Chromosome Synapsis and Recombination in Male-Sterile and Female-Fertile Interspecies Hybrids of the Dwarf Hamsters (Phodopus, Cricetidae)
Abstract
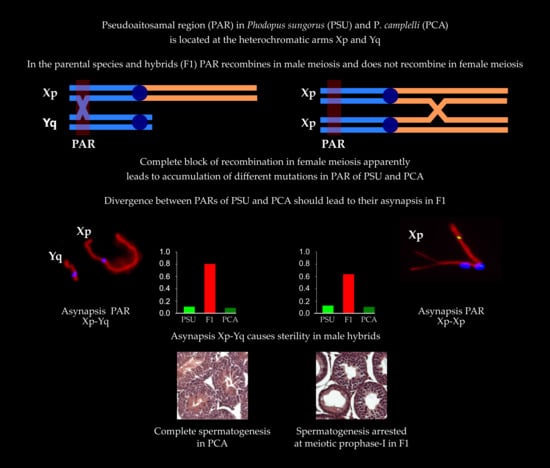
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Histological Analysis
2.3. Synaptonemal Complex Spreading and Immunostaining
2.4. Fluorescence In Situ Hybridization
2.5. Image Analysis
3. Results
3.1. Spermatogenesis
3.2. Autosome Pairing and Recombination
3.3. XY Chromosome Pairing and Recombination
3.4. XX Chromosome Pairing and Recombination
4. Discussion
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Mack, K.L.; Nachman, M.W. Gene regulation and speciation. Trends Genet. 2017, 33, 68–80. [Google Scholar] [CrossRef] [PubMed]
- Coyne, J.A.; Orr, H.A. Speciation; Sinauer Associates: Sunderland, MA, USA, 2004. [Google Scholar]
- Benirschke, K. Sterility and Fertility of Interspecific Mammalian Hybrids. In Comparative Aspects of Reproductive Failure; Springer: Berlin/Heidelberg, Germany, 1967; pp. 218–234. [Google Scholar]
- Torgasheva, A.A.; Borodin, P.M. Cytological basis of sterility in male and female hybrids between sibling species of grey voles Microtus arvalis and M. levis. Sci. Rep. 2016, 6, 36564. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, T.; Gregorova, S.; Mihola, O.; Anger, M.; Sebestova, J.; Denny, P.; Simecek, P.; Forejt, J. Mechanistic basis of infertility of mouse intersubspecific hybrids. Proc. Natl. Acad. Sci. USA 2013, 110, E468–E477. [Google Scholar] [CrossRef] [PubMed]
- Chandley, A.C.; Short, R.V.; Allen, W.R. Cytogenetic studies of three equine hybrids. J. Reprod. Fertil. Suppl. 1975, 23, 356–370. [Google Scholar]
- Graves, J.A.M.; O’Neill, R.J. Sex chromosome evolution and Haldane’s rule. J. Hered. 1997, 88, 358–360. [Google Scholar] [CrossRef] [PubMed]
- Hale, D.W.; Washburn, L.L.; Eicher, E.M. Meiotic abnormalities in hybrid mice of the C57BL/6J × Mus spretus cross suggests a cytogenetic basis for Haldane’s rule of hybrid sterility. Cytogenet. Genome Res. 1993, 63, 221–234. [Google Scholar] [CrossRef] [PubMed]
- Dumont, B.L. Meiotic consequences of genetic divergence across the murine pseudoautosomal region. Genetics 2017, 205, 1089–1100. [Google Scholar] [CrossRef] [PubMed]
- Graves, J.A.M. Did sex chromosome turnover promote divergence of the major mammal groups? BioEssays 2016, 38, 734–743. [Google Scholar] [CrossRef] [PubMed]
- Ishishita, S.; Tsuboi, K.; Ohishi, N.; Tsuchiya, K.; Matsuda, Y. Abnormal pairing of X and Y sex chromosomes during meiosis I in interspecific hybrids of Phodopus campbelli and P. sungorus. Sci. Rep. 2015, 5, 9435. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Yamada, F.; Hashimoto, T.; Abe, S.; Matsuda, Y.; Kuroiwa, A. Centromere repositioning in the X chromosome of XO/XO mammals, Ryukyu spiny rat. Chromosom. Res 2008, 16, 587–593. [Google Scholar] [CrossRef] [PubMed]
- Neumann, K.; Michaux, J.; Lebedev, V.; Yigit, N.; Colak, E.; Ivanova, N.; Poltoraus, A.; Surov, A.; Markov, G.; Maak, S.; et al. Molecular phylogeny of the Cricetinae subfamily based on the mitochondrial cytochrome b and 12S rRNA genes and the nuclear vWF gene. Mol. Phylogenet. Evol. 2006, 39, 135–148. [Google Scholar] [CrossRef] [PubMed]
