The Human Tyrosyl-DNA Phosphodiesterase 1 (hTdp1) Inhibitor NSC120686 as an Exploratory Tool to Investigate Plant Tdp1 Genes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Treatments
2.2. DNA Diffusion Assay
2.3. Single Cell Gel Electrophoresis
2.4. RNA Extraction, Complementary DNA Synthesis, and Quantitative Real-Time Polymerase Chain Reaction Analysis
2.5. Bioinformatic and Statistical Analysis
3. Results
3.1. Effect of the hTdp1 Inhibitor NSC120686 on Medicago truncatula Cell Viability
3.2. Exposure of Medicago truncatula Calli to NSC120686 Results in Accumulation of DNA Damage
3.3. In Silico Mining for Putative Interacting Partners of Tdp1
- Golgin A1 (GOLGA1, #Q92805.3), probably involved in maintaining Golgi structure;
- X-ray repair complementing defective repair in Chinese hamster cells 5 and 6 (XRCC5, #P13010.3; XRCC6, #P12956.2), single-stranded DNA-dependent adenosine triphosphate (ATP)-dependent helicases involved in non-homologous end joining (NHEJ);
- Protein kinase, DNA-activated, catalytic polypeptide (PRKDC, #P78527.3), a serine/threonine-protein kinase that acts as a molecular sensor for DNA damage; involved in NHEJ;
- DNA Topoisomerase I (TOP1, #P11387.2), involved in the release of supercoiling and torsional tension of DNA introduced during the DNA replication and transcription by transiently cleaving and rejoining one strand of the DNA duplex;
- DNA cross-link repair 1C (DCLRE1C, #Q96SD1.2), involved in DSB repair mainly through NHEJ pathway;
- Ligase III, DNA, ATP-dependent (LIG3, #P49916.2), functions as a heterodimer with DNA-repair protein XRCC1 in the nucleus and can correct defective DNA strand-break repair and sister chromatid exchange following treatment with ionizing radiation and alkylating agents;
- Meiotic recombination 11 homolog A (MRE11A; #P49959.3), component of the MRN complex, plays a central role in DSB repair;
- APEX nuclease (multifunctional DNA repair enzyme) 1 (APEX1, #P27695.2), plays a central role in the cellular response to oxidative stress; functions as an apurinic/apyrimidinic (AP) endodeoxyribonuclease in the DNA base excision repair (BER) pathway;
- Polymerase (DNA directed) beta (POLB, #P06746.3), repair polymerase that plays a key role in BER pathway.
- MRE11 (#AT5G54260.1);
- DNAse I-like superfamily protein (#AT3G48425), apurinic/apyrimidinic (AP) endonuclease involved in active DNA demethylation and gene imprinting;
- Endonuclease 2 (#AT4G36050), exhibits apurinic/apyrimidinic (AP) endonuclease activity in vitro;
- Calcium-dependent lipid-binding (CaLB domain) family protein (#AT3G61030);
- C2 calcium/lipid-binding endonuclease/exonuclease/phosphatase (#AT3G60950);
- Apurinic endonuclease-redox protein (ARP, #AT2G41460.1), involved in the repair oxidative DNA damage and may act as a redox factor;
- DNA topoisomerase 1 alpha (TOP1ALPHA, #AED96613.1);
- DNA topoisomerase 1 beta (TOP1BETA, #AT5G55310.1);
- DNA binding (#AT4G26701), putative DNA topoisomerase type I;
- Aprataxin (APTX, #AT5G01310.1), DNA-binding protein involved in SSB repair, DSB repair, and BER.
