The Methylome of Vertebrate Sex Chromosomes
Abstract
:1. Introduction
2. Sex Chromosome Evolution
3. Dosage Compensation: A Shared Problem
4. X Chromosome Inactivation in Mammals
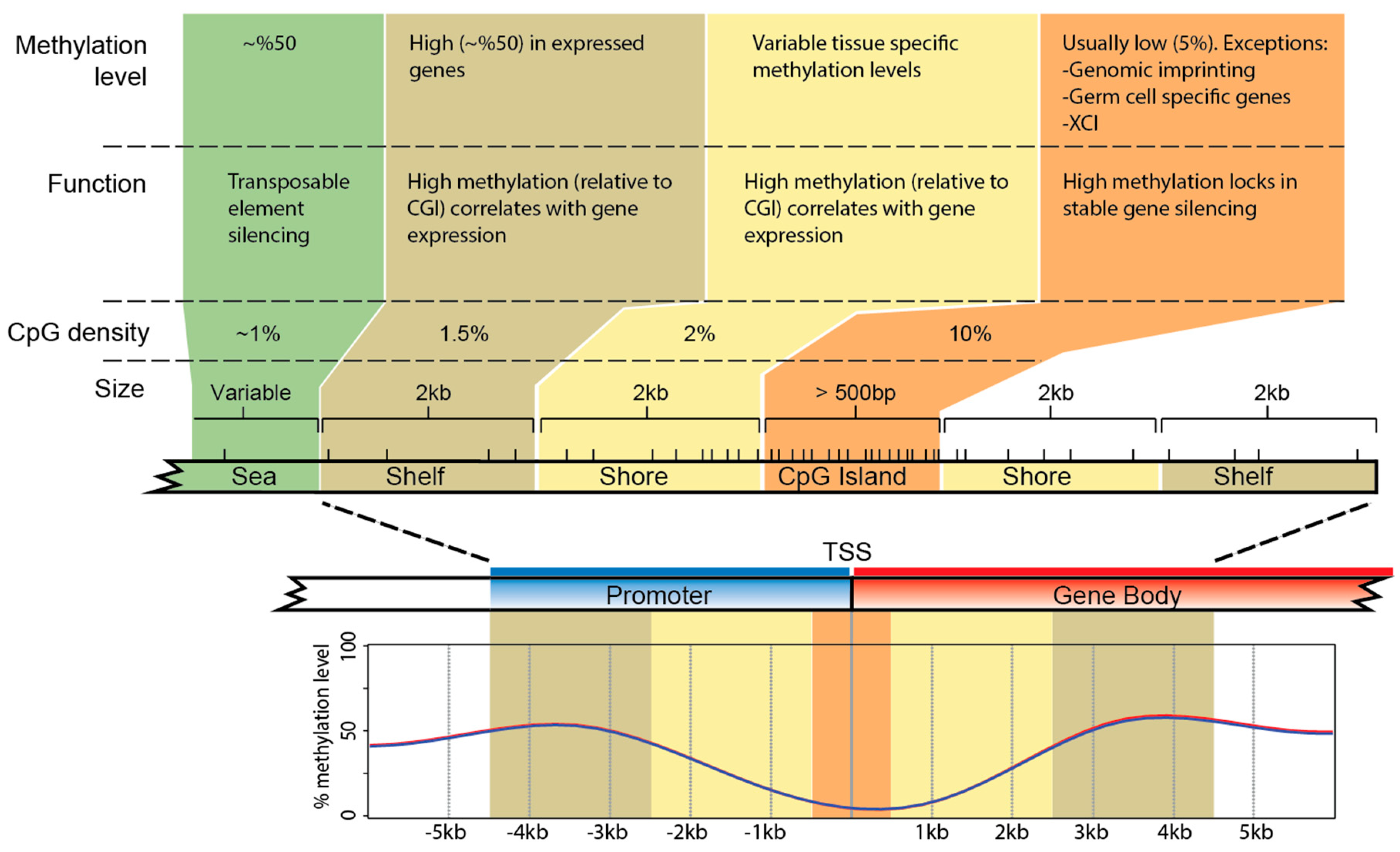
5. DNA Methylation
6. DNA Methylation and Eutherian X Chromosome Inactivation
7. DNA Methylation and Marsupial X Chromosome Inactivation
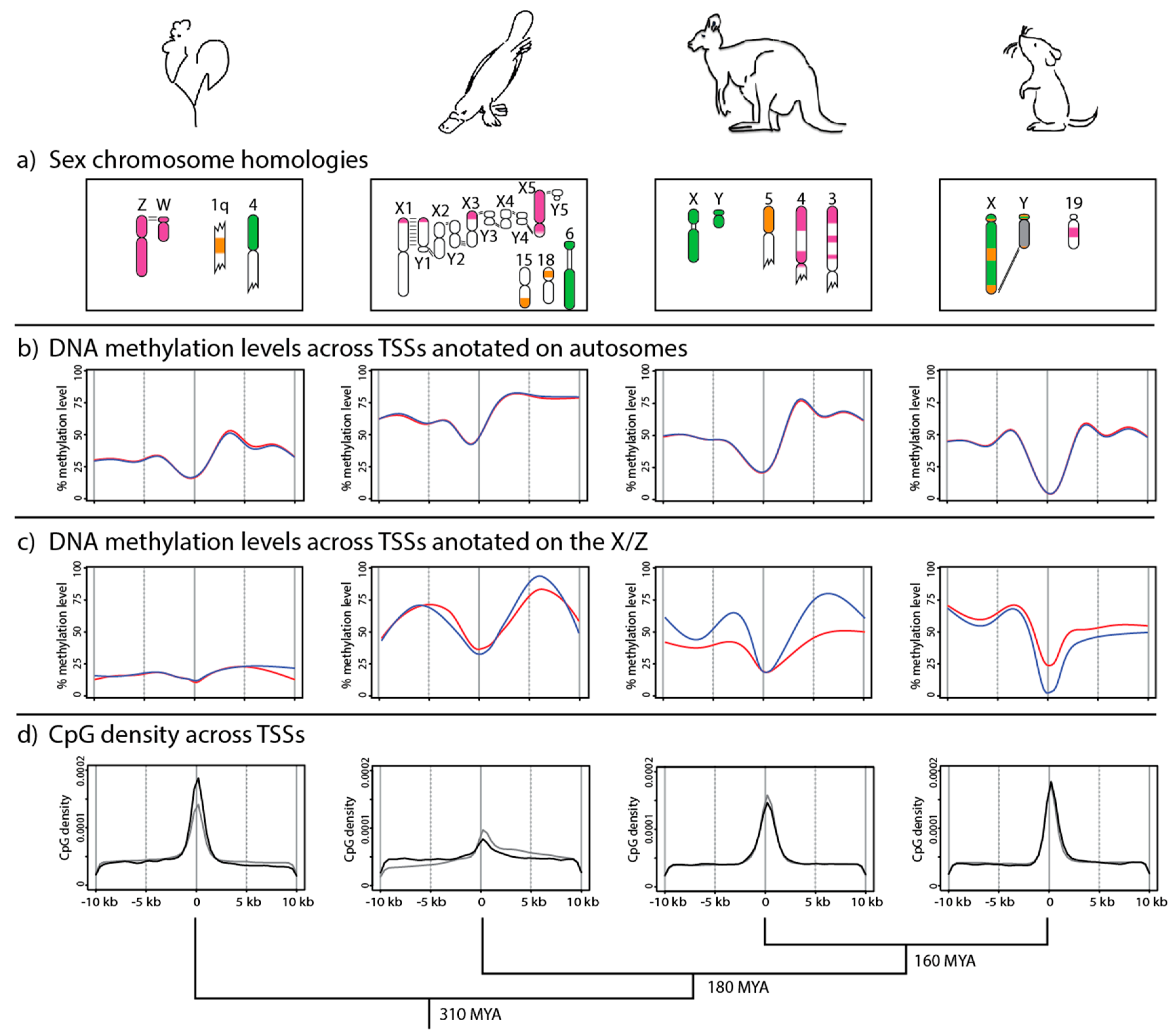
8. DNA Methylation Patterns in Monotremes and Birds
9. Evolution of DNA Methylation Patterns on Sex Chromosomes
10. Summary
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Sinclair, A.H.; Berta, P.; Palmer, M.S.; Hawkins, J.R.; Griffiths, B.L.; Smith, M.J.; Foster, J.W.; Frischauf, A.M.; Lovell-Badge, R.; Goodfellow, P.N. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature 1990, 346, 240–244. [Google Scholar] [CrossRef] [PubMed]
- Koopman, P.; Gubbay, J.; Vivian, N.; Goodfellow, P.; Lovell-Badge, R. Male development of chromosomally female mice transgenic for Sry. Nature 1991, 351, 117–121. [Google Scholar] [CrossRef] [PubMed]