- Schmid, M.; Haaf, T.; Weis, H.; Schempp, W. Chromosomal homoeologies in hamster species of the genus Phodopus (Rodentia, Cricetinae). Cytogenet. Cell Genet. 1986, 43, 168–173. [Google Scholar] [CrossRef] [PubMed]
- Romanenko, S.A.; Volobouev, V.T.; Perelman, P.L.; Lebedev, V.S.; Serdukova, N.A.; Trifonov, V.A.; Biltueva, L.S.; Nie, W.; O’Brien, P.C.M.; Bulatova, N.S.; et al. Karyotype evolution and phylogenetic relationships of hamsters (Cricetidae, Muroidea, Rodentia) inferred from chromosomal painting and banding comparison. Chromosom. Res. 2007, 15, 283–298. [Google Scholar] [CrossRef] [PubMed]
- Vistorin, G.; Gamperl, R.; Rosenkranz, W. Studies on sex chromosomes of four hamster species: Cricetus cricetus, Cricetulus griseus, Mesocricetus auratus, and Phodopus sungorus. Cytogenet. Cell Genet. 1977, 18, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Brekke, T.D.; Henry, L.A.; Good, J.M. Genomic imprinting, disrupted placental expression, and speciation. Evolution 2016, 70, 2690–2703. [Google Scholar] [CrossRef] [PubMed]
- Sokolov, V.E.; Vasil’eva, N.Y. Hybridological analysis confirms the species specificity of Phodopus sungorus (Pallas, 1773) and Phodopus campbelli (Thomas, 1905). Dokl. Biol. Sci. 1993, 332, 120–123. [Google Scholar]
- Safronova, L.D.; Cherepanova, E.V.; Vasil’eva, N.Y. Specific features of the first meiotic division in hamster hybrids obtained by backcrossing Phodopus sungorus and Phodopus campbelli. Dokl. Biol. Sci. 1999, 35, 184–188. [Google Scholar]
- Safronova, L.D.; Malygin, V.M.; Levenkova, E.S.; Orlov, V.N. Cytogenetic results of hybridization of hamsters Phodopus sungorus and Phodopus campbelli. Dokl. Biol. Sci. 1991, 327, 266–271. [Google Scholar]
- Safronova, L.D.; Vasil’eva, N.Y. Meiotic anomalies in interspecific hamster hybrids Phodopus sungorus (Pallas, 1773) and Ph. campbelli (Thomas, 1905). Genetika 1996, 32, 186–194. [Google Scholar]
- Anderson, L.K.; Reeves, A.; Webb, L.M.; Ashley, T. Distribution of crossing over on mouse synaptonemal complexes using immunofluorescent localization of MLH1 protein. Genetics 1999, 151, 1569–1579. [Google Scholar] [PubMed]
- Turinetto, V.; Giachino, C. Multiple facets of histone variant H2AX: A DNA double-strand-break marker with several biological functions. Nucleic Acids Res. 2015, 43, 2489–2498. [Google Scholar] [CrossRef] [PubMed]
- Leblond, C.P.; Clermont, Y. Spermiogenesis of rat, mouse, hamster and guinea pig as revealed by the “periodic acid-fuchsin sulfurous acid” technique. Am. J. Anat. 1952, 90, 167–215. [Google Scholar] [CrossRef] [PubMed]
- Wing, T.Y.; Christensen, A.K. Morphometric studies on rat seminiferous tubules. Am. J. Anat. 1982, 165, 13–25. [Google Scholar] [CrossRef] [PubMed]
- Wyrobek, A.J.; Bruce, W.R. Chemical induction of sperm abnormalities in mice. Proc. Natl. Acad. Sci. USA 1975, 72, 4425–4429. [Google Scholar] [CrossRef] [PubMed]
- Peters, A.H.; Plug, A.W.; van Vugt, M.J.; de Boer, P. A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome Res. 1997, 5, 66–68. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Graphodatsky, A.S. Animal probes and ZOO-FISH. In Fluorescence In Situ Hybridization (FISH)—Application Guide; Liehr, T., Ed.; Springer: Berlin/Heidelberg, Germany, 2017; pp. 395–415. [Google Scholar]
- Trifonov, V.; Vorobieva, N.; Serdyukova, N.; Rens, W. FISH with and without COT1 DNA. In Fluorescence In Situ Hybridization (FISH)—Application Guide; Liehr, T., Ed.; Springer: Berlin/Heidelberg, Germany, 2017; pp. 123–133. [Google Scholar]
