, D.; Martí-Bonmatí, L.; Aliño, S.F. Human AAT gene transfer to pig liver improved by using a perfusion isolated organ endovascular procedure. Eur. Radiol. 2016, 26, 95–102. [Google Scholar] [CrossRef]
- Jacobs, F.; Wisse, E.; De Geest, B. The role of liver sinusoidal cells in hepatocyte-directed gene transfer. Am. J. Pathol. 2010, 176, 14–21. [Google Scholar] [CrossRef]
- Sendra, L.; Miguel, A.; Pérez-Enguix, D.; Herrero, M.J.; Montalvá, E.; García-Gimeno, M.A.; Noguera, I.; Díaz, A.; Pérez, J.; Sanz, P.; et al. Studying Closed Hydrodynamic Models of “In Vivo” DNA Perfusion in Pig Liver for Gene Therapy Translation to Humans. PLoS ONE 2016, 11, e0163898. [Google Scholar] [CrossRef]
- Zhang, G.; Wooddell, C.I.; Hegge, J.O.; Griffin, J.B.; Huss, T.; Braun, S.; Wolff, J.A. Functional efficacy of dystrophin expression from plasmids delivered to mdx mice by hydrodynamic limb vein injection. Hum. Gene Ther. 2010, 21, 221–237. [Google Scholar] [CrossRef]
- Zhang, G.; Marshall, A.L.; Thomas, A.L.; Kernan, K.A.; Su, Y.; LeBoeuf, R.C.; Dong, X.R.; Tchao, B.N. In vivo knockdown of nicotinic acetylcholine receptor alpha1 diminishes aortic atherosclerosis. Atherosclerosis 2011, 215, 34–42. [Google Scholar] [CrossRef]
- Boström, P.; Wu, J.; Jedrychowski, M.P.; Korde, A.; Ye, L.; Lo, J.C.; Rasbach, K.A.; Boström, E.A.; Choi, J.H.; Long, J.Z.; et al. A PGC1-alpha-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature 2012, 481, 463–468. [Google Scholar] [CrossRef] [Green Version]
- Guess, M.G.; Barthel, K.K.; Pugach, E.K.; Leslie, A. Measuring microRNA reporter activity in skeletal muscle using hydrodynamic limb vein injection of plasmid DNA combined with in vivo imaging. Skelet Muscle 2013, 3, 19. [Google Scholar] [CrossRef]
- Mukumoto, H.; Takahashi, Y.; Ando, M.; Nishikawa, M.; Takakura, Y. Expression profile-dependent improvement of insulin sensitivity by gene delivery of interleukin-6 in a mouse model of type II diabetes. Mol. Pharm. 2013, 10, 3812–3821. [Google Scholar] [CrossRef]
- Nagata, K.; Itaka, K.; Baba, M.; Uchida, S.; Ishii, T.; Kataoka, K. Muscle-targeted hydrodynamic gene introduction of insulin-like growth factor-1 using polyplex nanomicelle to treat peripheral nerve injury. J. Control. Release 2014, 183, 27–34. [Google Scholar] [CrossRef]
- Aliño, S.F.; Bobadilla, M.; Garcia-Sanz, M.; Lejarreta, M.; Unda, F.; Hilario, E. In vivo delivery of human alpha 1-antitrypsin gene to mouse hepatocytes by liposomes. Biochem. Biophys. Res. Commun. 1993, 192, 174–181. [Google Scholar] [CrossRef]
- Aliño, S.F.; Bobadilla, M.; Crespo, J.; Lejarreta, M. Human alpha 1-antitrypsin gene transfer to in vivo mouse hepatocytes. Hum. Gene Ther. 1996, 7, 531–536. [Google Scholar] [CrossRef]
- Zhang, G.; Budker, V.; Wolff, J.A. High levels of foreign gene expression in hepatocytes after tail vein injections of naked plasmid DNA. Hum. Gene Ther. 1999, 10, 1735–1737. [Google Scholar] [CrossRef]
- Herrero, M.J.; Monleon, D.; Morales, J.M.; Mata, M.; Serna, E.; Aliño, S.F. Analysis of metabolic and gene expression changes after hydrodynamic DNA injection into mouse liver. Biol. Pharm. Bull. 2011, 34, 167–172. [Google Scholar] [CrossRef]
- Schüttrumpf, J.; Milanov, P.; Roth, S.; Seifried, E.; Tonn, T. Non-viral gene transfer results in therapeutic factor IX levels in haemophilia B mice. Hamostaseologie 2008, 28 (Suppl. 1), S92–S95. [Google Scholar]
- Keravala, A.; Chavez, C.L.; Hu, G.; Woodard, L.E.; Monahan, P.E.; Calos, M.P. Long-term phenotypic correction in factor IX knockout mice by using PhiC31 integrase-mediated gene therapy. Gene Ther. 2011, 18, 842–848. [Google Scholar] [CrossRef]
- Kim, H.S.; Kim, J.C.; Lee, Y.K.; Kim, J.S.; Park, Y.S. Hepatic control elements promote long-term expression of human coagulation factor IX gene in hydrodynamically transfected mice. J. Gene Med. 2011, 13, 365–372. [Google Scholar] [CrossRef] [PubMed]
- Schüttrumpf, J.; Milanov, P.; Abriss, D.; Roth, S.; Tonn, T.; Seifried, E. Transgene loss and changes in the promoter methylation status as determinants for expression duration in nonviral gene transfer for factor IX. Hum. Gene Ther. 2011, 22, 101–106. [Google Scholar] [CrossRef]
- Matsui, H.; Fujimoto, N.; Sasakawa, N.; Ohinata, Y.; Shima, M.; Yamanaka, S.; Sugimoto, M.; Hotta, A. Delivery of full-length factor VIII using a piggyBac transposon vector to correct a mouse model of hemophilia A. PLoS ONE 2014, 9, e104957. [Google Scholar] [CrossRef]
- Holm, D.A.; Dagnaes-Hansen, F.; Simonsen, H.; Gregersen, N.; Bolund, L.; Jensen, T.G.; Corydon, T.J. Expression of short-chain acyl-CoA dehydrogenase (SCAD) proteins in the liver of SCAD deficient mice after hydrodynamic gene transfer. Mol. Genet. Metab. 2003, 78, 250–258. [Google Scholar] [CrossRef]
- Turunen, T.A.; Kurkipuro, J.; Heikura, T.; Vuorio, T.; Hytönen, E.; Izsvák, Z.; Ylä-Herttuala, S. Sleeping Beauty Transposon Vectors in Liver-directed Gene Delivery of LDLR and VLDLR for Gene Therapy of Familial Hypercholesterolemia. Mol. Ther. 2016, 24, 620–635. [Google Scholar] [CrossRef]
- Jiang, J.; Yamato, E.; Miyazaki, J. Long-term control of food intake and body weight by hydrodynamics-based delivery of plasmid DNA encoding leptin or CNTF. J. Gene Med. 2003, 5, 977–983. [Google Scholar] [CrossRef]
- González-Muniesa, P.; Milagro, F.I.; Campión, J.; Martínez, J.A. Reduction in energy efficiency induced by expression of the uncoupling protein, UCP1, in mouse liver mitochondria. Int. J. Mol. Med. 2006, 17, 591–597. [Google Scholar] [CrossRef]
- He, C.X.; Shi, D.; Wu, W.J.; Ding, Y.F.; Feng, D.M.; Lu, B.; Chen, H.M.; Yao, J.H.; Shen, Q.; Lu, D.R.; et al. Insulin expression in livers of diabetic mice mediated by hydrodynamics-based administration. World J. Gastroenterol. 2004, 10, 567–572. [Google Scholar] [CrossRef]
- Fukushima, M.; Hattori, Y.; Tsukada, H.; Koga, K.; Kajiwara, E.; Kawano, K.; Kobayashi, T.; Kamata, K.; Maitani, Y. Adiponectin gene therapy of streptozotocin-induced diabetic mice using hydrodynamic injection. J. Gene Med. 2007, 9, 976–985. [Google Scholar] [CrossRef]
- Ma, Y.; Liu, D. Hydrodynamic delivery of adiponectin and adiponectin receptor 2 gene blocks high-fat diet-induced obesity and insulin resistance. Gene Ther. 2013, 20, 846–852. [Google Scholar] [CrossRef]
- Vakili, S.; Ebrahimi, S.S.; Sadeghi, A.; Gorgani-Firuzjaee, S.; Beigy, M.; Pasalar, P.; Meshkani, R. Hydrodynamic-based delivery of PTP1B shRNA reduces plasma glucose levels in diabetic mice. Mol. Med. Rep. 2013, 7, 211–216. [Google Scholar] [CrossRef]