- Foster, J.W.; Brennan, F.E.; Hampikian, G.K.; Goodfellow, P.N.; Sinclair, A.H.; Lovell-Badge, R.; Selwood, L.; Renfree, M.B.; Cooper, D.W.; Graves, J.A.M. Evolution of sex determination and the Y chromosome: Sry-related sequences in marsupials. Nature 1992, 359, 531–533. [Google Scholar] [CrossRef] [PubMed]
- Graves, J.A.M. The origin and function of the mammalian Y chromosome and Y-borne genes -an evolving understanding. Bioessays 1995, 17, 311–320. [Google Scholar] [CrossRef] [PubMed]
- Waters, P.D.; Delbridge, M.L.; Deakin, J.E.; El-Mogharbel, N.; Kirby, P.J.; Carvalho-Silva, D.R.; Graves, J.A.M. Autosomal location of genes from the conserved mammalian X in the platypus (Ornithorhynchus anatinus): Implications for mammalian sex chromosome evolution. Chromosome Res 2005, 13, 401–410. [Google Scholar] [CrossRef] [PubMed]
- Rens, W.; O'Brien, P.C.; Grutzner, F.; Clarke, O.; Graphodatskaya, D.; Tsend-Ayush, E.; Trifonov, V.A.; Skelton, H.; Wallis, M.C.; Johnston, S.; et al. The multiple sex chromosomes of platypus and echidna are not completely identical and several share homology with the avian Z. Genome Biol. 2007, 8, R243. [Google Scholar] [CrossRef] [PubMed]
- Veyrunes, F.; Waters, P.D.; Miethke, P.; Rens, W.; McMillan, D.; Alsop, A.E.; Grutzner, F.; Deakin, J.E.; Whittington, C.M.; Schatzkamer, K.; et al. Bird-like sex chromosomes of platypus imply recent origin of mammal sex chromosomes. Genome Res. 2008, 18, 965–973. [Google Scholar] [CrossRef] [PubMed]
- Graves, J.A.M.; Peichel, C.L. Are homologies in vertebrate sex determination due to shared ancestry or to limited options? Genome Biol. 2010, 11, 205. [Google Scholar] [CrossRef] [PubMed]
- Waters, S.A.; Livernois, A.M.; Patel, H.; O'Meally, D.; Craig, J.M.; Marshall Graves, J.A.; Suter, C.M.; Waters, P.D. Landscape of DNA methylation on the marsupial X. Mol. Biol. Evol. 2018, 35, 431–439. [Google Scholar] [CrossRef] [PubMed]
- Ensembl X Chromosome Summary. Available online: https://www.ensembl.org/Homo_sapiens/Location/Chromosome?redirect=no;r=X (accessed on 30 April 2018).