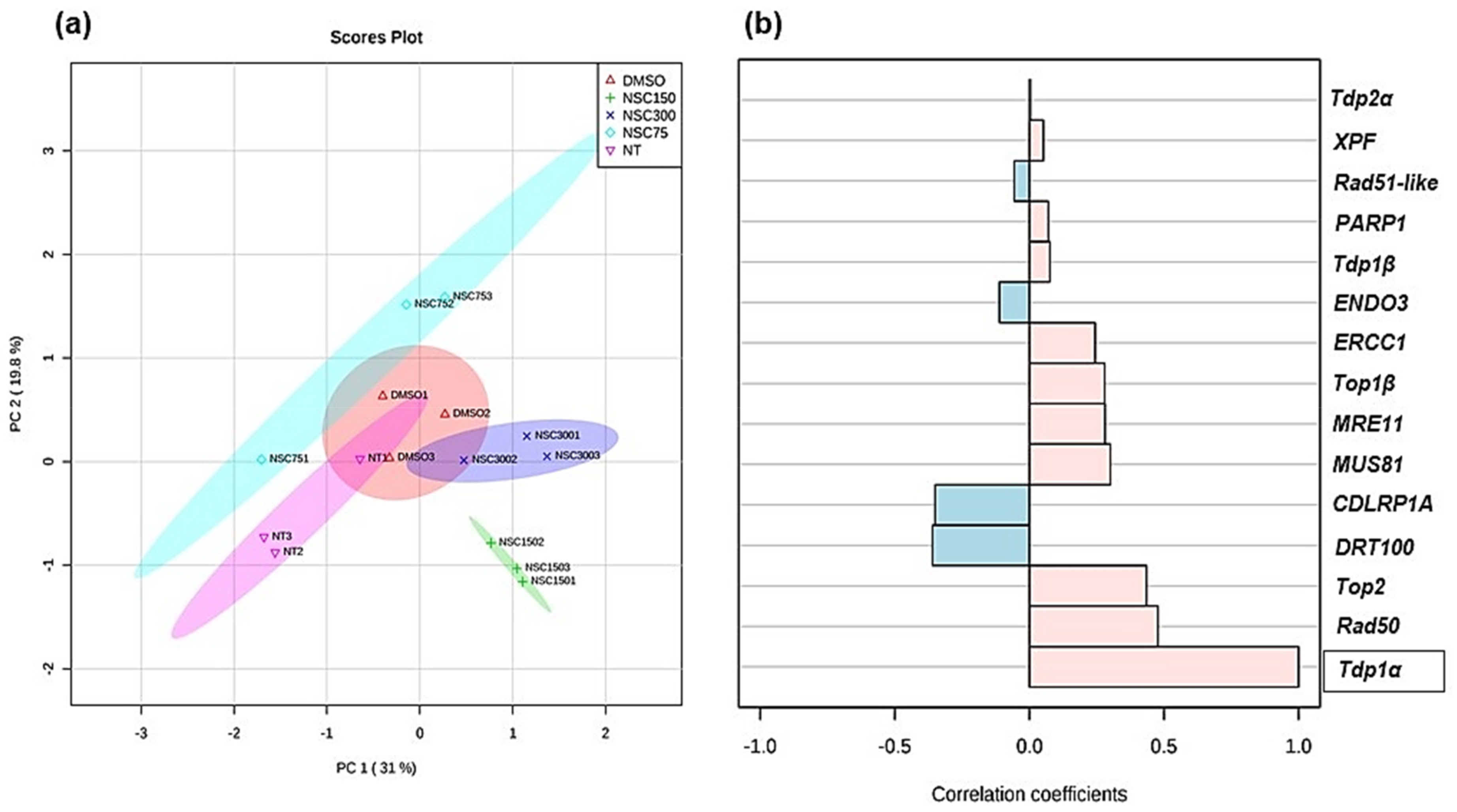
3.4. Expression Profiling of Medicago truncatula Tdp1 and Putative Interactors in Response to NSC120686 Treatment
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Xu, Y.; Her, C. Inhibition of topoisomerase (DNA) I (TOP1): DNA damage repair and anticancer therapy. Biomolecules 2015, 5, 1652–1670. [Google Scholar] [CrossRef] [PubMed]
- Pouliot, J.J.; Yao, K.C.; Robertson, C.A.; Nash, H.A. Yeast gene for a Tyr-DNA phosphodiesterase that repairs topoisomerase I covalent complexes. Science 1999, 286, 552–555. [Google Scholar] [CrossRef] [PubMed]
- Deng, C.; Brown, J.A.; You, D.; Brown, J.M. Multiple endonucleases function to repair covalent topoisomerase I complexes in Saccharomyces cerevisiae. Genetics 2005, 170, 591–600. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-W.; Regairaz, M.; Seiler, J.A.; Agama, K.K.; Doroshow, J.H.; Pommier, Y. Poly(ADP-ribose) polymerase and XPF-ERCC1 participate in distinct pathways for the repair of topoisomerase I-induced DNA damage in mammal cells. Nucleic Acids Res. 2011, 39, 3607–3620. [Google Scholar] [CrossRef] [PubMed]
- Pommier, Y. Topoisomerase I inhibitors: Camptothecins and beyond. Nat. Rev. Cancer 2006, 6, 789–802. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.N.; Pommier, Y.; Marchand, C. Tyrosyl-DNA phosphodiesterase 1 (Tdp1) inhibitors. Expert Opin. Ther. Pat. 2011, 21, 1285–1292. [Google Scholar] [CrossRef] [PubMed]
- Weidlich, I.E.; Dexheimer, T.; Marchand, C.; Antony, S.; Pommier, Y.; Nicklaus, M.C. Inhibitors of human tyrosyl-DNA phospodiesterase (hTdp1) developed by virtual screening using ligand-based pharmacophores. Bioorg. Med. Chem. 2010, 18, 182–189. [Google Scholar] [CrossRef] [PubMed]
- Perego, P.; Cossa, G.; Tinelli, S.; Corna, E.; Carenini, N.; Gatti, L.; De Cesare, M.; Ciusani, E.; Zunino, F.; Luison, E.; et al. Role of tyrosyl-DNA phosphodiesterase 1 and inter-players in regulation of tumor cell sensitivity to topoisomerase I inhibition. Biochem. Pharmacol. 2012, 83, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Al-Keilani, M.S.A. The Role of Tyrosyl-DNA Phosphodiesterase I (TDP1) as a Prognostic and Predictive Factor in Malignant Glioma. Ph.D. Thesis, University of Iowa, Iowa City, IA, USA, 2013. [Google Scholar]
- Macovei, A.; Balestrazzi, A.; Confalonieri, M.; Carbonera, D. The tyrosyl-DNA phosphodiesterase gene family in Medicago truncatula Gaertn: Bioinformatic investigation and expression profiles in response to copper- and PEG-mediated stress. Planta 2010, 232, 303–407. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-Y.; Kim, H.; Hwang, H.-J.; Jeong, Y.-M.; Na, S.H.; Woo, J.-C.; Kim, S.-G. Identification of tyrosyl-DNA phosphodiesterase as a novel DNA damage repair enzyme in Arabidopsis. Plant Physiol. 2010, 154, 1460–1469. [Google Scholar] [CrossRef] [PubMed]
