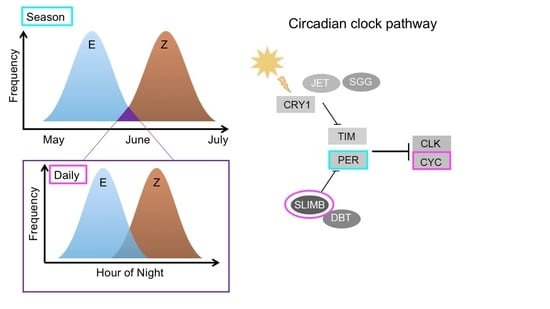
Non-Pleiotropic Coupling of Daily and Seasonal Temporal Isolation in the European Corn Borer
Abstract
:1. Introduction
2. Methods
2.1. Mating Trials
2.2. Sampling Time Points
2.3. Library Sequencing
2.4. Transcriptome Assembly
2.5. Differential Expression Analysis
2.6. Annotation
3. Results
3.1. Mating Trials
3.2. Transcriptome Assembly
3.3. Differential Expression Analysis
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Coyne, J.A.; Orr, H.A. Speciation; Sinauer Associates: Sunderland, MA, USA, 2004; Volume 37. [Google Scholar]
- Sobel, J.M.; Chen, G.F.; Watt, L.R.; Schemske, D.W. The Biology of Speciation. Evolution 2010, 64, 295–315. [Google Scholar] [CrossRef] [PubMed]
- Butlin, R.K.; Smadja, C.M. Coupling, Reinforcement, and Speciation. Am. Nat. 2017, 191, 155–172. [Google Scholar] [CrossRef] [PubMed]
- Dopman, E.B.; Robbins, P.S.; Seaman, A. Components of reproductive isolation between North American pheromone strains of the European corn borer. Evolution 2010, 64, 881–902. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Barrientos, D.; Grealy, A.; Nosil, P. The Genetics and Ecology of Reinforcement. Ann. N. Y. Acad. Sci. 2009, 1168, 156–182. [Google Scholar] [CrossRef] [PubMed]
- Emerson, K.J.; Bradshaw, W.E.; Holzapfel, C.M. Complications of complexity: Integrating environmental, genetic and hormonal control of insect diapause. Trends Genet. 2009, 25, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Arnold, S.J. Constraints on phenotypic evolution. Am. Nat. 1992, 140, S85–S107. [Google Scholar] [CrossRef] [PubMed]
- Fraser, H.B.; Hirsh, A.E.; Steinmetz, L.M.; Scharfe, C.; Feldman, M.W. Evolutionary rate in the protein interaction network. Science 2002, 296, 750–752. [Google Scholar] [CrossRef] [PubMed]
- Barton, N.H. Pleiotropic models of quantitative variation. Genetics 1990, 124, 773–782. [Google Scholar] [PubMed]
- Griswold, C.K.; Whitlock, M.C. The Genetics of Adaptation: The Roles of Pleiotropy, Stabilizing Selection and Drift in Shaping the Distribution of Bidirectional Fixed Mutational Effects. Genetics 2003, 165, 2181–2192. [Google Scholar] [PubMed]
- Seehausen, O.; Butlin, R.K.; Keller, I.; Wagner, C.E.; Boughman, J.W.; Hohenlohe, P.A.; Peichel, C.L.; Sætre, G.-P.; Bank, C.; Brännström, Å. Genomics and the origin of species. Nat. Rev. Genet. 2014, 15, 176–192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maes, G.E.; Pujolar, J.M.; Hellemans, B.; Volckaert, F.A.M. Evidence for isolation by time in the European eel (Anguilla anguilla L.). Mol. Ecol. 2006, 15, 2095–2107. [Google Scholar] [CrossRef] [PubMed]
- O’Malley, K.G.; Camara, M.D.; Banks, M.A. Candidate loci reveal genetic differentiation between temporally divergent migratory runs of Chinook salmon (Oncorhynchus tshawytscha). Mol. Ecol. 2007, 16, 4930–4941. [Google Scholar] [CrossRef] [PubMed]
- Fukami, H.; Omori, M.; Shimoike, K.; Hayashibara, T.; Hatta, M. Ecological and genetic aspects of reproductive isolation by different spawning times in Acropora corals. Mar. Biol. 2003, 142, 679–684. [Google Scholar] [CrossRef]