- Baribault, H.; Majeti, J.Z.; Ge, H.; Wang, J.; Xiong, Y.; Gardner, J.; Yang, L.; Gupte, J.; Gong, Y.; Pan, Z.; et al. Advancing therapeutic discovery through phenotypic screening of the extracellular proteome using hydrodynamic intravascular injection. Expert. Opin. Ther. Targets 2014, 18, 1253–1264. [Google Scholar] [CrossRef] [PubMed]
- Camassola, M.; Braga, L.M.; Delgado-Cañedo, A.; Dalberto, T.P.; Matte, U.; Burin, M.; Giugliani, R.; Nardi, N.B. Nonviral in vivo gene transfer in the mucopolysaccharidosis I murine model. J. Inherit. Metab. Dis. 2005, 28, 1035–1043. [Google Scholar] [CrossRef]
- Richard, M.; Arfi, A.; Seguin, J.; Gandolphe, C.; Scherman, D. Widespread biochemical correction of murine mucopolysaccharidosis type VII pathology by liver hydrodynamic plasmid delivery. Gene Ther. 2009, 16, 746–756. [Google Scholar] [CrossRef]
- Quiviger, M.; Arfi, A.; Mansard, D.; Delacotte, L.; Pastor, M.; Scherman, D.; Marie, C. High and prolonged sulfamidase secretion by the liver of MPS-IIIA mice following hydrodynamic tail vein delivery of antibiotic-free pFAR4 plasmid vector. Gene Ther. 2014, 21, 1001–1007. [Google Scholar] [CrossRef]
- Pergolizzi, R.G.; Jin, G.; Chan, D.; Pierre, L.; Bussel, J.; Ferris, B.; Leopold, P.L.; Crystal, R.G. Correction of a murine model of von Willebrand disease by gene transfer. Blood 2006, 108, 862–869. [Google Scholar] [CrossRef]
- Belcher, J.D.; Vineyard, J.V.; Bruzzone, C.M.; Chen, C.; Beckman, J.D.; Nguyen, J.; Steer, C.J.; Vercellotti, G.M. Heme oxygenase-1 gene delivery by Sleeping Beauty inhibits vascular stasis in a murine model of sickle cell disease. J. Mol. Med. 2010, 88, 665–675. [Google Scholar] [CrossRef]
- Viecelli, H.M.; Harbottle, R.P.; Wong, S.P.; Schlegel, A.; Chuah, M.K.; VandenDriessche, T.; Harding, C.O.; Thöny, B. Treatment of phenylketonuria using minicircle-based naked-DNA gene transfer to murine liver. Hepatology 2014, 60, 1035–1043. [Google Scholar] [CrossRef]
- Grisch-Chan, H.M.; Schlegel, A.; Scherer, T.; Allegri, G.; Heidelberger, R.; Tsikrika, P.; Schmeer, M.; Schleef, M.; Harding, C.O.; Häberle, J.; et al. Low-Dose Gene Therapy for Murine PKU Using Episomal Naked DNA Vectors Expressing PAH from Its Endogenous Liver Promoter. Mol. Ther. Nucleic Acids 2017, 7, 339–349. [Google Scholar] [CrossRef]
- Yang, P.L.; Althage, A.; Chung, J.; Chisari, F.V. Hydrodynamic injection of viral DNA: a mouse model of acute hepatitis B virus infection. Proc. Natl. Acad. Sci. USA 2002, 99, 13825–13830. [Google Scholar] [CrossRef]
- McCaffrey, A.P.; Ohashi, K.; Meuse, L.; Shen, S.; Lancaster, A.M.; Lukavsky, P.J.; Sarnow, P.; Kay, M.A. Determinants of hepatitis C translational initiation in vitro, in cultured cells and mice. Mol. Ther. 2002, 5, 676–684. [Google Scholar] [CrossRef]
- Kim, S.I.; Shin, D.; Lee, H.; Ahn, B.Y.; Yoon, Y.; Kim, M. Targeted delivery of siRNA against hepatitis C virus by apolipoprotein A-I-bound cationic liposomes. J. Hepatol. 2009, 50, 479–488. [Google Scholar] [CrossRef]
- Zender, L.; Hutker, S.; Liedtke, C.; Tillmann, H.L.; Zender, S.; Mundt, B.; Waltemathe, M.; Gosling, T.; Flemming, P.; Malek, N.P.; et al. Caspase 8 small interfering RNA prevents acute liver failure in mice. Proc. Natl. Acad. Sci. USA 2003, 100, 7797–7802. [Google Scholar] [CrossRef]
- Saito, Y.; Kon, S.; Fujiwara, Y.; Nakayama, Y.; Kurotaki, D.; Fukuda, N.; Kimura, C.; Kanayama, M.; Ito, K.; Diao, H.; et al. Osteopontin small interfering RNA protects mice from fulminant hepatitis. Hum. Gene Ther. 2007, 18, 1205–1214. [Google Scholar] [CrossRef]
- Xu, J.F.; Xiao, H.; Hu, G.Y.; Zheng, S.H.; Liu, W.; Yuan, C.L.; Yang, H.; Lü, J.; Zheng, F.; Wang, C.Y.; et al. Ectopic B7-H4-Ig expression attenuates concanavalin A-induced hepatic injury. Clin. Immunol. 2010, 136, 30–41. [Google Scholar] [CrossRef]
- Bulau, A.-M.; Fink, M.; Maucksch, C.; Kappler, R.; Mayr, D.; Wagner, K.; Bufler, P. In vivo expression of interleukin-37 reduces local and systemic inflammation in concanavalin A-induced hepatitis. ScientificWorldJournal 2011, 11, 2480–2490. [Google Scholar] [CrossRef]
- Shashidharamurthy, R.; Machiah, D.; Bozeman, E.N.; Srivatsan, S.; Patel, J.; Cho, A.; Jacob, J.; Selvaraj, P. Hydrodynamic delivery of plasmid DNA encoding human FcgammaR-Ig dimers blocks immune-complex mediated inflammation in mice. Gene Ther. 2012, 19, 877–885. [Google Scholar] [CrossRef]
- Anavi, S.; Hahn-Obercyger, M.; Margalit, R.; Madar, Z.; Tirosh, O. A novel antihypoglycemic role of inducible nitric oxide synthase in liver inflammatory response induced by dietary cholesterol and endotoxemia. Antioxid. Redox Signal. 2013, 19, 1889–1901. [Google Scholar] [CrossRef]
- Huang, M.; Sun, R.; Wei, H.; Tian, Z. Simultaneous knockdown of multiple ligands of innate receptor NKG2D prevents natural killer cell-mediated fulminant hepatitis in mice. Hepatology 2013, 57, 277–288. [Google Scholar] [CrossRef]
- Görtz, D.; Braun, G.S.; Maruta, Y.; Djudjaj, S.; van Roeyen, C.R.; Martin, I.V.; Küster, A.; Schmitz-Van de Leur, H.; Scheller, J.; Ostendorf, T.; et al. Anti-interleukin-6 therapy through application of a monogenic protein inhibitor via gene delivery. Sci. Rep. 2015, 5, 14685. [Google Scholar] [CrossRef]
- Tsai, S.M.; Wang, W.P. Expression and function of fibroblast growth factor (FGF) 7 during liver regeneration. Cell. Physiol. Biochem. 2011, 27, 641–652. [Google Scholar] [CrossRef]
- Wu, X.; He, Y.; Falo, L.D., Jr.; Hui, K.M.; Huang, L. Regression of human mammary adenocarcinoma by systemic administration of a recombinant gene encoding the hFlex-TRAIL fusion protein. Mol. Ther. 2001, 3, 368–374. [Google Scholar] [CrossRef]
- Yazawa, H.; Murakami, T.; Li, H.M.; Back, T.; Kurosaka, K.; Suzuki, Y.; Shorts, L.; Akiyama, Y.; Maruyama, K.; Parsoneault, E.; et al. Hydrodynamics-based gene delivery of naked DNA encoding fetal liver kinase-1 gene effectively suppresses the growth of pre-existing tumors. Cancer Gene Ther. 2006, 13, 993–1001. [Google Scholar] [CrossRef]
- Miyakawa, N.; Nishikawa, M.; Takahashi, Y.; Ando, M.; Misaka, M.; Watanabe, Y.; Takakura, Y. Prolonged circulation half-life of interferon gamma activity by gene delivery of interferon gamma-serum albumin fusion protein in mice. J. Pharm. Sci. 2011, 100, 2350–2357. [Google Scholar] [CrossRef]
- Ando, M.; Takahashi, Y.; Nishikawa, M.; Takakura, Y. Control of spatiotemporal distribution of interferon gamma by genetically fusing functional peptides. Yakugaku Zasshi 2012, 132, 1399–1406. [Google Scholar] [CrossRef]
- Miyakawa, N.; Nishikawa, M.; Takahashi, Y.; Ando, M.; Misaka, M.; Watanabe, Y.; Takakura, Y. Gene delivery of albumin binding peptide-interferon-gamma fusion protein with improved pharmacokinetic properties and sustained biological activity. J. Pharm. Sci. 2013, 102, 3110–3118. [Google Scholar] [CrossRef]