- Reeves, A. MicroMeasure: A new computer program for the collection and analysis of cytogenetic data. Genome 2001, 44, 439–443. [Google Scholar] [CrossRef] [PubMed]
- Capilla, L.; Garcia Caldés, M.; Ruiz-Herrera, A. Mammalian meiotic recombination: A toolbox for genome evolution. Cytogenet. Genome Res. 2016, 150, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Segura, J.; Ferretti, L.; Ramos-Onsins, S.; Capilla, L.; Farré, M.; Reis, F.; Oliver-Bonet, M.; Fernández-Bellón, H.; Garcia, F.; Garcia-Caldés, M.; Robinson, T.J.; Ruiz-Herrera, A. Evolution of recombination in eutherian mammals: Insights into mechanisms that affect recombination rates and crossover interference. Proc. R. Soc. B Biol. Sci. 2013, 280, 1945. [Google Scholar] [CrossRef] [PubMed]
- Basheva, E.A.; Bidau, C.J.; Borodin, P.M. General pattern of meiotic recombination in male dogs estimated by MLH1 and RAD51 immunolocalization. Chromosome Res. 2008, 16, 709–719. [Google Scholar] [CrossRef] [PubMed]
- Mary, N.; Barasc, H.; Ferchaud, S.; Billon, Y.; Meslier, F.; Robelin, D.; Calgaro, A.; Loustau-Dudez, A.-M. M.; Bonnet, N.; Yerle, M.; et al. Meiotic recombination analyses of individual chromosomes in male domestic pigs (Sus scrofa domestica). PLoS ONE 2014, 9, e99123. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Mikhaail-Philips, M.; Oliver-Bonet, M.; Ko, E.; Rademaker, A.; Turek, P.; Martin, R.H. Reduced meiotic recombination on the XY bivalent is correlated with an increased incidence of sex chromosome aneuploidy in men with non-obstructive azoospermia. Mol. Hum. Reprod. 2008, 14, 399–404. [Google Scholar] [CrossRef] [PubMed]
- Romanenko, S.A.; Perelman, P.L.; Serdukova, N.A.; Trifonov, V.A.; Biltueva, L.S.; Wang, J.; Li, T.; Nie, W.; O’Brien, P.C.M.; Volobouev, V.T.; et al. Reciprocal chromosome painting between three laboratory rodent species. Mamm. Genome 2006, 17, 1183–1192. [Google Scholar] [CrossRef] [PubMed]
- Brekke, T.D.; Steele, K.A.; Mulley, J.F. Inbred or Outbred? Genetic Diversity in Laboratory Rodent Colonies. G3 Genes Genomes Genet. 2017, 8, g3.300495.2017. [Google Scholar] [CrossRef]
- Moses, M.J. Synaptonemal complex karyotyping in spermatocytes of the Chinese hamster (Cricetulus griseus). II. Morphology of the XY pair in spread preparations. Chromosoma 1977, 60, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Ashley, T.; Gaeth, A.P.; Creemers, L.B.; Hack, A.M.; de Rooij, D.G. Correlation of meiotic events in testis sections and microspreads of mouse spermatocytes relative to the mid-pachytene checkpoint. Chromosoma 2004, 113, 126–136. [Google Scholar] [CrossRef] [PubMed]
- Mangs, H.A.; Morris, B.J. The human pseudoautosomal region (PAR): Origin, function and future. Curr. Genom. 2007, 8, 129–136. [Google Scholar] [CrossRef]
- Miklos, G.L.G. Sex-chromosome pairing and male fertility. Cytogenet. Genome Res. 1974, 13, 558–577. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, T.A.; Burgoyne, P.S. Evidence that sex chromosome asynapsis, rather than excess Y gene dosage, is responsible for the meiotic impairment of XYY mice. Cytogenet. Genome Res. 2000, 89, 38–43. [Google Scholar] [CrossRef] [PubMed]
- Burgoyne, P.S.; Mahadevaiah, S.K. Unpaired sex chromosomes and gametogenic failure. Chromosom. Today 1993, 11, 243–263. [Google Scholar]