- Levitan, D.R.; Fukami, H.; Jara, J.; Kline, D.; McGovern, T.M.; McGhee, K.E.; Swanson, C.A.; Knowlton, N. Mechanisms of reproductive isolation among sympatric broadcast-spawning corals of the Montastraea annularis species complex. Evolution 2004, 58, 308–323. [Google Scholar] [CrossRef] [PubMed]
- Sawadogo, S.P.; Costantini, C.; Pennetier, C.; Diabaté, A.; Gibson, G.; Dabiré, R.K. Differences in timing of mating swarms in sympatric populations of Anopheles coluzzii and Anopheles gambiae s.s. (formerly A. gambiae M and S molecular forms) in Burkina Faso, West Africa. Parasit. Vectors 2013, 6, 275. [Google Scholar] [CrossRef] [PubMed]
- Pashley, D.P.; Hammond, A.M.; Hardy, T.N. Reproductive isolating mechanisms in fall armyworm host strains (Lepidoptera: Noctuidae). Ann. Entomol. Soc. Am. 1992, 85, 400–405. [Google Scholar] [CrossRef]
- Monti, L.; Genermont, J.; Malosse, C.; Lalanne Cassou, B. A genetic analysis of some components of reproductive isolation between two closely related species, Spodoptera latifascia (Walker) and S. descoinsi (Lalanne-Cassou and Silvain) (Lepidoptera: Noctuidae). J. Evol. Biol. 1997, 10, 121–134. [Google Scholar] [CrossRef]
- Ueno, H.; Furukawa, S.; Tsuchida, K. Difference in the time of mating activity between host-associated populations of the rice stem borer, Chilo suppressalis (Walker). Entomol. Sci. 2006, 9, 255–259. [Google Scholar] [CrossRef]
- Huang, C.; Hu, B.; Li, J.; Wang, Y. Water-oats harbors two strains of the striped stem borer Chilo suppressalis (Lepidoptera: Crambidae) with temporal divergence in mating behavior. Appl. Entomol. Zool. 2016, 51, 457–463. [Google Scholar] [CrossRef]
- Hendry, A.P.; Day, T. Population structure attributable to reproductive time: Isolation by time and adaptation by time. Mol. Ecol. 2005, 14, 901–916. [Google Scholar] [CrossRef] [PubMed]
- Taylor, R.S.; Friesen, V.L. The role of allochrony in speciation. Mol. Ecol. 2017, 26, 3330–3342. [Google Scholar] [CrossRef] [PubMed]
- Denlinger, D.L.; Hahn, D.A.; Merlin, C.; Holzapfel, C.M.; Bradshaw, W.E. Keeping time without a spine: What can the insect clock teach us about seasonal adaptation? Philos. Trans. R. Soc. B Biol. Sci. 2017, 372. [Google Scholar] [CrossRef] [PubMed]
- Krupp, J.J.; Billeter, J.-C.; Wong, A.; Choi, C.; Nitabach, M.N.; Levine, J.D. Pigment-dispersing factor modulates pheromone production in clock cells that influence mating in Drosophila. Neuron 2013, 79, 54–68. [Google Scholar] [CrossRef] [PubMed]
- Dauwalder, B.; Tsujimoto, S.; Moss, J.; Mattox, W. The Drosophila takeout gene is regulated by the somatic sex-determination pathway and affects male courtship behavior. Genes Dev. 2002, 16, 2879–2892. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Sehgal, A. Probing the relative importance of molecular oscillations in the circadian clock. Genetics 2008, 178, 1147–1155. [Google Scholar] [CrossRef] [PubMed]
- Hardin, P.E. The circadian timekeeping system of Drosophila. Curr. Biol. 2005, 15, R714–R722. [Google Scholar] [CrossRef] [PubMed]