- Ando, M.; Takahashi, Y.; Yamashita, T.; Fujimoto, M.; Nishikawa, M.; Watanabe, Y.; Takakura, Y. Prevention of adverse events of interferon gamma gene therapy by gene delivery of interferon gamma-heparin-binding domain fusion protein in mice. Mol. Ther. Methods Clin. Dev. 2014, 1, 14023. [Google Scholar] [CrossRef]
- Ochoa, M.C.; Fioravanti, J.; Duitman, E.H.; Medina-Echeverz, J.; Palazon, A.; Arina, A.; Dubrot, J.; Alfaro, C.; Morales-Kastresana, A.; Murillo, O.; et al. Liver gene transfer of interkeukin-15 constructs that become part of circulating high density lipoproteins for immunotherapy. PLoS ONE 2012, 7, e52370. [Google Scholar] [CrossRef]
- Barao, I.; Alvarez, M.; Redelman, D.; Weiss, J.M.; Ortaldo, J.R.; Wiltrout, R.H.; Murphy, W.J. Hydrodynamic delivery of human IL-15 cDNA increases murine natural killer cell recovery after syngeneic bone marrow transplantation. Biol. Blood Marrow Transplant. 2011, 17, 1754–1764. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Liu, D. IL-15/sIL-15Ralpha gene transfer suppresses Lewis lung cancer growth in the lungs, liver and kidneys. Cancer Gene Ther. 2016, 23, 54–60. [Google Scholar] [CrossRef]
- Qiu, C.; Li, Y.; Zhou, M.; Liu, J.; Li, M.; Wu, Y.; Xu, D.; Li, M. Hydrodynamic delivery of IL-28B (IFN-lambda3) gene ameliorates lung inflammation induced by cigarette smoke exposure in mice. Biochem. Biophys. Res. Commun. 2014, 447, 513–519. [Google Scholar] [CrossRef]
- Sondergaard, M.; Dagnaes-Hansen, F.; Flyvbjerg, A.; Jensen, T.G. Normalization of growth in hypophysectomized mice using hydrodynamic transfer of the human growth hormone gene. Am. J. Physiol. Endocrinol. Metab. 2003, 285, E427–E432. [Google Scholar] [CrossRef]
- Lee, S.; Hong, S.W.; Choi, H.S.; Lee, L.Y.; Nam, C.; Rhee, Y.; Chung, U.-I.; Lim, S.-K. Experimental parathyroid hormone gene therapy using OC31 integrase. Endocr. J. 2008, 55, 1033–1041. [Google Scholar] [CrossRef]
- Okumura, A.; Saito, T.; Otani, I.; Kojima, K.; Yamada, Y.; Ishida-Okawara, A.; Nakazato, K.; Asano, M.; Kanayama, K.; Iwakura, Y.; et al. Suppressive role of leukocyte cell-derived chemotaxin 2 in mouse anti-type II collagen antibody-induced arthritis. Arthritis Rheum. 2008, 58, 413–421. [Google Scholar] [CrossRef]
- Watcharanurak, K.; Nishikawa, M.; Takahashi, Y.; Kabashima, K.; Takahashi, R.; Takakura, Y. Regulation of immunological balance by sustained interferon-gamma gene transfer for acute phase of atopic dermatitis in mice. Gene Ther. 2013, 20, 538–544. [Google Scholar] [CrossRef]
- Wesche-Soldato, D.E.; Chung, C.S.; Lomas-Neira, J.; Doughty, L.A.; Gregory, S.H.; Ayala, A. In vivo delivery of caspase-8 or Fas siRNA improves the survival of septic mice. Blood 2005, 106, 2295–2301. [Google Scholar] [CrossRef] [PubMed]
- Tompkins, S.M.; Lo, C.Y.; Tumpey, T.M.; Epstein, S.L. Protection against lethal influenza virus challenge by RNA interference in vivo. Proc. Natl. Acad. Sci. USA 2004, 101, 8682–8686. [Google Scholar] [CrossRef]
- Lu, S.L.; Tsai, C.Y.; Luo, Y.H.; Kuo, C.F.; Lin, W.C.; Chang, Y.T.; Wu, J.J.; Chuang, W.J.; Liu, C.C.; Chao, L.; et al. Kallistatin modulates immune cells and confers anti-inflammatory response to protect mice from group A streptococcal infection. Antimicrob. Agents Chemother. 2013, 57, 5366–5372. [Google Scholar] [CrossRef]
- Nakamura, G.; Maruyama, H.; Ishii, S.; Shimotori, M.; Kameda, S.; Kono, T.; Miyazaki, J.; Kulkarni, A.B.; Gejyo, F. Naked plasmid DNA-based alpha-galactosidase A gene transfer partially reduces systemic accumulation of globotriaosylceramide in Fabry mice. Mol. Biotechnol. 2008, 38, 109–119. [Google Scholar] [CrossRef]
- Bell, J.B.; Podetz-Pedersen, K.M.; Aronovich, E.L.; Belur, L.R.; McIvor, R.S.; Hackett, P.B. Preferential delivery of the Sleeping Beauty transposon system to livers of mice by hydrodynamic injection. Nat. Protoc. 2007, 2, 3153–3165. [Google Scholar] [CrossRef]
- Hackett, P.B., Jr.; Aronovich, E.L.; Hunter, D.; Urness, M.; Bell, J.B.; Kass, S.J.; Cooper, L.J.; McIvor, S. Efficacy and safety of Sleeping Beauty transposon-mediated gene transfer in preclinical animal studies. Curr. Gene Ther. 2011, 11, 341–349. [Google Scholar] [CrossRef]
- Doherty, J.E.; Huye, L.E.; Yusa, K.; Zhou, L.; Craig, N.L.; Wilson, M.H. Hyperactive piggyBac gene transfer in human cells and in vivo. Hum. Gene Ther. 2012, 23, 311–320. [Google Scholar] [CrossRef]
- Chen, I.Y.; Paulmurugan, R.; Nielsen, C.H.; Wang, D.S.; Chow, V.; Robbins, R.C.; Gambhir, S.S. A titratable two-step transcriptional amplification strategy for targeted gene therapy based on ligand-induced intramolecular folding of a mutant human estrogen receptor. Mol. Imaging Biol. 2014, 16, 224–234. [Google Scholar] [CrossRef]
- Chu, Q.; Joseph, M.; Przybylska, M.; Yew, N.S.; Scheule, R.K. Transient siRNA-mediated attenuation of liver expression from an alpha-galactosidase A plasmid reduces subsequent humoral immune responses to the transgene product in mice. Mol. Ther. 2005, 12, 264–273. [Google Scholar] [CrossRef]
- Wesche-Soldato, D.E.; Lomas-Neira, J.; Perl, M.; Chung, C.S.; Ayala, A. Hydrodynamic delivery of siRNA in a mouse model of sepsis. Methods Mol. Biol. 2008, 442, 67–73. [Google Scholar] [PubMed]
- Magnusson, T.; Haase, R.; Schleef, M.; Wagner, E.; Ogris, M. Sustained, high transgene expression in liver with plasmid vectors using optimized promoter-enhancer combinations. J. Gene Med. 2011, 13, 382–391. [Google Scholar] [CrossRef]
- Yan, S.; Fu, Q.; Zhou, Y.; Wang, J.; Liu, Y.; Duan, X.; Jia, S.; Peng, J.; Gao, B.; Du, J. High levels of gene expression in the hepatocytes of adult mice, neonatal mice and tree shrews via retro-orbital sinus hydrodynamic injections of naked plasmid DNA. J. Control. Release 2012, 161, 763–771. [Google Scholar] [CrossRef]
- Shigekawa, M.; Hikita, H.; Kodama, T.; Shimizu, S.; Li, W.; Uemura, A.; Miyagi, T.; Hosui, A.; Kanto, T.; Hiramatsu, N. Pancreatic STAT3 protects mice against caerulein-induced pancreatitis via PAP1 induction. Am. J. Pathol. 2012, 181, 2105–2113. [Google Scholar] [CrossRef]
- Dagnaes-Hansen, F.; Holst, H.U.; Søndergaard, M.; Vorup-Jensen, T.; Flyvbjerg, A.; Jensen, U.B.; Jensen, T.G. Physiological effects of human growth hormone produced after hydrodynamic gene transfer of a plasmid vector containing the human ubiquitin promotor. J. Mol. Med. 2002, 80, 665–670. [Google Scholar] [CrossRef]
- Hagstrom, J.E.; Hegge, J.; Zhang, G.; Noble, M.; Budker, V.; Lewis, D.L.; Herweijer, H.; Wolff, J.A. A facile nonviral method for delivering genes and siRNAs to skeletal muscle of mammalian limbs. Mol. Ther. 2004, 10, 386–398. [Google Scholar] [CrossRef]