- Skaletsky, H.; Kuroda-Kawaguchi, T.; Minx, P.J.; Cordum, H.S.; Hillier, L.; Brown, L.G.; Repping, S.; Pyntikova, T.; Ali, J.; Bieri, T.; et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 2003, 423, 825–837. [Google Scholar] [CrossRef] [PubMed]
- Graves, J.A.M. Sex chromosome specialization and degeneration in mammals. Cell 2006, 124, 901–914. [Google Scholar] [CrossRef] [PubMed]
- Bellott, D.W.; Hughes, J.F.; Skaletsky, H.; Brown, L.G.; Pyntikova, T.; Cho, T.J.; Koutseva, N.; Zaghlul, S.; Graves, T.; Rock, S.; et al. Mammalian Y chromosomes retain widely expressed dosage-sensitive regulators. Nature 2014, 508, 494–499. [Google Scholar] [CrossRef] [PubMed]
- Cortez, D.; Marin, R.; Toledo-Flores, D.; Froidevaux, L.; Liechti, A.; Waters, P.D.; Grutzner, F.; Kaessmann, H. Origins and functional evolution of Y chromosomes across mammals. Nature 2014, 508, 488–493. [Google Scholar] [CrossRef] [PubMed]
- Charlesworth, B. The evolution of sex chromosomes. Science 1991, 251, 1030–1033. [Google Scholar] [CrossRef] [PubMed]
- Lahn, B.T.; Page, D.C. Four evolutionary strata on the human X chromosome. Science 1999, 286, 964–967. [Google Scholar] [CrossRef] [PubMed]
- Bellott, D.W.; Skaletsky, H.; Cho, T.J.; Brown, L.; Locke, D.; Chen, N.; Galkina, S.; Pyntikova, T.; Koutseva, N.; Graves, T.; et al. Avian W and mammalian Y chromosomes convergently retained dosage-sensitive regulators. Nat. Genet. 2017, 49, 387–394. [Google Scholar] [CrossRef] [PubMed]
- Rens, W.; Grützner, F.; O'Brien, P.C.; Fairclough, H.; Graves, J.A.M.; Ferguson-Smith, M.A. Resolution and evolution of the duck-billed platypus karyotype with an X1Y1X2Y2X3Y3X4Y4X5Y5 male sex chromosome constitution. Proc. Natl. Acad. Sci. USA 2004, 101, 16257–16261. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Terai, Y.; Mizoiri, S.; Aibara, M.; Nishihara, H.; Watanabe, M.; Kuroiwa, A.; Hirai, H.; Hirai, Y.; Matsuda, Y.; et al. B chromosomes have a functional effect on female sex determination in Lake Victoria cichlid fishes. PLoS Genet. 2011, 7, e1002203. [Google Scholar] [CrossRef] [PubMed]
- Julien, P.; Brawand, D.; Soumillon, M.; Necsulea, A.; Liechti, A.; Schutz, F.; Daish, T.; Grutzner, F.; Kaessmann, H. Mechanisms and evolutionary patterns of mammalian and avian dosage compensation. PLoS Biol. 2012, 10, e1001328. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, A.S.; Beermann, W. Synthesis of ribonucleic acid by the X-chromosomes of Drosophila melanogaster and the problem of dosage compensation. Nature 1965, 207, 785–786. [Google Scholar] [CrossRef] [PubMed]
- Lyon, M.F. Gene action in the X-chromosome of the mouse. Nature 1961, 190, 372–373. [Google Scholar] [CrossRef] [PubMed]
- Ohno, S. Sex Chromosomes and Sex-Linked Genes; Springer: New York, NY, USA, 1967. [Google Scholar]
- Pessia, E.; Makino, T.; Bailly-Bechet, M.; McLysaght, A.; Marais, G.A. Mammalian X chromosome inactivation evolved as a dosage-compensation mechanism for dosage-sensitive genes on the X chromosome. Proc. Natl. Acad. Sci. USA 2012, 109, 5346–5351. [Google Scholar] [CrossRef] [PubMed]
- Graves, J.A. Evolution of vertebrate sex chromosomes and dosage compensation. Nat. Rev. Genet. 2016, 17, 33–46. [Google Scholar] [CrossRef] [PubMed]