- Emery, P.; So, W.V.; Kaneko, M.; Hall, J.C.; Rosbash, M. CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell 1998, 95, 669–679. [Google Scholar] [CrossRef]
- Zhan, S.; Merlin, C.; Boore, J.L.; Reppert, S.M. The monarch butterfly genome yields insights into long-distance migration. Cell 2011, 147, 1171–1185. [Google Scholar] [CrossRef] [PubMed]
- Sandrelli, F.; Costa, R.; Kyriacou, C.P.; Rosato, E. Comparative analysis of circadian clock genes in insects. Insect Mol. Biol. 2008, 17, 447–463. [Google Scholar] [CrossRef] [PubMed]
- Sakai, T.; Ishida, N. Circadian rhythms of female mating activity governed by clock genes in Drosophila. Proc. Natl. Acad. Sci. USA 2001, 98, 9221–9225. [Google Scholar] [CrossRef] [PubMed]
- Tauber, E.; Roe, H.; Costa, R.; Hennessy, J.M.; Kyriacou, C.P. Temporal mating isolation driven by a behavioral gene in Drosophila. Curr. Biol. 2003, 13, 140–145. [Google Scholar] [CrossRef]
- Fuchikawa, T.; Sanada, S.; Nishio, R.; Matsumoto, A.; Matsuyama, T.; Yamagishi, M.; Tomioka, K.; Tanimura, T.; Miyatake, T. The clock gene cryptochrome of Bactrocera cucurbitae (Diptera: Tephritidae) in strains with different mating times. Heredity 2010, 104, 387–392. [Google Scholar] [CrossRef] [PubMed]
- Miyatake, T.; Matsumoto, A.; Matsuyama, T.; Ueda, H.R.; Toyosato, T.; Tanimura, T. The period gene and allochronic reproductive isolation in Bactrocera cucurbitae. Proc. R. Soc. B Biol. Sci. 2002, 269, 2467–2472. [Google Scholar] [CrossRef] [PubMed]
- Hänniger, S.; Dumas, P.; Schöfl, G.; Gebauer-Jung, S.; Vogel, H.; Unbehend, M.; Heckel, D.G.; Groot, A.T. Genetic basis of allochronic differentiation in the fall armyworm. BMC Evol. Boil. 2017, 17, 68. [Google Scholar] [CrossRef] [PubMed]
- Poehn, B.; Szkiba, D.; Preussner, M.; Sedlazeck, F.J.; Zrim, A.; Neumann, T.; Nguyen, L.-T.; Betancourt, A.J.; Hummel, T.; Vogel, H.; et al. The genomic basis of circadian and circalunar timing adaptations in a midge. Nature 2016, 540, 69–73. [Google Scholar]
- Koštál, V. Eco-physiological phases of insect diapause. J. Insect Physiol. 2006, 52, 113–127. [Google Scholar] [CrossRef] [PubMed]
- Meuti, M.E.; Denlinger, D.L. Evolutionary links between circadian clocks and photoperiodic diapause in insects. Integr. Comp. Biol. 2013, 53, 131–143. [Google Scholar] [CrossRef] [PubMed]
- Saunders, D.S.; Lewis, R.D.; Warman, G.R. Photoperiodic induction of diapause: Opening the black box. Physiol. Entomol. 2004, 29, 1–15. [Google Scholar] [CrossRef]
- Ikeno, T.; Numata, H.; Goto, S.G. Circadian clock genes period and cycle regulate photoperiodic diapause in the bean bug Riptortus pedestris males. J. Insect Physiol. 2011, 57, 935–938. [Google Scholar] [CrossRef] [PubMed]
- Bradshaw, W.E.; Holzapfel, C.M.; Mathias, D. Circadian rhythmicity and photoperiodism in the pitcher-plant mosquito: Can the seasonal timer evolve independently of the circadian clock? Am. Nat. 2006, 167, 601–605. [Google Scholar] [PubMed]
- Bradshaw, W.E.; Emerson, K.J.; Holzapfel, C.M. Genetic correlations and the evolution of photoperiodic time measurement within a local population of the pitcher-plant mosquito, Wyeomyia smithii. Heredity 2012, 108, 473–479. [Google Scholar] [CrossRef] [PubMed]