- Zhang, G.; Gao, X.; Song, Y.K.; Vollmer, R.; Stolz, D.B.; Gasiorowski, J.Z.; Dean, D.A.; Liu, D. Hydroporation as the mechanism of hydrodynamic delivery. Gene Ther. 2004, 11, 675–682. [Google Scholar] [CrossRef]
- Li, W.; Ma, N.; Ong, L.L.; Kaminski, A.; Skrabal, C.; Ugurlucan, M.; Lorenz, P.; Gatzen, H.H.; Lützow, K.; Lendlein, A. Enhanced thoracic gene delivery by magnetic nanobead-mediated vector. J. Gene Med. 2008, 10, 897–909. [Google Scholar] [CrossRef]
- Suda, T.; Suda, K.; Liu, D. Computer-assisted hydrodynamic gene delivery. Mol. Ther. 2008, 16, 1098–1104. [Google Scholar] [CrossRef]
- Podetz-Pedersen, K.M.; Bell, J.B.; Steele, T.W.; Wilber, A.; Shier, W.T.; Belur, L.R.; McIvor, R.S.; Hackett, P.B. Gene expression in lung and liver after intravenous infusion of polyethylenimine complexes of Sleeping Beauty transposons. Hum. Gene Ther. 2010, 21, 210–220. [Google Scholar] [CrossRef]
- Wooddell, C.I.; Hegge, J.O.; Zhang, G.; Sebestyen, M.G.; Noble, M.; Griffin, J.B.; Pfannes, L.V.; Herweijer, H.; Hagstrom, J.E.; Braun, S.; et al. Dose response in rodents and nonhuman primates after hydrodynamic limb vein delivery of naked plasmid DNA. Hum. Gene Ther. 2011, 22, 889–903. [Google Scholar] [CrossRef]
- Bu, X.; Zhou, Y.; Zhang, H.; Qiu, W.; Chen, L.; Cao, H.; Fang, L.; Wen, P.; Tan, R.; Yang, J. Systemic administration of naked plasmid encoding HGF attenuates puromycin aminonucleoside-induced damage of murine glomerular podocytes. Am. J. Physiol. Renal Physiol. 2011, 301, F784–F792. [Google Scholar] [CrossRef]
- Duguid, J.G.; Li, C.; Shi, M.; Logan, M.J.; Alila, H.; Rolland, A.; Tomlinson, E.; Sparrow, J.T.; Smith, L.C. A physicochemical approach for predicting the effectiveness of peptide-based gene delivery systems for use in plasmid-based gene therapy. Biophys. J. 1998, 74, 2802–2814. [Google Scholar] [CrossRef]
- Moret, I.; Esteban Peris, J.; Guillem, V.M.; Benet, M.; Revert, F.; Dasí, F.; Crespo, A.; Aliño, S.F. Stability of PEI-DNA and DOTAP-DNA complexes: effect of alkaline pH, heparin and serum. J. Control. Release 2001, 76, 169–181. [Google Scholar] [CrossRef]
- Alino, S.F.; Escriga, E.; Reverta, F.; Guillema, V.M.; Crespoa, A. Pharmacodynamic approach to study the gene transfer process employing non-viral vectors. Biochem. Pharmacol. 2000, 60, 1845–1853. [Google Scholar] [CrossRef]
- Xu, Z.X.; Chen, J.Z.; Yue, Y.B.; Zhang, J.Q.; Li, Z.H.; Feng, D.M.; Ruan, Z.C.; Tian, L.; Xue, J.L.; Wang, Q.J. A 16-bp RBE element mediated Rep-dependent site-specific integration in AAVS1 transgenic mice for expression of hFIX. Gene Ther. 2009, 16, 589–595. [Google Scholar] [CrossRef]
- Hibbitt, O.C.; Harbottle, R.P.; Waddington, S.N.; Bursill, C.A.; Coutelle, C.; Channon, K.M.; Wade-Martins, R. Delivery and long-term expression of a 135 kb LDLR genomic DNA locus in vivo by hydrodynamic tail vein injection. J. Gene Med. 2007, 9, 488–497. [Google Scholar] [CrossRef]
- Shahaf, G.; Moser, H.; Ozeri, E.; Mizrahi, M.; Abecassis, A.; Lewis, E.C. alpha-1-antitrypsin gene delivery reduces inflammation, increases T-regulatory cell population size and prevents islet allograft rejection. Mol. Med. 2011, 17, 1000–1011. [Google Scholar] [CrossRef]
- Hibbitt, O.; Wade-Martins, R. High capacity extrachromosomal gene expression vectors. Methods Mol. Biol. 2011, 738, 19–40. [Google Scholar]
- Wooddell, C.I.; Reppen, T.; Wolff, J.A.; Herweijer, H. Sustained liver-specific transgene expression from the albumin promoter in mice following hydrodynamic plasmid DNA delivery. J. Gene Med. 2008, 10, 551–563. [Google Scholar] [CrossRef]
- Higuchi, N.; Maruyama, H.; Kuroda, T.; Kameda, S.; Iino, N.; Kawachi, H.; Nishikawa, Y.; Hanawa, H.; Tahara, H.; Miyazaki, J.; et al. Hydrodynamics-based delivery of the viral interleukin-10 gene suppresses experimental crescentic glomerulonephritis in Wistar-Kyoto rats. Gene Ther. 2003, 10, 1297–1310. [Google Scholar] [CrossRef]
- Inoue, S.; Hakamata, Y.; Kaneko, M.; Kobayashi, E. Gene therapy for organ grafts using rapid injection of naked DNA: application to the rat liver. Transplantation 2004, 77, 997–1003. [Google Scholar] [CrossRef]
- Miki, Y.; Maruyama, S.; Liu, D.; Kobayashi, T.; Sato, F.; Shimizu, H.; Kato, S.; Sato, W.; Morita, Y.; Yuzawa, Y. In vivo gene transfer of endo-beta-galactosidase C removes alphaGal antigen on erythrocytes and endothelial cells of the organs. Xenotransplantation 2004, 11, 444–451. [Google Scholar] [CrossRef]
- Wang, C.H.; Liang, C.L.; Huang, L.T.; Liu, J.K.; Hung, P.H.; Sun, A.; Hung, K.S. Single intravenous injection of naked plasmid DNA encoding erythropoietin provides neuroprotection in hypoxia-ischemia rats. Biochem. Biophys. Res. Commun. 2004, 314, 1064–1071. [Google Scholar] [CrossRef]
- Yokoi, H.; Mukoyama, M.; Nagae, T.; Mori, K.; Suganami, T.; Sawai, K.; Yoshioka, T.; Koshikawa, M.; Nishida, T.; Takigawa, M. Reduction in connective tissue growth factor by antisense treatment ameliorates renal tubulointerstitial fibrosis. J. Am. Soc. Nephrol. 2004, 15, 1430–1440. [Google Scholar] [CrossRef]
- Zhang, X.; Dong, X.; Sawyer, G.J.; Collins, L.; Fabre, J.W. Regional hydrodynamic gene delivery to the rat liver with physiological volumes of DNA solution. J. Gene Med. 2004, 6, 693–703. [Google Scholar] [CrossRef]
- Liu, H.; Hanawa, H.; Yoshida, T.; Elnaggar, R.; Hayashi, M.; Watanabe, R.; Toba, K.; Yoshida, K.; Chang, H.; Okura, Y. Effect of hydrodynamics-based gene delivery of plasmid DNA encoding interleukin-1 receptor antagonist-Ig for treatment of rat autoimmune myocarditis: possible mechanism for lymphocytes and noncardiac cells. Circulation 2005, 111, 1593–1600. [Google Scholar] [CrossRef]
- Chang, H.; Hanawa, H.; Yoshida, T.; Hayashi, M.; Liu, H.; Ding, L.; Otaki, K.; Hao, K.; Yoshida, K.; Kato, K. Alteration of IL-17 related protein expressions in experimental autoimmune myocarditis inhibition of IL-17 by IL-10-Ig fusion gene transfer. Circ. J. 2008, 72, 813–819. [Google Scholar] [CrossRef]
- Chang, H.; Wang, Y.; Li, G.; Zhang, L.; Zhang, G.W.; Liao, Y.C.; Hanawa, H.; Zou, J. Effect of hydrodynamics-based delivery of IL-18BP fusion gene on rat experimental autoimmune myocarditis. Clin. Exp. Med. 2014, 14, 397–408. [Google Scholar] [CrossRef]
- Tsoulfas, G.; Takahashi, Y.; Liu, D.; Yagnik, G.; Wu, T.; Murase, N.; Geller, D.A. Hydrodynamic plasmid DNA gene therapy model in liver transplantation. J. Surg. Res. 2006, 135, 242–249. [Google Scholar] [CrossRef]
- Chen, S.W.; Zhang, X.R.; Wang, C.Z.; Chen, W.Z.; Xie, W.F.; Chen, Y.X. RNA interference targeting the platelet-derived growth factor receptor beta subunit ameliorates experimental hepatic fibrosis in rats. Liver Int. 2008, 28, 1446–1457. [Google Scholar] [CrossRef]