- Balestrazzi, A.; Confalonieri, M.; Macovei, A.; Carbonera, D. Seed imbibition in Medicago truncatula Gaertn: Expression profiles of DNA repair genes in relation to PEG-mediated stress. J. Plant Physiol. 2011, 168, 706–713. [Google Scholar] [CrossRef] [PubMed]
- Donà, M.; Confalonieri, M.; Minio, A.; Biggiogera, M.; Buttafava, A.; Raimondi, E.; Delledonne, M.; Ventura, L.; Sabatini, M.E.; Macovei, A.; et al. RNA-Seq analysis discloses early senescence and nucleolar dysfunction triggered by Tdp1α depletion in Medicago truncatula. J. Exp. Bot. 2013, 64, 1941–1951. [Google Scholar] [CrossRef] [PubMed]
- Sabatini, M.E.; Donà, M.; Leonetti, P.; Minio, A.; Delledonne, M.; Carbonera, D.; Confalonieri, M.; Giraffa, G.; Balestrazzi, A. Depletion of tyrosyl-DNA phosphodiesterase 1α (MtTdp1α) affects transposon expression in Medicago truncatula. J. Integr. Plant Biol. 2015, 58, 618–622. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Na, S.H.; Lee, S.-Y.; Jeong, Y.-M.; Hwang, H.-J.; Hur, J.Y.; Park, S.-H.; Woo, J.-C.; Kim, A.-G. Structure-function studies of a plant tyrosyl-DNA phosphodiesterase provide novel insights into DNA repair mechanisms of Arabidopsis thaliana. Biochem. J. 2012, 443, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Davies, D.R.; Interthal, H.; Champoux, J.J.; Hol, W.G. Insights into substrate binding and catalytic mechanism of human tyrosyl-DNA phosphodiesterase (Tdp1) from vanadate and tungstate-inhibited structures. J. Mol. Biol. 2002, 324, 917–932. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Faè, M.; Balestrazzi, A.; Confalonieri, M.; Donà, M.; Macovei, A.; Valassi, A.; Giraffa, G.; Carbonera, D. Copper-mediated genotoxic stress is attenuated by the overexpression of the DNA repair gene MtTdp2α (tyrosyl-DNA phosphodiesterase 2) in Medicago truncatula plants. Plant Cell Rep. 2014, 33, 1071–1080. [Google Scholar] [CrossRef] [PubMed]
- Ventura, L.; Macovei, A.; Donà, M.; Paparella, S.; Buttafava, A.; Giovannini, A.; Carbonera, D.; Balestrazzi, A. Genotoxic effects due to in vitro culture and H2O2 treatments in Petunia × hybrida cells monitored through DNA diffusion assay, FPG-SCGE and gene expression profile analyses. Acta Physiol. Plant. 2014, 36, 331–341. [Google Scholar] [CrossRef]
- Singh, N.P. Apoptosis by DNA diffusion assay. In Methods in Molecular Medicine-Chemiosensitivity; Blumenthal, R., Ed.; Humana Press: New York, NY, USA, 2003; pp. 78–94. [Google Scholar]