- Kochansky, J.; Cardé, R.T.; Liebherr, J.; Roelofs, W.L. Sex pheromone of the European corn borer, Ostrinia nubilalis (Lepidoptera: Pyralidae), in New York. J. Chem. Ecol. 1975, 1, 225–231. [Google Scholar] [CrossRef]
- Glover, T.J.; Knodel, J.J.; Robbins, P.S.; Eckenrode, C.J.; Roelofs, W.L. Gene flow among three races of European corn norers (Lepidoptera: Pyralidae) in New York State. Environ. Entomol. 1991, 20, 1356–1362. [Google Scholar] [CrossRef]
- Liebherr, J.; Roelofs, W.L. Laboratory Hybridization and Mating Period Studies Using Two Pheromones Strains of Ostrinia nubilalis. Ann. Entomol. Soc. Am. 1975, 68, 305–309. [Google Scholar] [CrossRef]
- Kárpáti, Z.; Molnar, B.; Szőcs, G. Pheromone titer and mating frequency of E- and Z-strains of the European corn borer, Ostrinia nubilalis: Fluctuation during scotophase and age dependence. Acta Phytopathol. Entomol. Hung. 2007, 42, 331–341. [Google Scholar] [CrossRef]
- Glover, T.J.; Robbins, P.S.; Eckenrode, C.J.; Roelofs, W.L. Genetic control of voltinism characteristics in European corn borer races assessed with a marker gene. Arch. Insect Biochem. Physiol. 1992, 20, 107–117. [Google Scholar] [CrossRef]
- McLeod, D.G.R.; Beck, S.D. Photoperiodic termination of diapause in an insect. Biol. Bull. 1963, 124, 84–96. [Google Scholar] [CrossRef]
- Dopman, E.B.; Perez, L.; Bogdanowicz, S.M.; Harrison, R.G. Consequences of reproductive barriers for genealogical discordance in the European corn borer. Proc. Natl. Acad. Sci. USA 2005, 102, 14706–14711. [Google Scholar] [CrossRef] [PubMed]
- Wadsworth, C.B.; Woods, W.A., Jr.; Hahn, D.A.; Dopman, E.B. One phase of the dormancy developmental pathway is critical for the evolution of insect seasonality. J. Evol. Biol. 2013, 26, 2359–2368. [Google Scholar] [CrossRef] [PubMed]
- Dopman, E.B.; Bogdanowicz, S.M.; Harrison, R.G. Genetic mapping of sexual isolation between E and Z pheromone strains of the European corn norer (Ostrinia nubilalis). Genetics 2004, 167, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Levy, R.C.; Kozak, G.M.; Wadsworth, C.B.; Coates, B.S.; Dopman, E.B. Explaining the sawtooth: Latitudinal periodicity in a circadian gene correlates with shifts in generation number. J. Evol. Biol. 2015, 28, 40–53. [Google Scholar] [CrossRef] [PubMed]
- Wadsworth, C.B.; Li, X.; Dopman, E.B. A recombination suppressor contributes to ecological speciation in Ostrinia moths. Heredity 2015, 114, 593–600. [Google Scholar] [CrossRef] [PubMed]
- Wadsworth, C.B.; Dopman, E.B. Transcriptome profiling reveals mechanisms for the evolution of insect seasonality. J. Exp. Biol. 2015, 218, 3611–3622. [Google Scholar] [CrossRef] [PubMed]
- Giebultowicz, J.M. Molecular mechanism and cellular distribution of insect circadian clocks. Annu. Rev. Entomol. 2000, 45, 769–793. [Google Scholar] [CrossRef] [PubMed]