- Xing, Y.; Pua, E.C.; Lu, X.; Zhong, P. Low-amplitude ultrasound enhances hydrodynamic-based gene delivery to rat kidney. Biochem. Biophys. Res. Commun. 2009, 386, 217–222. [Google Scholar] [CrossRef]
- Sawyer, G.J.; Zhang, X.; Fabre, J.W. Technical requirements for effective regional hydrodynamic gene delivery to the left lateral lobe of the rat liver. Gene Ther. 2010, 17, 560–564. [Google Scholar] [CrossRef]
- Cim, A.; Sawyer, G.J.; Zhang, X.; Su, H.; Collins, L.; Jones, P.; Antoniou, M.; Reynes, J.P.; Lipps, H.J.; Fabre, J.W. In vivo studies on non-viral transdifferentiation of liver cells towards pancreatic beta cells. J. Endocrinol. 2012, 214, 277–288. [Google Scholar] [CrossRef]
- Zhao, M.D.; Sun, Y.M.; Fu, G.F.; Du, Y.Z.; Chen, F.Y.; Yuan, H.; Zheng, C.H.; Zhang, X.M.; Hu, F.Q. Gene therapy of endometriosis introduced by polymeric micelles with glycolipid-like structure. Biomaterials 2012, 33, 634–643. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Li, Y.; Li, W.; Wu, Q.; Gao, R. Gene transfer of c-met confers protection against D-galactosamine/lipopolysaccharide-induced acute liver failure. Dig. Dis. Sci. 2012, 57, 925–934. [Google Scholar] [CrossRef] [PubMed]
- Yasuzaki, Y.; Yamada, Y.; Kanefuji, T.; Harashima, H. Localization of exogenous DNA to mitochondria in skeletal muscle following hydrodynamic limb vein injection. J. Control. Release 2013, 172, 805–811. [Google Scholar] [CrossRef]
- Eastman, S.J.; Baskin, K.M.; Hodges, B.L.; Chu, Q.; Gates, A.; Dreusicke, R.; Anderson, S.; Scheule, R.K. Development of catheter-based procedures for transducing the isolated rabbit liver with plasmid DNA. Hum. Gene Ther. 2002, 13, 2065–2077. [Google Scholar] [CrossRef]
- Kamimura, K.; Kanefuji, T.; Yokoo, T.; Abe, H.; Suda, T.; Kobayashi, Y.; Zhang, G.; Aoyagi, Y.; Liu, D. Safety assessment of liver-targeted hydrodynamic gene delivery in dogs. PLoS ONE 2014, 9, e107203. [Google Scholar] [CrossRef]
- Yoshino, H.; Hashizume, K.; Kobayashi, E. Naked plasmid DNA transfer to the porcine liver using rapid injection with large volume. Gene Ther. 2006, 13, 1696–1702. [Google Scholar] [CrossRef]
- Aliño, S.F.; Herrero, M.J.; Noguera, I.; Dasí, F.; Sánchez, M. Pig liver gene therapy by noninvasive interventionist catheterism. Gene Ther. 2007, 14, 334–343. [Google Scholar] [CrossRef]
- Fabre, J.W.; Grehan, A.; Whitehorne, M.; Sawyer, G.J.; Dong, X.; Salehi, S.; Eckley, L.; Zhang, X.; Seddon, M.; Shah, A.M. Hydrodynamic gene delivery to the pig liver via an isolated segment of the inferior vena cava. Gene Ther. 2008, 15, 452–462. [Google Scholar] [CrossRef]
- Kamimura, K.; Zhang, G.; Liu, D. Image-guided, intravascular hydrodynamic gene delivery to skeletal muscle in pigs. Mol. Ther. 2010, 18, 93–100. [Google Scholar] [CrossRef]
- Fabre, J.W.; Whitehorne, M.; Grehan, A.; Sawyer, G.J.; Zhang, X.; Davenport, M.; Rela, M. Critical physiological and surgical considerations for hydrodynamic pressurization of individual segments of the pig liver. Hum. Gene Ther. 2011, 22, 879–887. [Google Scholar] [CrossRef]
- Carreño, O.; Sendra, L.; Montalvá, E.; Miguel, A.; Orbis, F.; Herrero, M.J.; Noguera, I.; Aliño, S.F.; Lopez-Andujar, R. A surgical model for isolating the pig liver in vivo for gene therapy. Eur. Surg. Res. 2013, 51, 47–57. [Google Scholar] [CrossRef]
- Sendra, L.; Carreño, O.; Miguel, A.; Montalvá, E.; Herrero, M.J.; Orbis, F.; Noguera, I.; Barettino, D.; López-Andújar, R.; Aliño, S.F. Low RNA translation activit limits the efficacy of hydrodynamic gene transfer to pig liver in vivo. J. Gene Med. 2014, 16, 179–192. [Google Scholar]
- de Groot, G.H.; Reuvers, C.B.; Schalm, S.W.; Boks, A.L.; Terpstra, O.T.; Jeekel, H.; ten Kate, F.W.; Bruinvels, J. A reproducible model of acute hepatic failure by transient ischemia in the pig. J. Surg. Res. 1987, 42, 92–100. [Google Scholar] [CrossRef]
- Zacharoulis, D.; Rountas, C.; Katsimpoulas, M.; Morianos, J.; Chatziandreou, I.; Vassilopoulos, G. Efficient liver gene transfer with foamy virus vectors. Med. Sci. Monit. Basic Res. 2013, 19, 214–220. [Google Scholar] [CrossRef]
- Gruntman, A.M.; Flotte, T.R. Progress with Recombinant Adeno-Associated Virus Vectors for Gene Therapy of Alpha-1 Antitrypsin Deficiency. Hum. Gene Ther. Methods 2015, 26, 77–81. [Google Scholar] [CrossRef]
- Brunetti-Pierri, N.; Ng, T.; Iannitti, D.; Cioffi, W.; Stapleton, G.; Law, M.; Breinholt, J.; Palmer, D.; Grove, N.; Rice, K. Transgene expression up to 7 years in nonhuman primates following hepatic transduction with helper-dependent adenoviral vectors. Hum. Gene Ther. 2013, 24, 761–765. [Google Scholar] [CrossRef]
- Romero-Vásquez, F.; Chávez, M.; Pérez, M.; Arcaya, J.L.; García, A.J.; Rincón, J.; Rodríguez-Iturbe, B. Overexpression of HGF transgene attenuates renal inflammatory mediators, Na(+)-ATPase activity and hypertension in spontaneously hypertensive rats. Biochim. Biophys. Acta 2012, 1822, 1590–1599. [Google Scholar] [CrossRef] [PubMed]
- Corridon, P.R.; Rhodes, G.J.; Leonard, E.C.; Basile, D.P.; Gattone, V.H.; Bacallao, R.L.; Atkinson, S.J. A method to facilitate and monitor expression of exogenous genes in the rat kidney using plasmid and viral vectors. Am. J. Physiol. Renal Physiol. 2013, 304, F1217–F1229. [Google Scholar] [CrossRef]
- De La Vega, J.; Braak, B.T.; Azzoni, A.R.; Monteiro, G.A.; Prazeres, D.M. Impact of plasmid quality on lipoplex-mediated transfection. J. Pharm. Sci. 2013, 102, 3932–3941. [Google Scholar] [CrossRef]
- Kamimura, K.; Yokoo, T.; Abe, H.; Kobayashi, Y.; Ogawa, K.; Shinagawa, Y.; Inoue, R.; Terai, S. Image-Guided Hydrodynamic Gene Delivery: Current Status and Future Directions. Pharmaceutics 2015, 7, 213–223. [Google Scholar] [CrossRef]
- Konry, T.; Sarkar, S.; Sabhachandani, P.; Cohen, N. Innovative Tools and Technology for Analysis of Single Cells and Cell-Cell Interaction. Annu. Rev. Biomed. Eng. 2016, 18, 259–284. [Google Scholar] [CrossRef]
- Beck, M.; Schmidt, A.; Malmstroem, J.; Claassen, M.; Ori, A.; Szymborska, A.; Herzog, F.; Rinner, O.; Ellenberg, J.; Aebersold, R. The quantitative proteome of a human cell line. Mol. Syst. Biol. 2011, 7, 549. [Google Scholar] [CrossRef]
- Han, F.; Lillard, S.J. In-situ sampling and separation of RNA from individual mammalian cells. Anal. Chem. 2000, 72, 4073–4079. [Google Scholar] [CrossRef]
- Herrero, M.J.; Sabater, L.; Guenechea, G.; Sendra, L.; Montilla, A.I.; Abargues, R.; Navarro, V.; Aliño, S.F. DNA delivery to ’ex vivo’ human liver segments. Gene Ther. 2012, 19, 504–512. [Google Scholar] [CrossRef]
- Sendra Gisbert, L.; Miguel Matas, A.; Sabater Ortí, L.; Herrero, M.J.; Sabater Olivas, L.; Montalvá Orón, E.M.; Frasson, M.; Abargues López, R.; López-Andújar, R.; García-Granero Ximénez, E.; et al. Efficacy of hydrodynamic interleukin 10 gene transfer in human liver segments with interest in transplantation. Liver Transpl. 2017, 23, 50–62. [Google Scholar] [CrossRef]