- Al Nadaf, S.; Waters, P.D.; Koina, E.; Deakin, J.E.; Jordan, K.S.; Graves, J.A. Activity map of the tammar X chromosome shows that marsupial X inactivation is incomplete and escape is stochastic. Genome Biol. 2010, 11, R122. [Google Scholar] [CrossRef] [PubMed]
- Livernois, A.M.; Waters, S.A.; Deakin, J.E.; Marshall Graves, J.A.; Waters, P.D. Independent evolution of transcriptional inactivation on sex chromosomes in birds and mammals. PLoS Genet. 2013, 9, e1003635. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Mank, J.E.; Li, J.; Yang, N.; Qu, L. Allele-specific expression analysis does not support sex chromosome inactivation on the chicken Z chromosome. Genome Biol. Evol. 2017, 9, 619–626. [Google Scholar] [CrossRef] [PubMed]
- Warnefors, M.; Mossinger, K.; Halbert, J.; Studer, T.; VandeBerg, J.L.; Lindgren, I.; Fallahshahroudi, A.; Jensen, P.; Kaessmann, H. Sex-biased microRNA expression in mammals and birds reveals underlying regulatory mechanisms and a role in dosage compensation. Genome Res. 2017, 27, 1961–1973. [Google Scholar] [CrossRef] [PubMed]
- Brown, C.J.; Ballabio, A.; Rupert, J.L.; Lafreniere, R.G.; Grompe, M.; Tonlorenzi, R.; Willard, H.F. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 1991, 349, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Grant, J.; Mahadevaiah, S.K.; Khil, P.; Sangrithi, M.N.; Royo, H.; Duckworth, J.; McCarrey, J.R.; VandeBerg, J.L.; Renfree, M.B.; Taylor, W.; et al. Rsx is a metatherian RNA with Xist-like properties in X-chromosome inactivation. Nature 2012, 487, 254–258. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.; Zhang, Q.C.; da Rocha, S.T.; Flynn, R.A.; Bharadwaj, M.; Calabrese, J.M.; Magnuson, T.; Heard, E.; Chang, H.Y. Systematic discovery of Xist RNA binding proteins. Cell 2015, 161, 404–416. [Google Scholar] [CrossRef] [PubMed]
- McHugh, C.A.; Chen, C.K.; Chow, A.; Surka, C.F.; Tran, C.; McDonel, P.; Pandya-Jones, A.; Blanco, M.; Burghard, C.; Moradian, A.; et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 2015, 521, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Minajigi, A.; Froberg, J.E.; Wei, C.; Sunwoo, H.; Kesner, B.; Colognori, D.; Lessing, D.; Payer, B.; Boukhali, M.; Haas, W.; et al. Chromosomes. A comprehensive Xist interactome reveals cohesin repulsion and an RNA-directed chromosome conformation. Science 2015, 349, 282. [Google Scholar] [CrossRef] [PubMed]
- Heard, E. Delving into the diversity of facultative heterochromatin: The epigenetics of the inactive X chromosome. Curr. Opin. Genet. Dev. 2005, 15, 482–489. [Google Scholar] [CrossRef] [PubMed]
- Balaton, B.P.; Cotton, A.M.; Brown, C.J. Derivation of consensus inactivation status for X-linked genes from genome-wide studies. Biol. Sex Differ. 2015, 6, 35. [Google Scholar] [CrossRef] [PubMed]
- Carrel, L.; Willard, H.F. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature 2005, 434, 400–404. [Google Scholar] [CrossRef] [PubMed]
- Al Nadaf, S.; Deakin, J.E.; Gilbert, C.; Robinson, T.J.; Graves, J.A.; Waters, P.D. A cross-species comparison of escape from X inactivation in eutheria: Implications for evolution of X chromosome inactivation. Chromosoma 2012, 121, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Berletch, J.B.; Ma, W.; Yang, F.; Shendure, J.; Noble, W.S.; Disteche, C.M.; Deng, X. Escape from X inactivation varies in mouse tissues. PLoS Genet. 2015, 11, e1005079. [Google Scholar] [CrossRef] [PubMed]