- Schuckel, J.; Siwicki, K.K.; Stengl, M. Putative circadian pacemaker cells in the antenna of the hawkmoth Manduca sexta. Cell Tissue Res. 2007, 330, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Merlin, C.; Lucas, P.; Rochat, D.; François, M.-C.; Maïbèche-Coisne, M.; Jacquin-Joly, E. An antennal circadian clock and circadian rhythms in peripheral pheromone reception in the moth Spodoptera littoralis. J. Biol. Rhythm. 2007, 22, 502–514. [Google Scholar] [CrossRef] [PubMed]
- Kobelková, A.; Závodská, R.; Sauman, I.; Bazalová, O.; Doležel, D. Expression of clock genes period and timeless in the central nervous system of the Mediterranean flour moth, Ephestia kuehniella. J. Biol. Rhythm. 2015, 30, 104–116. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Charif, D.; Lobry, J.R. SeqinR 1.0-2: A contributed package to the R project for statistical computing devoted to biological sequences retrieval and analysis. In Structural Approaches to Sequence Evolution; Springer: Berlin/Heidelberg, Germany, 2007; pp. 207–232. [Google Scholar]
- Risso, D.; Schwartz, K.; Sherlock, G.; Dudoit, S. GC-content normalization for RNA-seq data. BMC Bioinform. 2011, 12, 480. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Al-Wathiqui, N.; Lewis, S.M.; Dopman, E.B. Using RNA sequencing to characterize female reproductive genes between Z and E Strains of European corn borer moth (Ostrinia nubilalis). BMC Genom. 2014, 15, 189. [Google Scholar] [CrossRef] [PubMed]
- Shimomura, M.; Minami, H.; Suetsugu, Y.; Ohyanagi, H.; Satoh, C.; Antonio, B.; Nagamura, Y.; Kadono-Okuda, K.; Kajiwara, H.; Sezutsu, H.; et al. KAIKObase: An integrated silkworm genome database and data mining tool. BMC Genom. 2009, 10, 486. [Google Scholar] [CrossRef] [PubMed]
- Zhan, S.; Reppert, S.M. MonarchBase: The monarch butterfly genome database. Nucleic Acids Res. 2012, 41, D758–D763. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, G.; Schroeder, A.J.; Goodman, J.L.; Strelets, V.B.; Crosby, M.A.; Thurmond, J.; Emmert, D.B.; Gelbart, W.M.; Consortium, F. FlyBase: Introduction of the Drosophila melanogaster Release 6 reference genome assembly and large-scale migration of genome annotations. Nucleic Acids Res. 2014, 43, D690–D697. [Google Scholar] [CrossRef] [PubMed]
- Eden, E.; Navon, R.; Steinfeld, I.; Lipson, D.; Yakhini, Z. GOrilla: A tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinf. 2009, 10, 48. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Coates, B.S.; Ge, X.; Bai, S.; He, K.; Wang, Z. Male-and female-biased gene expression of olfactory-related genes in the antennae of Asian corn borer, Ostrinia furnacalis (Guenée) (Lepidoptera: Crambidae). PLoS ONE 2015, 10, e0128550. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Ozaki, K.; Ishikawa, Y.; Matsuo, T. Identification of candidate odorant receptors in Asian corn borer Ostrinia furnacalis. PLoS ONE 2015, 10, e0121261. [Google Scholar] [CrossRef] [PubMed]
- Wei, T.; Simko, V. corrplot Visualization of a Correlation Matrix; R Package Version 0.77; CRAN: Vienna, Austria, 2016. [Google Scholar]
- Groot, A.T. Circadian rhythms of sexual activities in moths: A review. Front. Ecol. Evol. 2014, 2, 43. [Google Scholar] [CrossRef]