- Sendra, L.; Miguel, A.; Herrero, M.J.; Forteza, M.J.; Chaustre, F.L.; Noguera, I.; Diaz, A.; Bodi, V.; Alino, S.F. Human Interleukin-10 Naked DNA Delivery to Infarcted Pig Heart by Catheter Mediated Retrograde Injection in Coronary Sinus. J. Clin. Exp. Cardiol. 2014, 5, 315. [Google Scholar] [CrossRef]
- Seldon, P.M.; Barnes, P.J.; Giembycz, M.A. Interleukin-10 does not mediate the inhibitory effect of PDE-4 inhibitors and other cAMP-elevating drugs on lipopolysaccharide-induced tumors necrosis factor-alpha generation from human peripheral blood monocytes. Cell Biochem. Biophys. 1998, 29, 179–201. [Google Scholar] [CrossRef]
- Frasson, M.; Sendra, L.; Miguel, A.; Herrero, M.J.; Montalvá, E.; López-Andújar, R.; Martínez-Pastor, J.; Martí-Bonmatí, L.; Granero, E.G.; Aliño, S. Hydrodynamic Il10 Gene Transfer In Human Colon: Results From An "Ex-Vivo" Study With Potential Clinical Application In Crohn’s Disease. Inflamm. Bowel Dis. 2017, 23, 1360–1370. [Google Scholar] [CrossRef]
- Guillem, V.M.; Tormo, M.; Revert, F.; Benet, I.; García-Conde, J.; Crespo, A.; Aliño, S.F. Polyethyleneimine-based immunopolyplex for targeted gene transfer in human lymphoma cell lines. J. Gene Med. 2002, 4, 170–182. [Google Scholar] [CrossRef]
- Guillem, V.M.; Tormo, M.; Moret, I.; Benet, I.; García-Conde, J.; Crespo, A.; Aliño, S.F. Targeted oligonucleotide delivery in human lymphoma cell lines using a polyethyleneimine based immunopolyplex. J. Control. Release 2002, 83, 133–146. [Google Scholar] [CrossRef]
- Lledo, S.; Alfonso, R.; Alino, S.F. Antisense gene therapy using anti-k-ras and antitelomerase oligonucleotides in colorectal cancer. Rev. Esp. Enferm. Dig. 2005, 97, 472–480. [Google Scholar] [CrossRef]
- Díaz-Moscoso, A.; Guilloteau, N.; Bienvenu, C.; Méndez-Ardoy, A.; Blanco, J.L.; Benito, J.M.; Le Gourriérec, L.; Di Giorgio, C.; Vierling, P.; Defaye, J. Mannosyl-coated nanocomplexes from amphiphilic cyclodextrins and pDNA for site-specific gene delivery. Biomaterials 2011, 32, 7263–7273. [Google Scholar] [CrossRef]
- Taniyama, Y.; Azuma, J.; Kunugiza, Y.; Iekushi, K.; Rakugi, H.; Morishita, R. Therapeutic option of plasmid-DNA based gene transfer. Curr. Top. Med. Chem. 2012, 12, 1630–1637. [Google Scholar] [CrossRef]
- Balbino, T.A.; Azzoni, A.R.; de la Torre, L.G. Microfluidic devices for continuous production of pDNA/cationic liposome complexes for gene delivery and vaccine therapy. Colloids Surf. B Biointerfaces 2013, 111, 203–210. [Google Scholar] [CrossRef]
- Sevimli, S.; Sagnella, S.; Kavallaris, M.; Bulmus, V.; Davis, T.P. Assessment of cholesterol-derived ionic copolymers as potential vectors for gene delivery. Biomacromolecules 2013, 14, 4135–4149. [Google Scholar] [CrossRef]
- Heller, R.; Heller, L.C. Gene electrotransfer clinical trials. Adv. Genet. 2015, 89, 235–262. [Google Scholar]
- Mendrek, B.; Mendrek, B.; Sieroń, Ł.; Żymełka-Miara, I.; Binkiewicz, P.; Libera, M.; Smet, M.; Trzebicka, B.; Sieroń, A.L.; Kowalczuk, A.; et al. Nonviral Plasmid DNA Carriers Based on N,N’-Dimethylaminoethyl Methacrylate and Di(ethylene glycol) Methyl Ether Methacrylate Star Copolymers. Biomacromolecules 2015, 16, 3275–3285. [Google Scholar] [CrossRef]
- Zhang, Z.B.; Gao, W.; Shi, Y.; Liu, L.; Ma, N.; Chen, J.; Zhu, Z.J. Protective role of normothermic machine perfusion during reduced-size liver transplantation in pigs. Liver Transpl. 2016, 22, 968–978. [Google Scholar] [CrossRef]
| Author | Year | Organ | Gene | Gene Construct | HD Variants | Experimental Aim | Disease | Long-Term Expression |
|---|---|---|---|---|---|---|---|---|
| Bell, J.B. [83] | 2007 | Liver | HemOxigenase-1, LDLR/VLDLR | SB Transposon | HD | Transgene expression, qRT-PCR, Integration, Plasma presence, Clinic variables | Method, Sickle-cell disease, Familial hypercholesterolemia | up to >6 months |
| Belcher, JD [50] | 2010 | |||||||
| Hackett, P.B. [84] | 2011 | |||||||
| Turunen TA [38] | 2016 | |||||||
| Yang, PL [53] | 2002 | Liver | HBV | Sleeping beauty transposon-super genomic DNA | HD-HBV induction | Histology, titration, southern blot, northern blot, PCR, ELISA | Hepatitis B | 20 days |
| Doherty J [85] | 2012 | Liver/HEK293, HeLa, T cells | Neomycin resistance, full length Factor VIII | PiggyBac Transposon Vector cDNA | HD | Transgene expression, qPCR, clinical assays | Method, Haemophilia A | up to >300 days |
| Matsui H [36] | 2014 | |||||||
| Chen, I.Y. [86] | 2014 | Liver/heart | Luc | Titrable two-step transcriptional amplification vector strategy | HD/intramyocardial injection | Fluc exp modulation by raloxifene-mediated activator | ||
| Camassola M [46] | 2005 | Liver/syst | IUDA | Superfect complexes cDNA | HD caudal vs. intraperitoneal | Activity, transgene expression, PCR | Muccopolysacharidosis I | 14 days |
| Alino SF [1] | 1994 | Liver | hAAT (pTG7101) | Small liposomes | single & multiple dose + Partial HTx | ELISA & Liver Cytophotometry | 15 days | |
| Zender, L [56] | 2003 | Liver, heart, vascular | Cas8, alpha-gal, antiviral, Fas, Cas8, GFP, OPN, nAchRalpha1, Apo-LP | siRNA | Intraportal, aortic, in vitro | Luciferase, expression, Western Blot, ELISA, immunofluorescence, survival, virus titre, histochemistry | Liver failure, Influenza, sepsis, hepatitis, transplantation, atherosclerosis | up to 4 months |
| Tompkins, SM [80] | 2004 | |||||||
| Chu [87] | 2005 | |||||||
| Wesche-Soldato, DE [79] | 2005 | |||||||
| Saito Y [57] | 2007 | |||||||
| Zhang G [23] | 2011 | |||||||
| Kim, S.I. [55] | 2009 | |||||||
| Wesche-Soldato DE [88] | 2008 | |||||||
| Huang M [62] | 2013 | Liver | NKG2D, PTP1B | shRNA | HD | Cell count, histology, cytometry, Luciferase, Western Blot, qPCR, glucose levels | Hepatitis, diabetes | up to 10 days |
| Vakili S [44] | 2013 | |||||||
| Magnusson T. [89] | 2011 | Liver | Luc | Promoter: CMV-EF1a | HD-tail vein | Luciferase, qPCR | up to 2 months | |
| Schuttrumpf J [35] | 2011 | Liver | FIX human | Plasmid vs. minicircle | HD | Expression, function, methylation | Haemophilia B | 100 days |
| Ando M [70] | 2014 | Liver, kidney, muscle, lung, cells, tumour | IFN-heparin binding domain, hAAT, hGF, PTH, alpha-Gal, hFIX, IFN-alb, hIL37, Luc, FcgammaR-Ig, PAP1, mIL10, mKATE, hKS, IL28B, mIL15+mIL15R, hGH, EPO, Distr, LacZ, VWF, Flk-1, UCP, Adiponectin, IGF, IFN-albumin binding peptide | Plasmid DNA | HD, retro-orbital, hind limb, im, kidney injection, saphenous vein | Serum concentration, expression, ELISA, qPCR, Western Blot, histology, Luciferase, glucose levels, injury, electron microscopy | Cancer, hAAT deficiency, hypophysectomised, hypoparathyroidism, Fabry disease, Haemophilia B, metastasis, hepatitis, pancreatitis, obesity, dystrophy, inflammation, hGH deficiency, streptococcus infection, method, Von Willebrand disease, diabetes, nerve injury | up to >8 months |