- Collins, A.R. The comet assay for DNA damage and repair. Mol. Biotechnol. 2004, 26, 249–261. [Google Scholar] [CrossRef]
- Oñate-Sánchez, L.; Vicente-Carbajosa, J. DNA-free RNA isolation protocols for Arabidopsis thaliana, including seeds and siliques. BMC Res. Notes 2008, 1, 93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Confalonieri, M.; Faè, M.; Balestrazzi, A.; Donà, M.; Macovei, A.; Valassi, A.; Giraffa, G.; Carbonera, D. Enhanced osmotic stress tolerance in Medicago truncatula plants overexpressing the DNA repair gene MtTdp2α (tyrosyl-DNA phosphodiesterase 2). Plant Cell Tissue Organ Cult. 2014, 116, 187–203. [Google Scholar] [CrossRef]
- Sabatini, M.E.; Pagano, A.; Araùjo, S.; Balestrazzi, A.; Macovei, A. The Tyrosyl-DNA phosphodiesterase 1β (Tdp1β) gene discloses an early response to abiotic stresses. Genes 2017, 8, 305. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W.; Horgan, G.W.; Dempfle, L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002, 9, e36. [Google Scholar] [CrossRef]
- Warde-Farley, D.; Donaldson, S.L.; Comes, O.; Zuberi, K.; Badrawi, R.; Chao, P.; Franz, M.; Grouios, C.; Kazi, F.; Lopes, C.T.; et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010, 38, W214–W220. [Google Scholar] [CrossRef] [PubMed]
- Ventura, L.; Giovannini, A.; Savio, M.; Donà, M.; Macovei, A.; Buttafava, A.; Carbonera, D.; Balestrazzi, A. Single Cell Gel Electrophoresis (Comet) assay with plants: Research on DNA repair and ecogenotoxicity testing. Chemosphere 2013, 92, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Pradillo, M.; Knoll, A.; Oliver, C.; Varas, J.; Corredor, E.; Puchta, H.; Santos, J.L. Involvement of the Cohesin Cofactor PDS5 (SPO76) during meiosis and DNA repair in Arabidopsis thaliana. Front. Plant Sci. 2015, 6, 1034. [Google Scholar] [CrossRef] [PubMed]
- Al-Keilani, M.; Agarwal, S.; Alqudah, M.; Sibenaller, Z.; Ryken, T.; Assem, M. Tyrosyl DNA phosphodiesterase I is a prognostic factor and its inhibition synergizes response to topoisomerase poisons in malignant glioma. In Proceedings of the 104th Annual Meeting of the American Association for Cancer Research, Washington, DC, USA, 6–10 April 2013. [Google Scholar]
- Araújo, S.; Balestrazzi, A.; Faè, M.; Morano, M.; Carbonera, D.; Macovei, A. MtTdp2α-overexpression boosts the growth phase of Medicago truncatula cell suspension and increases the expression of key genes involved in the antioxidant response and genome stability. Plant Cell Tissue Organ Cult. 2016, 127, 675–680. [Google Scholar] [CrossRef]