- Hanin, O.; Azrielli, A.; Zakin, V.; Applebaum, S.; Rafaeli, A. Identification and differential expression of a sex-peptide receptor in Helicoverpa armigera. Insect Biochem. Mol. Biol. 2011, 41, 537–544. [Google Scholar] [CrossRef] [PubMed]
- Hao, H.; Allen, D.L.; Hardin, P.E. A circadian enhancer mediates PER-dependent mRNA cycling in Drosophila melanogaster. Mol. Cell. Biol. 1997, 17, 3687–3693. [Google Scholar] [CrossRef] [PubMed]
- Glossop, N.R.; Houl, J.H.; Zheng, H.; Ng, F.S.; Dudek, S.M.; Hardin, P.E. VRILLE feeds back to control circadian transcription of Clock in the Drosophila circadian oscillator. Neuron 2003, 37, 249–261. [Google Scholar] [CrossRef]
- Ko, H.W.; Jiang, J.; Edery, I. Role for Slimb in the degradation of Drosophila Period protein phosphorylated by Doubletime. Nature 2002, 420, 673–678. [Google Scholar] [CrossRef] [PubMed]
- Nikhil, K.L.; Abhilash, L.; Sharma, V.K. Molecular correlates of circadian clocks in fruit fly Drosophila melanogaster populations exhibiting early and late emergence chronotypes. J. Biol. Rhythm. 2016, 31, 125–141. [Google Scholar] [CrossRef] [PubMed]
- Koolman, J. Control of ecdysone biosynthesis in insects. Neth. J. Zool. 1994, 45, 83–88. [Google Scholar] [CrossRef]
- Beckstead, R.B.; Lam, G.; Thummel, C.S. The genomic response to 20-hydroxyecdysone at the onset of Drosophila metamorphosis. Genome Biol. 2005, 6, R99. [Google Scholar] [CrossRef] [PubMed]
- Ables, E.T.; Hwang, G.H.; Finger, D.S.; Hinnant, T.D.; Drummond-Barbosa, D. A genetic mosaic screen reveals ecdysone-responsive genes regulating Drosophila oogenesis. G3 Genes Genomes Genet. 2016, 6, 2629–2642. [Google Scholar] [CrossRef] [PubMed]
- Nitabach, M.N.; Blau, J.; Holmes, T.C. Electrical silencing of Drosophila pacemaker neurons stops the free-running circadian clock. Cell 2002, 109, 485–495. [Google Scholar] [CrossRef]
- Ragland, G.J.; Keep, E. Comparative transcriptomics support evolutionary convergence of diapause responses across Insecta. Physiol. Entomol. 2017. [Google Scholar] [CrossRef]
- Bloch, G.; Hazan, E.; Rafaeli, A. Circadian rhythms and endocrine functions in adult insects. J. Insect Physiol. 2013, 59, 56–69. [Google Scholar] [CrossRef] [PubMed]
- Cusson, M.; Tobe, S.S.; McNeil, J.N. Juvenile hormones: Their role in the regulation of the pheromonal communication system of the armyworm moth, Pseudaletia unipuncta. Arch. Insect Biochem. Physiol. 1994, 25, 329–345. [Google Scholar] [CrossRef]
- Yagi, S.; Fukaya, M. Juvenile hormone as a key factor regulating larval diapause of the rice stem borer, Chilo suppressalis (Lepidoptera: Pyralidae). Appl. Entomol. Zool. 1974, 9, 247–255. [Google Scholar] [CrossRef]
- Nijhout, H.F.; Williams, C.M. Control of moulting and metamorphosis in the tobacco hornworm, Manduca sexta (L.): Growth of the last-instar larva and the decision to pupate. J. Exp. Biol. 1974, 61, 481–491. [Google Scholar] [PubMed]
- So, W.V.; Sarov-Blat, L.; Kotarski, C.K.; McDonald, M.J.; Allada, R.; Rosbash, M. Takeout, a novel Drosophila gene under circadian clock transcriptional regulation. Mol. Cell. Biol. 2000, 20, 6935–6944. [Google Scholar] [CrossRef] [PubMed]