| Zhang G [4] | 2000 | |||||||
| Sondergaard, M [75] | 2003 | |||||||
| Lee S [76] | 2008 | |||||||
| Nakamura G [82] | 2008 | |||||||
| Kim H.S. [34] | 2011 | |||||||
| Miyakawa N. [67] | 2011 | |||||||
| Bulau, AM [59] | 2011 | |||||||
| Yan S [90] | 2012 | |||||||
| Shashidharamurthy, R [60] | 2012 | |||||||
| Shigekawa, M [91] | 2012 | |||||||
| Gao M [10] | 2013 | |||||||
| Guess, MG [25] | 2013 | |||||||
| Lu S.L. [81] | 2013 | |||||||
| Qiu C [74] | 2014 | |||||||
| Sun H [73] | 2016 | |||||||
| Wolff, JA [6] | 1990 | |||||||
| Dagnaes-Hansen, F [92] | 2002 | |||||||
| Alino, SF [5] | 2003 | |||||||
| Hagstrom, JE [93] | 2004 | |||||||
| Zhang, G. [94] | 2004 | |||||||
| Crespo, A [17] | 2005 | |||||||
| Pergolizzi, RG [49] | 2006 | |||||||
| Yazawa, H [66] | 2006 | |||||||
| Gonzalez-Muniesa, P [40] | 2006 | |||||||
| Fukushima, M [42] | 2007 | |||||||
| Li, W. [95] | 2008 | |||||||
| Schuttrumpf, H [32] | 2008 | |||||||
| Suda T [96] | 2008 | |||||||
| Podetz-Pedersen, KM [97] | 2010 | |||||||
| Xu, JF. [58] | 2010 | |||||||
| Herrero, M.J. [31] | 2011 | |||||||
| Ma, Y [43] | 2013 | |||||||
| Miyakawa, N. [69] | 2013 | |||||||
| Wooddell, C [98] | 2011 | |||||||
| Nagata, K [27] | 2014 | |||||||
| He, C [41] | 2004 | Liver/Skeletal Muscle | Insulin, hGF, IL6, IFNg, mFGF21, SGSH, IL6-RFP-Fc, IL6 | Plasmid cDNA | HD, im | Expression, plasma protein, immunohistology, clinical analysis, pathology, qPCR, WB, ELISA | Diabetes type 1 and 2, obesity, glomerulonephritis, dermatitis, MPSIIIA, inflammation | up to 120 days |
| Bu, X [99] | 2011 | |||||||
| Mukumoto, H [26] | 2013 | |||||||
| Watcharanurak, K [78] | 2013 | |||||||
| Baribault, H. [45] | 2014 | |||||||
| Gao, M [11] | 2014 | |||||||
| Quiviger, M [48] | 2014 | |||||||
| Gortz, D [63] | 2015 | |||||||
| Ma, Y [12] | 2015 | |||||||
| Duguid, JG [100] | 1998 | Cell lines | b-Gal, hGH, eGFP | Peptide/DNA pH sensitive, PEI & DOTAP/DNA complexes | in vitro | Cytochemistry, g-gal chemoluminiscence, fluorimetry, electrofluorescence, TEM, cytofluorescence, dynamics of gene transfer | 14 days | |
| Moret, I [101] | 2001 | |||||||
| Alino, SF [102] | 2000 | PD:D-R, Em, EC50, Pot, Afin | ||||||
| Alino, SF [28] | 1993 | Liver | hAAT | Large/small liposomes, Liposomes (-)vs(+) plus | iv, HTx | Cytophotometry, DNA, Size Distribution, ELISA | up to 5 months | |
| Alino, SF [5] | 1993 | |||||||
| Alino, SF [29] | 1996 | |||||||
| Crespo, J [2] | 1996 | |||||||
| Budker, V [7] | 1996 | Liver | b-Gal, hGH | Naked | DNA, hypertonic solution-portal injection, hepatic vein occlusion | ELISA & histology | 2 days | |
| Xu, Z.X. [103] | 2009 | Liver | hFIX, hAAT | Integrative DNA plasmid | HD | Specific insertion, plasma concentration, toxicity, expression, IHC | Haemophilia B | up to 250 days |
| Keravala, A. [33] | 2011 | |||||||
| Ando, M [68] | 2012 | Liver | IFN | pDNA varying CpG motifs number | HD | Expression | Cancer | |
| Viecelli HM [51] | 2014 | Liver | mPAH | Minicircular cDNA | HD | Expression serum and tissue, qPCR, histology | Phenylketonuria | >1 year |
| Liver | HCV, others | Genomic RNA-HCV internal ribosome entry site firefly luciferase, Non-viral | HD-HCV model | Histology, Luciferase | Hepatitis C | 10 days | ||
| McCaffrey, AP [54] | 2002 | |||||||
| Habbitt, OC [104] | 2007 | Liver | gDNA (100 kb), eGFP, LDLR | gDNA, GenomicGenes, BAC | HD | Efficacy vs. DNA copy number | Cholestrolemia | 4 months |
| Okumura, A [77] | 2008 | Liver | LECT2 | Expression vector non-viral | HD | Inflammatory expression, histopathology, PCR | Arthritis | 12 days |
| Zhang, G [22] | 2010 | Muscle | full-length Dystrophin Gene | Full length DNA | HD-limb vein | Distribution, expression, myofibres damage, Western Blot | Duchene | |
| Shahaf, G [105] | 2011 | Liver | hAAT | Epstein Bar Virus-plasmid | HD | Islet function, Treg, macrophage, IL1 | Islet allogenic transplant | up to 100 days |
| Ochoa, M [71] | 2012 | Liver | IL15+ApoA1+IL15Ra | Expression plasmid cDNA | HD | Cell count, pathology, Western Blot, PCR, cytometry | Cancer | 60 days |
| Holm, DA [37] | 2003 | Liver | SCAD, promoter genomic elements | cDNA | HD, in vitro | Plasma protein, NK reconstitution, toxicity | Metabolic disease | 31 days |
| Barao, I [72] | 2011 | Immunodeficiency and transplantation | 18 days | |||||
| Hibbit, O. [106] | 2011 | |||||||
| Dasi, F [3] | 2001 | Liver, Plasma | hAAT | ASF-Lp, PS, DOTAP, NLS | iv + Partial HTx | ELISA, PCR, Sequencing | hAAT deficiency | 6 months & 12 months |
| Wooddell, CI [107] | 2008 | Liver | Alkaline Phosphatase Reporter gene | Albumin promoter | HD | Plasma protein | Method | Albumin 1 year vs. CMV 1 day |
| Author | Year | Species | Organ | Gene | Gene Construct | Methodology | Variables | Disease | Long-Term Expression |
|---|---|---|---|---|---|---|---|---|---|
| Budker, V [8] | 1998 | Rat | Muscle | b-Gal, luciferase | Naked, Solution hypo/hypertonic | Artery injection High pressure (hind-limb) | Histochemistry, Luciferase | 2 days | |
| Eastman, SJ [125] | 2002 | Rabbit | Liver | Alkaline Phosphatase Reporter gene | DNA | HD catheter lobar and whole liver | Plasma Alkaline Phosphatases | Model | 2 days |
| Hagstrom, JE [93] | 2004 | Mouse, Rat, Dog, Primate | Muscle | DNA Luc vs. Ad; EPO; Distr | DNA, siRNA, Ad | HD vein limb | Luciferase | 30 days | |
| Inoue, S [109] | 2004 | Rat | Liver | b-Gal, luc-image, CTLA4Ig | DNA dosing CTLA4Ig | HD system and local- catheter | Transplantation | 2 days | |
| Zhang, X [113] | 2004 | Rat | Liver | Luciferase | DNA | HD vs. regional Portal | Luciferase | Method | short |
| Tosoulfas, G [117] | 2006 | hAAT, Luc | DNA | HD ex vivo DNA injection IVC closed | Injury, histopathology, physiology, efficacy | Transplantation | >5 days | ||
| Chang, H. [115] | 2008 | IL10-Ig fusion gene | DNA | IL-17, IL1beta, TNFa, IL1… | Myocarditis | ||||
| Suda, T [96] | 2008 | Mouse, Rat, Pig | Liver, Kidney, Muscle | Luc, GFP, Ad-GFP | DNA, Ad | HD computer assisted | Pressure, gene delivery/expression | ||
| Xing, Y [119] | 2009 | Rat | Kidney | Luc, EPO | HD and ultrasound combination | Method | |||
| Sawyer, GJ [120] | 2010 | Rat | Liver | Luc | DNA | HD-Regional Lobe without occlusion | Efficacy, luciferase activity | Method | |
| Wooddell, C [98] | 2011 | Mouse, Rat, Rhesus monkey | Muscle | LacZ | Plasmid DNA complexes | HD hind limb | Expression and delivery | null | 49 weeks |