- Wakefield, M.J.; Keohane, A.M.; Turner, B.M.; Graves, J.A. Histone underacetylation is an ancient component of mammalian X chromosome inactivation. Proc. Natl. Acad. Sci. USA 1997, 94, 9665–9668. [Google Scholar] [CrossRef] [PubMed]
- Chaumeil, J.; Waters, P.D.; Koina, E.; Gilbert, C.; Robinson, T.J.; Graves, J.A. Evolution from Xist-independent to Xist-controlled X-chromosome inactivation: Epigenetic modifications in distantly related mammals. PLoS ONE 2011, 6, e19040. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rens, W.; Wallduck, M.S.; Lovell, F.L.; Ferguson-Smith, M.A.; Ferguson-Smith, A.C. Epigenetic modifications on X chromosomes in marsupial and monotreme mammals and implications for evolution of dosage compensation. Proc. Natl. Acad. Sci. USA 2010, 107, 17657–17662. [Google Scholar] [CrossRef] [PubMed]
- Mahadevaiah, S.K.; Royo, H.; VandeBerg, J.L.; McCarrey, J.R.; Mackay, S.; Turner, J.M. Key features of the X inactivation process are conserved between marsupials and eutherians. Curr. Biol. 2009, 19, 1478–1484. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, M.M.; Bird, A. DNA methylation landscapes: Provocative insights from epigenomics. Nat. Rev. Genet. 2008, 9, 465–476. [Google Scholar] [CrossRef] [PubMed]
- Graves, J.A.M. 5-azacytidine-induced re-expression of alleles on the inactive X chromosome in a hybrid mouse cell line. Exp. Cell Res. 1982, 141, 99–105. [Google Scholar] [CrossRef]
- Cooper, D.W.; Johnston, P.G.; Watson, J.M.; Graves, J.A.M. X inactivation in marsupials and monotremes. Semin. Dev. Biol. 1993, 4, 117–128. [Google Scholar] [CrossRef]
- Holliday, R.; Pugh, J.E. DNA modification mechanisms and gene activity during development. Science 1975, 187, 226–232. [Google Scholar] [CrossRef] [PubMed]
- Bird, A.P.; Wolffe, A.P. Methylation-induced repression—Belts, braces, and chromatin. Cell 1999, 99, 451–454. [Google Scholar] [CrossRef]
- Charlton, J.; Downing, T.L.; Smith, Z.D.; Gu, H.; Clement, K.; Pop, R.; Akopian, V.; Klages, S.; Santos, D.P.; Tsankov, A.M.; et al. Global delay in nascent strand DNA methylation. Nat. Struct. Mol. Biol. 2018, 25, 327–332. [Google Scholar] [CrossRef] [PubMed]
- Duncan, B.K.; Miller, J.H. Mutagenic deamination of cytosine residues in DNA. Nature 1980, 287, 560–561. [Google Scholar] [CrossRef] [PubMed]
- Bird, A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002, 16, 6–21. [Google Scholar] [CrossRef] [PubMed]
- Antequera, F.; Bird, A. CpG islands. EXS 1993, 64, 169–185. [Google Scholar] [PubMed]
- Edgar, R.; Tan, P.P.; Portales-Casamar, E.; Pavlidis, P. Meta-analysis of human methylomes reveals stably methylated sequences surrounding CpG islands associated with high gene expression. Epigenet. Chromatin 2014, 7, 28. [Google Scholar] [CrossRef] [PubMed]
- Irizarry, R.A.; Ladd-Acosta, C.; Wen, B.; Wu, Z.; Montano, C.; Onyango, P.; Cui, H.; Gabo, K.; Rongione, M.; Webster, M.; et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat. Genet. 2009, 41, 178–186. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Xia, Y.; Li, L.; Gong, D.; Yao, Y.; Luo, H.; Lu, H.; Yi, N.; Wu, H.; Zhang, X.; et al. Double restriction-enzyme digestion improves the coverage and accuracy of genome-wide CpG methylation profiling by reduced representation bisulfite sequencing. BMC Genom. 2013, 14, 11. [Google Scholar] [CrossRef] [PubMed]