- Locato, V.; Balestrazzi, A.; De Gara, L.; Carbonera, D. Reduced expression of top1β gene induces programmed cell death and alters ascorbate metabolism in Daucus carota cultured cells. J. Exp. Bot. 2006, 57, 1667–1676. [Google Scholar] [CrossRef] [PubMed]
- Van Doorn, W.G.; Beers, E.P.; Dangl, J.L.; Franklin-Tong, V.E.; Gallois, P.; Hata-Nishimura, I.; Jones, A.M.; Kawai-Yamada, M.; Lam, E.; Mundy, J.; et al. Morphological classification of plant cell deaths. Cell Death Differ. 2011, 18, 1241–1246. [Google Scholar] [CrossRef] [PubMed]
- Locato, V.; De Gara, L. Programmed cell death in plants: An overview. Methods Mol. Biol. 2018, 1743, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Pommier, Y.; Huang, S.N.; Gao, R.; Das, B.B.; Murai, J.; Marchand, C. Tyrosyl-DNA-phosphodiesterases (TDP1 and TDP2). DNA Repair 2014, 19, 114–129. [Google Scholar] [CrossRef] [PubMed]
- Lebedeva, N.A.; Rechkunova, N.I.; Lavrik, O.I. AP-site cleavage activity of tyrosyl-DNA phosphodiesterase 1. FEBS Lett. 2011, 585, 683–686. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.N.; Sopory, S.K.; Reddy, M.K. Plant DNA topoisomerases: Structure, function, and cellular roles in plant development. Crit. Rev. Plant Sci. 2004, 23, 251–269. [Google Scholar] [CrossRef]
- Gaillard, P.-H.L.; Wood, R.D. Activity of individual ERCC1 and XPF subunits in DNA nucleotide excision repair. Nucleic Acids Res. 2001, 29, 872–879. [Google Scholar] [CrossRef] [PubMed]
- Lamarche, B.J.; Orazio, N.I.; Weitzman, M.D. The MRN complex in double-strand break repair and telomere maintenance. FEBS Lett. 2010, 584, 3682–3695. [Google Scholar] [CrossRef] [PubMed]
- Mannuss, A.; Dukowic-Schulze, S.; Suer, S.; Hartung, F.; Pacher, M.; Puchta, H. RAD5A, RECQ4A, and MUS81 have specific functions in homologous recombination and define different pathways of DNA repair in Arabidopsis thaliana. Plant Cell 2010, 22, 3318–3330. [Google Scholar] [CrossRef] [PubMed]
- Bleuyard, J.Y.; Gallego, M.E.; Savigny, F.; White, C.I. Differing requirements for the Arabidopsis Rad51 paralogs in meiosis and DNA repair. Plant J. 2005, 41, 533–545. [Google Scholar] [CrossRef] [PubMed]
- Pachkowski, B.F.; Tano, K.; Afonin, V.; Elder, R.H.; Takeda, S.; Watanabe, M.; Swenberg, J.A.; Nakamura, J. Cells deficient in PARP1 show an accelerated accumulation of DNA single strand breaks, but not AP sites, over the PARP1-proficient cells exposed to MMS. Mutat. Res. 2009, 671, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Fujimori, N.; Suzuki, N.; Nakajima, Y.; Suzuki, S. Plant DNA-damage repair/toleration 100 protein repairs UV-B-induced DNA damage. DNA Repair 2014, 21, 171–176. [Google Scholar] [CrossRef] [PubMed]