- Burgoyne, P.S.; Sutcliffe, M.J.; Mahadevaiah, S.K. The role of unpaired sex chromosomes in spermatogenic failure. Andrologia 1992, 24, 17–20. [Google Scholar] [CrossRef] [PubMed]
- Turner, J. Meiotic sex chromosome inactivation. Development 2007, 134, 1823–1831. [Google Scholar] [CrossRef] [PubMed]
- Inagaki, A.; Schoenmakers, S.; Baarends, W.M. DNA double strand break repair, chromosome synapsis and transcriptional silencing in meiosis. Epigenetics 2010, 5, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Berletch, J.B.; Nguyen, D.K.; Disteche, C.M. X chromosome regulation: Diverse patterns in development, tissues and disease. Nat. Rev. Genet. 2014, 15, 367–378. [Google Scholar] [CrossRef] [PubMed]
- Raman, R.; Das, P. Mammalian sex chromosomes III. Activity of pseudoautosomal steroid sulfatase enzyme during spermatogenesis in Mus musculus. Somat. Cell Mol. Genet. 1991, 17, 429–433. [Google Scholar] [CrossRef] [PubMed]
- Palmer, S.; Perry, J.; Kipling, D.; Ashworth, A. A gene spans the pseudoautosomal boundary in mice. Proc. Natl. Acad. Sci. USA 1997, 94, 12030–12035. [Google Scholar] [CrossRef] [PubMed]
- Raudsepp, T.; Chowdhary, B.P. The eutherian pseudoautosomal region. Cytogenet. Genome Res. 2016, 147, 81–94. [Google Scholar] [CrossRef] [PubMed]
- Otto, S.P.; Pannell, J.R.; Peichel, C.L.; Ashman, T.L.; Charlesworth, D.; Chippindale, A.K.; Delph, L.F.; Guerrero, R.F.; Scarpino, S.V.; McAllister, B.F. About PAR: The distinct evolutionary dynamics of the pseudoautosomal region. Trends Genet. 2011, 27, 358–367. [Google Scholar] [CrossRef] [PubMed]
- White, M.A.; Ikeda, A.; Payseur, B.A. A pronounced evolutionary shift of the pseudoautosomal region boundary in house mice. Mamm. Genome 2012, 23, 454–466. [Google Scholar] [CrossRef] [PubMed]
- Horn, A.; Basset, P.; Yannic, G.; Banaszek, A.; Borodin, P.M.; Bulatova, N.S.; Jadwiszczak, K.; Polyakov, A.V.; Ratkiewicz, M.; Searle, J.B.; et al. Chromosomal rearrangements do not seem to affect the gene flow in hybrid zones between karyotypic races of the common shrew (Sorex araneus). Evolution 2012, 66, 882–889. [Google Scholar] [CrossRef] [PubMed]
- De La Fuente, R.; Sanchez, A.; Marchal, J.A.; Viera, A.; Parra, M.T.; Rufas, J.S.; Page, J.; Sánchez, A.; Marchal, J.A.; Viera, A.; Parra, M.T.; et al. A synaptonemal complex-derived mechanism for meiotic segregation precedes the evolutionary loss of homology between sex chromosomes in arvicolid mammals. Chromosoma 2012, 121, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Hinch, A.G.; Altemose, N.; Noor, N.; Donnelly, P.; Myers, S.R. Recombination in the Human Pseudoautosomal Region PAR1. PLoS Genet. 2014, 10. [Google Scholar] [CrossRef] [PubMed]
- Flaquer, A.; Rappold, G.A.; Wienker, T.F.; Fischer, C. The human pseudoautosomal regions: A review for genetic epidemiologists. Eur. J. Hum. Genet. 2008, 16, 771–779. [Google Scholar] [CrossRef] [PubMed]
- Kauppi, L.; Barchi, M.; Baudat, F.; Romanienko, P.J.; Keeney, S.; Jasin, M. Distinct properties of the XY pseudoautosomal region crucial for male meiosis. Science 2011, 331, 916–920. [Google Scholar] [CrossRef] [PubMed]
- Ross, M.T.; Grafham, D.V.; Coffey, A.J.; Scherer, S.; McLay, K.; Muzny, D.; Platzer, M.; Howell, G.R.; Burrows, C.; Bird, C.P.; et al. The DNA sequence of the human X chromosome. Nature 2005, 434, 325–337. [Google Scholar] [CrossRef] [PubMed]