- Chedin, F. The DNMT3 family of mammalian de novo DNA methyltransferases. Prog. Mol. Biol. Transl. Sci. 2011, 101, 255–285. [Google Scholar] [PubMed]
- Monk, M.; Boubelik, M.; Lehnert, S. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development 1987, 99, 371–382. [Google Scholar] [PubMed]
- Cotton, A.M.; Price, E.M.; Jones, M.J.; Balaton, B.P.; Kobor, M.S.; Brown, C.J. Landscape of DNA methylation on the X chromosome reflects CpG density, functional chromatin state and X-chromosome inactivation. Hum. Mol. Genet. 2015, 24, 1528–1539. [Google Scholar] [CrossRef] [PubMed]
- Ziller, M.J.; Gu, H.; Muller, F.; Donaghey, J.; Tsai, L.T.; Kohlbacher, O.; De Jager, P.L.; Rosen, E.D.; Bennett, D.A.; Bernstein, B.E.; et al. Charting a dynamic DNA methylation landscape of the human genome. Nature 2013, 500, 477–481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chatterjee, A.; Stockwell, P.A.; Rodger, E.J.; Duncan, E.J.; Parry, M.F.; Weeks, R.J.; Morison, I.M. Genome-wide DNA methylation map of human neutrophils reveals widespread inter-individual epigenetic variation. Sci. Rep. 2015, 5, 17328. [Google Scholar] [CrossRef] [PubMed]
- Yoder, J.A.; Walsh, C.P.; Bestor, T.H. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997, 13, 335–340. [Google Scholar] [CrossRef]
- Mohandas, T.; Sparkes, R.S.; Shapiro, L.J. Reactivation of an inactive human X chromosome: Evidence for X inactivation by DNA methylation. Science 1981, 211, 393–396. [Google Scholar] [CrossRef] [PubMed]
- Venolia, L.; Gartler, S.M.; Wassman, E.R.; Yen, P.; Mohandas, T.; Shapiro, L.J. Transformation with DNA from 5-azacytidine-reactivated X chromosomes. Proc. Natl. Acad. Sci. USA 1982, 79, 2352–2354. [Google Scholar] [CrossRef] [PubMed]
- Hellman, A.; Chess, A. Gene body-specific methylation on the active X chromosome. Science 2007, 315, 1141–1143. [Google Scholar] [CrossRef] [PubMed]
- Weber, M.; Hellmann, I.; Stadler, M.B.; Ramos, L.; Paabo, S.; Rebhan, M.; Schubeler, D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat. Genet. 2007, 39, 457–466. [Google Scholar] [CrossRef] [PubMed]
- Sharp, A.J.; Stathaki, E.; Migliavacca, E.; Brahmachary, M.; Montgomery, S.B.; Dupre, Y.; Antonarakis, S.E. DNA methylation profiles of human active and inactive X chromosomes. Genome Res. 2011, 21, 1592–1600. [Google Scholar] [CrossRef] [PubMed]
- Lister, R.; Mukamel, E.A.; Nery, J.R.; Urich, M.; Puddifoot, C.A.; Johnson, N.D.; Lucero, J.; Huang, Y.; Dwork, A.J.; Schultz, M.D.; et al. Global epigenomic reconfiguration during mammalian brain development. Science 2013, 341, 1237905. [Google Scholar] [CrossRef] [PubMed]
- Schultz, M.D.; He, Y.; Whitaker, J.W.; Hariharan, M.; Mukamel, E.A.; Leung, D.; Rajagopal, N.; Nery, J.R.; Urich, M.A.; Chen, H.; et al. Human body epigenome maps reveal noncanonical DNA methylation variation. Nature 2015, 523, 212–216. [Google Scholar] [CrossRef] [PubMed]