- Saito, K.; Su, Z.H.; Emi, A.; Mita, K.; Takeda, M.; Fujiwara, Y. Cloning and expression analysis of takeout/JHBP family genes of silkworm, Bombyx mori. Insect Mol. Biol. 2006, 15, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Benito, J.; Hoxha, V.; Lama, C.; Lazareva, A.A.; Ferveur, J.-F.; Hardin, P.E.; Dauwalder, B. The circadian output gene takeout is regulated by Pdp1ε. Proc. Natl. Acad. Sci. USA 2010, 107, 2544–2549. [Google Scholar] [CrossRef] [PubMed]
- Wagner, G.P.; Pavlicev, M.; Cheverud, J.M. The road to modularity. Nat. Rev. Genet. 2007, 8, 921–931. [Google Scholar] [CrossRef] [PubMed]
- Fraser, H.B. Modularity and evolutionary constraint on proteins. Nat. Genet. 2005, 37, 351–352. [Google Scholar] [CrossRef] [PubMed]
- Dai, F.-Y.; Qiao, L.; Tong, X.-L.; Cao, C.; Chen, P.; Chen, J.; Lu, C.; Xiang, Z.-H. Mutations of an arylalkylamine-N-acetyltransferase, Bm-iAANAT, are responsible for silkworm melanism mutant. J. Biol. Chem. 2010, 285, 19553–19560. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, A.A.; Wang, Q.; Bembenek, J.; Ichihara, N.; Hiragaki, S.; Suzuki, T.; Takeda, M. N-acetyltransferase (nat) is a critical conjunct of photoperiodism between the circadian system and endocrine axis in Antheraea pernyi. PLoS ONE 2014, 9, e92680. [Google Scholar] [CrossRef] [PubMed]
- Sauman, I.; Reppert, S.M. Molecular characterization of prothoracicotropic hormone (PTTH) from the giant silkmoth Antheraea pernyi: Developmental appearance of PTTH-expressing cells and relationship to circadian clock cells in central brain. Dev. Biol. 1996, 178, 418–429. [Google Scholar] [CrossRef] [PubMed]
- Koutroumpa, F.A.; Groot, A.T.; Dekker, T.; Heckel, D.G. Genetic mapping of male pheromone response in the European corn borer identifies candidate genes regulating neurogenesis. Proc. Natl. Acad. Sci. USA 2016, 113, E6401–E6408. [Google Scholar] [CrossRef] [PubMed]
- Kozak, G.M.; Wadsworth, C.B.; Kahne, S.C.; Bogdanowicz, S.M.; Harrison, R.G.; Coates, B.S.; Dopman, E.B. A combination of sexual and ecological divergence contributes to rearrangement spread during initial stages of speciation. Mol. Ecol. 2017, 26, 2331–2347. [Google Scholar] [CrossRef] [PubMed]
- Lassance, J.-M.; Groot, A.T.; Liénard, M.A.; Antony, B.; Borgwardt, C.; Andersson, F.; Hedenström, E.; Heckel, D.G.; Lofstedt, C. Allelic variation in a fatty-acyl reductase gene causes divergence in moth sex pheromones. Nature 2010, 466, 486–489. [Google Scholar] [CrossRef] [PubMed]






© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Levy, R.C.; Kozak, G.M.; Dopman, E.B. Non-Pleiotropic Coupling of Daily and Seasonal Temporal Isolation in the European Corn Borer. Genes 2018, 9, 180. https://doi.org/10.3390/genes9040180
Levy RC, Kozak GM, Dopman EB. Non-Pleiotropic Coupling of Daily and Seasonal Temporal Isolation in the European Corn Borer. Genes. 2018; 9(4):180. https://doi.org/10.3390/genes9040180
Chicago/Turabian StyleLevy, Rebecca C., Genevieve M. Kozak, and Erik B. Dopman. 2018. "Non-Pleiotropic Coupling of Daily and Seasonal Temporal Isolation in the European Corn Borer" Genes 9, no. 4: 180. https://doi.org/10.3390/genes9040180