- Bussell, J.J.; Pearson, N.M.; Kanda, R.; Filatov, D.A.; Lahn, B.T. Human polymorphism and human-chimpanzee divergence in pseudoautosomal region correlate with local recombination rate. Gene 2006, 368, 94–100. [Google Scholar] [CrossRef] [PubMed]
- Stack, S.M. Heterochromatin, the synaptonemal complex and crossing over. J. Cell Sci. 1984, 71, 159–176. [Google Scholar] [PubMed]
- Dumont, B.L. Variation and evolution of the meiotic requirement for crossing over in mammals. Genetics 2017, 205, 155–168. [Google Scholar] [CrossRef] [PubMed]
- Ashley, T. G-band position effects on meiotic synapsis and crossing over. Genetics 1988, 118, 307–317. [Google Scholar] [PubMed]





| Genotype (n of Animals) | Stage of Seminiferous Epithelium Cycle | Empty Tubules | n of Tubules | Ratio of Cell Types | n of Tubules/Cells | ||||
|---|---|---|---|---|---|---|---|---|---|
| I–V | VI–XI | XII–XIV | SPG/SER 1 | P/SPG | SPTD/P | ||||
| P. sungorus (n = 4) | 0.18 ± 0.01 | 0.74 ± 0.01 | 0.07 ± 0.01 | 0.01 ± 0.01 | 1769 | 2.7 ± 1.5 | 1.3 ± 0.5 | 3.6 ± 0.5 | 84/10,094 |
| P. campbelli (n = 5) | 0.26 ± 0.05 | 0.67 ± 0.06 | 0.07 ± 0.03 | 0.00 ± 0.01 | 1871 | 2.8 ± 1.4 | 1.2 ± 0.4 | 3.8 ± 0.5 | 99/11,835 |
| F1 (n = 6) | 0.19 ± 0.01 * | 0.29 ± 0.01 * | 0.35 ± 0.01 * | 0.17 ± 0.01 * | 3877 | 1.7 ± 1.4 * | 2.8 ± 1.7 * | 0.2 ± 0.1 * | 152/17,265 |
| Group | Sex | n Animals | n Cells | SC Length, µm | MLH1 Focus Number |
|---|---|---|---|---|---|
| P. sungorus | f | 3 | 119 | 204.7 ± 31.5 | 19.2 ± 2.2 |
| m | 3 | 120 | 119.9 ± 9.5 | 19.1 ± 3.3 | |
| P. campbelli | f | 3 | 151 | 190.2 ± 40.7 | 18.5 ± 3.6 |
| m | 3 | 89 | 127.8 ± 12.0 | 19.2 ± 2.5 | |
| F1 | f | 4 | 86 | 176.2 ± 29.7 | 17.9 ± 3.6 |
| m | 2 | 177 | 133.3 ± 15.1 | 19.7 ± 2.4 |
| Group | XY Bivalent | XX Bivalent | |||||
|---|---|---|---|---|---|---|---|
| n Cells | Asynapsed | Synapsed without MLH1 | Synapsed with MLH1 | n Cells | Centromere Misalignment | Asynapsed Xp | |
| P. sungorus | 191 | 0.11 ± 0.02 | 0.36 ± 0.04 | 0.53 ± 0.04 | 151 | 0.52 ± 0.04 | 0.12 ± 0.02 |
| P. campbelli | 152 | 0.09 ± 0.02 | 0.26 ± 0.04 | 0.65 ± 0.04 | 321 | 0.37 ± 0.03 | 0.10 ± 0.02 |
| F1 | 213 | 0.78 ± 0.03 | 0.12 ± 0.02 | 0.01 ± 0.02 | 162 | 0.70 ± 0.04 | 0.62 ± 0.04 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bikchurina, T.I.; Tishakova, K.V.; Kizilova, E.A.; Romanenko, S.A.; Serdyukova, N.A.; Torgasheva, A.A.; Borodin, P.M. Chromosome Synapsis and Recombination in Male-Sterile and Female-Fertile Interspecies Hybrids of the Dwarf Hamsters (Phodopus, Cricetidae). Genes 2018, 9, 227. https://doi.org/10.3390/genes9050227
Bikchurina TI, Tishakova KV, Kizilova EA, Romanenko SA, Serdyukova NA, Torgasheva AA, Borodin PM. Chromosome Synapsis and Recombination in Male-Sterile and Female-Fertile Interspecies Hybrids of the Dwarf Hamsters (Phodopus, Cricetidae). Genes. 2018; 9(5):227. https://doi.org/10.3390/genes9050227
Chicago/Turabian StyleBikchurina, Tatiana I., Katerina V. Tishakova, Elena A. Kizilova, Svetlana A. Romanenko, Natalya A. Serdyukova, Anna A. Torgasheva, and Pavel M. Borodin. 2018. "Chromosome Synapsis and Recombination in Male-Sterile and Female-Fertile Interspecies Hybrids of the Dwarf Hamsters (Phodopus, Cricetidae)" Genes 9, no. 5: 227. https://doi.org/10.3390/genes9050227