- Ben-Yehoyada, M.; Wang, L.C.; Kozekov, I.D.; Rizzo, C.J.; Gottesman, M.E.; Gautier, J. Checkpoint signaling from a single DNA interstrand crosslink. Mol. Cell 2009, 35, 704–715. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, W.; Takami, T. Nucleases in higher plants and their possible involvement in DNA degradation during leaf senescence. J. Exp. Bot. 2014, 65, 3835–3843. [Google Scholar] [CrossRef] [PubMed]
- Das, B.B.; Huang, S.Y.; Murai, J.; Rehman, I.; Amé, J.C.; Sengupta, S.; Das, S.K.; Majumdar, P.; Zhang, H.; Biard, D.; et al. PARP1–TDP1 coupling for the repair of topoisomerase I—Induced DNA damage. Nucleic Acids Res. 2014, 42, 4435–4449. [Google Scholar] [CrossRef] [PubMed]
- Ray Chaudhuri, A.; Hashimoto, Y.; Herrador, R.; Neelsen, K.J.; Fachinetti, D.; Bermejo, R.; Cocito, A.; Costanzo, V.; Lopes, M. Topoisomerase I poisoning results in PARP-mediated replication fork reversal. Nat. Struct. Mol. Biol. 2012, 19, 419–423. [Google Scholar] [CrossRef] [PubMed]
- Elmaghrabi, A.M.; Rogers, H.J.; Francis, D.; Ochatt, S.J. PEG induces high expression of the cell cycle checkpoint gene WEE1 in embryogenic callus of Medicago truncatula: Potential link between cell cycle checkpoint regulation and osmotic stress. Front. Plant Sci. 2017, 8, 1479. [Google Scholar] [CrossRef] [PubMed]





| Gene | Accession No. | Forward Primer (5′-3′) | Reverse Primer (5′-3′) |
|---|---|---|---|
| Tdp1α | Medtr7g050860 | ACGAGTTGGGAGTGCTCTTT | GGGATTTATCCTTCGATTGTTT |
| Tdp1β | Medtr8g095490 | GGTTGGTTTGAGCCATCTTT | GCAGGCACATTGTGATTTCT |
| Tdp2α | Medtr8g146980 | CAGATGTTCAGCAAGGAACG | CCCGTCTTGCAAAGGATATT |
| Top1β | Medtr0172s0010 | ATACACGTGGGCTATTGTCG | TCACTTGGATGAATGCGTT |
| Top2 | Medtr3g103270 | AGGATCCGTGGGATTGTAAGGC | ACAACAGAGAGGCCAGCCATAG |
| Xpf | Medtr5g013480 | GGGTTCCGATGACGAAGTAT | CTCCACAGTCAAATCCTCCA |
| MRE11 | Medtr2g081100 | ATCCAAAGTGGTGCTGATGA | TGGATTCATTGTCCGAACTG |
| Rad50 | Medtr3g084300.1 | GGCGAGAAAGTTTGCCTTAG | GCCAATTTGCTTCATGTTGA |
| ERCC1 | Medtr1g082570.1 | CGTTCGTCAAATCCTCAGAA | TGAAGCTGCAGGAGCATTAT |
| MUS81 | Medtr3g022850.1 | AAGAAGCCACTGGATGTTCC | ATTTGGATGGCTTCTGGAAA |
| Rad51-like | Medtr4g124560.1 | ATGGCTCATGCAACCACGAC | AACCTTGCTTCGGCTTCAGC |
| PARP1 | Medtr1g088375 | AAACCCACCCTCCTTCGTAGT | GTCCCTCGGTCTCTTTCCAA |
| DRT100 | Medtr3g027940 | ACCCTACCACGGCATCTTCA | TCTTCTGACTCGCCACGGAG |
| DCLRP1A | Medtr3g105470.1 | TATGCGAGTCGGTTCAGCCT | AAGAAGGTGGCAGCAGGGTA |
| ENDO3 | Medtr5g056160.1 | CCTTGGTCGTCTGCTTTGCA | ATCGAGGAGCTGGTTGGTGT |
| ELF1α | Medtr6g021800 | GACAAGCGTGTGATCGAGAGATT | TTTCACGCTCAGCCTTAAGCT |
| Gene | Function; DNA Repair Pathways | NSC120686 | Tdp1α-2a [13] | IGROV-1/U87 [8,9] |
|---|---|---|---|---|