- Boers, R.; Boers, J.; de Hoon, B.; Kockx, C.; Ozgur, Z.; Molijn, A.; van, I.W.; Laven, J.; Gribnau, J. Genome-wide DNA methylation profiling using the methylation-dependent restriction enzyme LpnPI. Genome Res. 2018, 28, 88–99. [Google Scholar] [CrossRef] [PubMed]
- Brenet, F.; Moh, M.; Funk, P.; Feierstein, E.; Viale, A.J.; Socci, N.D.; Scandura, J.M. DNA methylation of the first exon is tightly linked to transcriptional silencing. PLoS ONE 2011, 6, e14524. [Google Scholar] [CrossRef] [PubMed]
- Kaslow, D.C.; Migeon, B.R. DNA methylation stabilizes X chromosome inactivation in eutherians but not in marsupials: Evidence for multistep maintenance of mammalian X dosage compensation. Proc. Natl. Acad. Sci. USA 1987, 84, 6210–6214. [Google Scholar] [CrossRef] [PubMed]
- Loebel, D.A.; Johnston, P.G. Methylation analysis of a marsupial X-linked CpG island by bisulfite genomic sequencing. Genome Res. 1996, 6, 114–123. [Google Scholar] [CrossRef] [PubMed]
- Hornecker, J.L.; Samollow, P.B.; Robinson, E.S.; Vandeberg, J.L.; McCarrey, J.R. Meiotic sex chromosome inactivation in the marsupial Monodelphis domestica. Genesis 2007, 45, 696–708. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Douglas, K.C.; Vandeberg, J.L.; Clark, A.G.; Samollow, P.B. Chromosome-wide profiling of X-chromosome inactivation and epigenetic states in fetal brain and placenta of the opossum, Monodelphis domestica. Genome Res. 2014, 24, 70–83. [Google Scholar] [CrossRef] [PubMed]
- Piper, A.A.; Bennett, A.M.; Noyce, L.; Swanton, M.K.; Cooper, D.W. Isolation of a clone partially encoding hill kangaroo X-linked hypoxanthine phosphoribosyltransferase: Sex differences in methylation in the body of the gene. Somat. Cell Mol. Genet. 1993, 19, 141–159. [Google Scholar] [CrossRef] [PubMed]
- Ingles, E.D.; Deakin, J.E. Global DNA methylation patterns on marsupial and devil facial tumour chromosomes. Mol. Cytogenet. 2015, 8, 74. [Google Scholar] [CrossRef] [PubMed]
- Mugal, C.F.; Arndt, P.F.; Holm, L.; Ellegren, H. Evolutionary consequences of DNA methylation on the GC content in vertebrate genomes. G3 2015, 5, 441–447. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.A. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012, 13, 484–492. [Google Scholar] [CrossRef] [PubMed]
- Vanderkraats, N.D.; Hiken, J.F.; Decker, K.F.; Edwards, J.R. Discovering high-resolution patterns of differential DNA methylation that correlate with gene expression changes. Nucleic Acids Res. 2013, 41, 6816–6827. [Google Scholar] [CrossRef] [PubMed]
- Kapourani, C.A.; Sanguinetti, G. Higher order methylation features for clustering and prediction in epigenomic studies. Bioinformatics 2016, 32, i405–i412. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Waters, S.A.; Capraro, A.; McIntyre, K.L.; Marshall Graves, J.A.; Waters, P.D. The Methylome of Vertebrate Sex Chromosomes. Genes 2018, 9, 230. https://doi.org/10.3390/genes9050230
Waters SA, Capraro A, McIntyre KL, Marshall Graves JA, Waters PD. The Methylome of Vertebrate Sex Chromosomes. Genes. 2018; 9(5):230. https://doi.org/10.3390/genes9050230
Chicago/Turabian StyleWaters, Shafagh A., Alexander Capraro, Kim L. McIntyre, Jennifer A. Marshall Graves, and Paul D. Waters. 2018. "The Methylome of Vertebrate Sex Chromosomes" Genes 9, no. 5: 230. https://doi.org/10.3390/genes9050230