| Cim, A [121] | 2012 | Rat | Liver | Pdx1, Ngn3, MafA | 5 different expression plasmids | HD | Expression, PCR, IHC | Diabetes type 1 | 28 days |
| Romero-Vasquez, F [138] | 2012 | Rat | Liver | hepatocyte growth factor | pCMV | HD | NFkB, RANTES, MCP1, IL6, oxidative stress | Renal hypertension | 6 weeks with weekly treatment |
| Zhao, M [122] | 2012 | Rat | Endometrium | pigment epithelium derived factor | Polymeric micelle | intravenous injection | Clinic observation of endometrium lesions | Endometriosis | |
| Corridon, PR [139] | 2013 | Rat | Kidney | eGFP, eGFP-actin/occluding/tubulin, tdTomato-H2B, RFP-actin | Plasmid, adenovirus, baculovirus | HD retrograde renal vein | Expression-intravital, confocal | 1 month | |
| De La Vega, J [140] | 2013 | Chinese hamster | Ovary cells | GFP | Plasmid lipofectamine lipoplexes | Methods of plasmid purification | Hydrodynamic diameter and zeta potential | ||
| Yasuzaki, Y [124] | 2013 | Rat | Muscle | Luc | DNA | HD-hindlimb | Expression, luminescence, qPCR, WB | Method | 24 h |
| Kamimura, K [126] | 2014 | Dog/Rat | Liver | Luc, hAAT, hFIX | Plasmid cDNA/DNA | HD-through hepatic veins of each 4 lobes with closed cava vein | Histology, physiological parameters | 6 weeks |
| Author | Year | Species | Organ | Gene | Gene Construct | Methodology | Variables | Disease | Long-Term Expression |
|---|---|---|---|---|---|---|---|---|---|
| Hagstrom, JE [93] | 2004 | Mouse, Rat, Dog, Primate | Muscle | DNA Luc vs. Ad; EPO; Distr | DNA, siRNA, Ad | HD vein limb | Luciferase | 30 days | |
| Yoshino, H [127] | 2006 | Pig | Liver | GFP, CTLA4-Ig | DNA | HD-cathe, closed (3 mg,150 mL, 5 mL/s) | Physiology, histology, fluorescence, plasma presence | Method | 1 day (161 ng/mL)-7 days |
| Alino, SF [128] | 2007 | Pig | Liver (small vs. Large) | hAAT | DNA | HD-Cathe, open (100 mL, 7.5 mL/s) | ELISA, IHC, injury, qRT-PCR | hAAT deficiency | 15 days (200 ng/mL) |
| Fabre, JW [129] | 2008 | Pig | Liver | pGL3 plasmid, Luc | DNA | HD-isolated segment of IVC | Pressure, ECG, heart rate, luciferase activity | Method | 1 day |
| Suda, T [96] | 2008 | Mouse, Rat, Pig | Liver, Kidney, Muscle | Luc, GFP, Ad-GFP | ADN, Ad | HD computer assisted | Pressure, gene delivery/expression | ||
| Aliño, SF [18] | 2010 | Pig | Heart | EGFP, GAPDH | Naked | HD Cath Coronary sinus | IHC, PCR, RT-PCR, copy number | Method | 1 day |
| Kamimura, K [130] | 2010 | Pig | Muscle | pCMV-Luc | DNA | HD hindlimb | Luciferase activity [95] | Method | 60 days |
| Fabre, JW [131] | 2011 | Pig | Liver segment | Luc | DNA | Surg-HD-LivSeg portal vs. hepat vein | Vascular pressure (>100 mmHg) | Method | Short |
| Hackett, PB [84] | 2011 | Small&Large animals/Rev | Liver | Luc | Sleeping Beauty Transposon | HD | Integration, Plasma presence | ||
| Wooddell, C [98] | 2011 | Mouse, Rat, Rhesus monkey | Muscle | LacZ | Plasmid DNA complexes | HD hind limb | Expression and delivery | 49 weeks | |
| Carreño, O [132] | 2013 | Pig | Liver | eGFP | Plasmid cDNA | Surgery isolation, HD simultaneous | Expression PCR | 1 day | |
| Zacharoulis, D [135] | 2013 | Pig | Liver | eGFP | Plasmid DNA vs. foamy virus vector-based | HD | Gene expression and qPCR | 1 week to 1 month | |
| Sendra, L [133] | 2014 | Pig | Liver | eGFP | Plasmid cDNA | HD-surgical isolation cava vs. porta | Gene and protein expression, qPCR, ELISA, TEM | 1 day | |
| Kamimura, K [141] | 2015 | Small and large animals | Liver | Various | Non-viral | HD | Various | ||
| Sendra, L [19] | 2016 | Pig | Liver | hAAT | Plasmid DNA | HD-open vs. closed catheterism | Tissue expression qPCR, ELISA, clinic observations | hAAT deficiency | 14 days |
| Human Liver | Total hAAT (copy/cell) | hAAT-flag (copy/cell) | hAAT-f/Total hAAT (%) |
|---|---|---|---|
| 1 | 7.16 × 105 | 3.89 × 105 | 54.31 |
| 2 | 8.98 × 105 | 5.31 × 105 | 59.18 |
| 3 | 9.62 × 105 | 1.97 × 105 | 20.46 |
| 4 | 4.03 × 105 | 2.43 × 105 | 60.13 |
| Average | 1.65 × 105 | 8.86 × 105 | 48.52 |
| sd | 1.59 × 105 | 1.04 × 105 | 18.88 |
| Author | Year | Model | Organ/Cell | Gene | Gene Construct | Methodology | Variables | Disease | Long-Term Expression |
|---|---|---|---|---|---|---|---|---|---|
| Guillem, V [150] | 2002 | Human | Lymphoid cell line | ODN-FITC | CD3-PEI/ODN- | In vitro | Fluorescence, Cells increase | Method | |
| Guillem, V [151] | 2002 | Human | Jurkat & Granta | eGFP | CD3-PEI/eGFP | In vitro | Selective gene delivery | Method | |
| Lledo, S [152] | 2005 | Human | Cell line SW480 | ASO-Kras | ASO phosphorotioates | In vitro | Cell viability | Cancer: colorectal | 72 h |
| Lee, S [76] | 2008 | mouse/human cell | Liver | PTH | Plasmid DNA | HD | Plasma protein, expression | Hypoparathyroidism | |
| Diaz-Moscoso, A [153] | 2011 | Human, Mouse | Macrophage | 80 nm manosilted cyclodextrin/DNAplex | In vitro | Delivery, FACS | |||
| Doherty, J [85] | 2012 | Mouse/Human cell | Liver/HEK293, HeLa, T cells | Neomycin resistance cassette | transposone-piggybac | HD | Transgene expression | 6 months | |
| Herrero, MJ [145] | 2012 | Human | Liver | eGFP | pCMV | HD | Expression, PCR, fluorescence, IHC | 2 days | |
| Taniyama, Y [154] | 2012 | Human | Heart | Various | Plasmid | physical procedures | Various | ||
| Balbino, TA [155] | 2013 | Human cells | HeLa | Cationic liposomes | Microfluidic systems comparison | Complex size, non-electrostatic bond, accessibility level | |||
| Sevimli, S [156] | 2013 | Human | Cells HepG2, H460, SHEP, MRC5 | GFP | Anionic and cationic polymers-siRNA | Transfection | Diameter, potential, stability, qPCR, WB, flow cytometry, confocal | ||
| Matsui, H [36] | 2014 | Mouse/Human cell | Liver/HEK293 | Full length Factor VIII | PiggyBac Transposon Vector cDNA | HD | Expression, PCR, qPCR, Coagulation assays | Haemophilia A | >300 days |
| Heller, R [157] | 2015 | Human | Various | Various | Non-viral | Electroporation | Clinical trials | Various | |
| Mendrek, B [158] | 2015 | Human | Cell line HT1080 (fibrosarcoma) | Plasmid-polyplex | Polyplexes DMAEMA (+) vs. DEGMA (0) | Hydrodynamic size, z potential, cytotoxicity, transfection efficacy |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sendra, L.; Herrero, M.J.; Aliño, S.F. Translational Advances of Hydrofection by Hydrodynamic Injection. Genes 2018, 9, 136. https://doi.org/10.3390/genes9030136
Sendra L, Herrero MJ, Aliño SF. Translational Advances of Hydrofection by Hydrodynamic Injection. Genes. 2018; 9(3):136. https://doi.org/10.3390/genes9030136
Chicago/Turabian StyleSendra, Luis, María José Herrero, and Salvador F. Aliño. 2018. "Translational Advances of Hydrofection by Hydrodynamic Injection" Genes 9, no. 3: 136. https://doi.org/10.3390/genes9030136