| Tdp1α | Repairs stalled TopI-DNA complexes; BER [34] | up-reg. | down-reg. | up-reg. |
| Tdp1β | Putatively repairs stalled topoisomerase I-DNA complexes; putative BER [10] | down-reg. | up-reg. | - |
| Tdp2α | Removal of DNA TopII-mediated DNA damage and cell signaling; NHEJ [34] | down-reg. | n.c. | n.d. |
| Top1β | Regulation of DNA topological state; cuts one of the two strands of double-stranded DNA [31] | n.c. | n.c. | n.d. |
| Top2 | Regulation of DNA topological state; cuts both strands of the DNA helix simultaneously in order to manage DNA tangles and supercoils [36] | n.c. | down-reg. | n.d. |
| Xpf | Endonuclease that excises dimers; NER [37] | down-reg. | n.c. | n.d. |
| MRE11 | Component of the MRN complex, central role in DSB repair; HR, NHEJ [38] | n.c. | n.c. | n.d. |
| Rad50 | Component of the MRN complex, central role in DSB repair; HR, NHEJ [38] | up-reg. | n.c. | n.d. |
| ERCC1 | Forms a catalytic complex with XPF; NER [37] | up-reg. | n.c. | n.d. |
| MUS81 | Resolves recombination intermediates during DNA repair after inter-strand cross-links, replication fork collapse, and DNA double-strand breaks; HR [39] | n.c. | n.c. | n.d. |
| Rad51-like | DNA repair during meiosis; HR [40] | n.c. | down-reg. | n.d. |
| PARP1 | Modifies various nuclear proteins by poly(ADP-ribosyl)ation, involved in the cell recovery from DNA damage; BER [41] | up-reg. | n.c. | n.d. |
| DRT100 | Repair of abasic sites and SSBs during UV-induced DNA damage; BER [42] | down-reg. | down-reg. | n.d. |
| DCLRP1A | DNA interstrand cross-link repair and checkpoint-mediated cell cycle arrest [43] | n.c. | down-reg. | n.d. |
| ENDO3 | T/G mismatch-specific endonuclease, nucleic acid binding, single-stranded DNA specific; MMR [44] | down-reg. | up-reg. | n.d. |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Macovei, A.; Pagano, A.; Sabatini, M.E.; Grandi, S.; Balestrazzi, A. The Human Tyrosyl-DNA Phosphodiesterase 1 (hTdp1) Inhibitor NSC120686 as an Exploratory Tool to Investigate Plant Tdp1 Genes. Genes 2018, 9, 186. https://doi.org/10.3390/genes9040186
Macovei A, Pagano A, Sabatini ME, Grandi S, Balestrazzi A. The Human Tyrosyl-DNA Phosphodiesterase 1 (hTdp1) Inhibitor NSC120686 as an Exploratory Tool to Investigate Plant Tdp1 Genes. Genes. 2018; 9(4):186. https://doi.org/10.3390/genes9040186
Chicago/Turabian StyleMacovei, Anca, Andrea Pagano, Maria Elisa Sabatini, Sofia Grandi, and Alma Balestrazzi. 2018. "The Human Tyrosyl-DNA Phosphodiesterase 1 (hTdp1) Inhibitor NSC120686 as an Exploratory Tool to Investigate Plant Tdp1 Genes" Genes 9, no. 4: 186. https://doi.org/10.3390/genes9040186





