Proteome-Wide Analysis of Trypanosoma cruzi Exponential and Stationary Growth Phases Reveals a Subcellular Compartment-Specific Regulation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Parasites and Sample Preparation
2.2. Protein Extraction and Digestion
2.3. Mass Spectrometry-Based Analysis of T. cruzi Tryptic Peptides
2.4. Protein Identification and Quantification
2.5. Bioinformatic Analysis
2.6. Sequence Search and Phylogenetic Analysis
2.7. TcPRODH and TcMDH Enzymatic Activities Assays
2.8. Statistical Analysis
3. Results and Discussion
3.1. Morphological and Molecular Characterization of T. cruzi in the Exponential and Stationary Phases
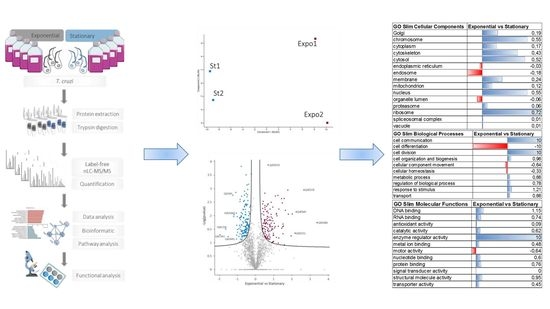
3.2. Differential Protein Expression in the T. cruzi Exponential and Stationary Phases
3.3. Membrane Proteins Regulation during Growth Phase Transitioning
3.4. The Uncharacterized Proteome Modulated during Transitioning from Exponential to Stationary Growth Phase
3.5. Protein N-Terminal Acetylation in T. cruzi Epimastigote during Transitioning from Exponential to Stationary Growth Phase
3.6. Methionine Oxidation in T. cruzi during Transitioning from Exponential to Stationary Growth Phase
3.7. T. cruzi Malate Dehydrogenase and Proline Dehydrogenase are Regulated during Transitioning from Exponential to Stationary Phase
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Tyson, J.J.; Novak, B. Control of cell growth, division and death: Information processing in living cells. Interface Focus 2014, 4. [Google Scholar] [CrossRef] [PubMed]
- Duronio, R.J.; Xiong, Y. Signaling pathways that control cell proliferation. Cold Spring Harb. Perspect. Biol. 2013, 5. [Google Scholar] [CrossRef] [PubMed]
- Herman, P.K. Stationary phase in yeast. Curr. Opin. Microbiol. 2002, 5, 602–607. [Google Scholar] [CrossRef]
- Cai, L.; Tu, B.P. Acetyl-CoA drives the transcriptional growth program in yeast. Cell Cycle 2011, 10, 3045–3046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, L.; Sutter, B.M.; Li, B.; Tu, B.P. Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol. Cell 2011, 42, 426–437. [Google Scholar] [CrossRef] [PubMed]
- Glick, D.; Barth, S.; Macleod, K.F. Autophagy: Cellular and molecular mechanisms. J. Pathol. 2010, 221, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Gray, J.V.; Petsko, G.A.; Johnston, G.C.; Ringe, D.; Singer, R.A.; Werner-Washburne, M. “Sleeping beauty”: Quiescence in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2004, 68, 187–206. [Google Scholar] [CrossRef] [PubMed]
- Rassi, A.J.; Rassi, A.; Marin-Neto, J.A. Chagas disease. Lancet 2010, 375, 1388–1402. [Google Scholar] [CrossRef]
- Tyler, K.M.; Engman, D.M. The life cycle of Trypanosoma cruzi revisited. Int. J. Parasitol. 2001, 31, 472–481. [Google Scholar] [CrossRef]
- Contreras, V.T.; Salles, J.M.; Thomas, N.; Morel, C.M.; Goldenberg, S. In vitro differentiation of Trypanosoma cruzi under chemically defined conditions. Mol. Biochem. Parasitol. 1985, 16, 315–327. [Google Scholar] [CrossRef]
- Barison, M.J.; Rapado, L.N.; Merino, E.F.; Furusho Pral, E.M.; Mantilla, B.S.; Marchese, L.; Nowicki, C.; Silber, A.M.; Cassera, M.B. Metabolomic profiling reveals a finely tuned, starvation-induced metabolic switch in Trypanosoma cruzi epimastigotes. J. Biol. Chem. 2017, 292, 8964–8977. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, R.; Cevallos, A.M.; Nepomuceno-Mejia, T.; Lopez-Villasenor, I. Stationary phase in Trypanosoma cruzi epimastigotes as a preadaptive stage for metacyclogenesis. Parasitol. Res. 2012, 111, 509–514. [Google Scholar] [CrossRef] [PubMed]
- Camargo, E.P. Growth and differentiation in Trypanosoma cruzi. I. Origin of metacyclic trypanosomes in liquid media. Rev. Inst. Med. Trop. Sao Paulo 1964, 6, 93–100. [Google Scholar] [PubMed]
- Parodi-Talice, A.; Monteiro-Goes, V.; Arrambide, N.; Avila, A.R.; Duran, R.; Correa, A.; Dallagiovanna, B.; Cayota, A.; Krieger, M.; Goldenberg, S.; et al. Proteomic analysis of metacyclic trypomastigotes undergoing Trypanosoma cruzi metacyclogenesis. J. Mass Spectrom. 2007, 42, 1422–1432. [Google Scholar] [CrossRef] [PubMed]
- De Godoy, L.M.; Marchini, F.K.; Pavoni, D.P.; Rampazzo Rde, C.; Probst, C.M.; Goldenberg, S.; Krieger, M.A. Quantitative proteomics of Trypanosoma cruzi during metacyclogenesis. Proteomics 2012, 12, 2694–2703. [Google Scholar] [CrossRef] [PubMed]
- Contreras, V.T.; Morel, C.M.; Goldenberg, S. Stage specific gene expression precedes morphological changes during Trypanosoma cruzi metacyclogenesis. Mol. Biochem. Parasitol. 1985, 14, 83–96. [Google Scholar] [CrossRef]
- Homsy, J.J.; Granger, B.; Krassner, S.M. Some factors inducing formation of metacyclic stages of Trypanosoma cruzi. J. Protozool. 1989, 36, 150–153. [Google Scholar] [CrossRef] [PubMed]
- Amorim, J.C.; Batista, M.; da Cunha, E.S.; Lucena, A.C.R.; Lima, C.V.P.; Sousa, K.; Krieger, M.A.; Marchini, F.K. Quantitative proteome and phosphoproteome analyses highlight the adherent population during Trypanosoma cruzi metacyclogenesis. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Brener, Z.; Chiari, E. Aspects of early growth of different Trypanosoma cruzi strains in culture medium. J. Parasitol. 1965, 51, 922–926. [Google Scholar] [CrossRef] [PubMed]
- Paes, L.S.; Suarez Mantilla, B.; Zimbres, F.M.; Pral, E.M.; Diogo de Melo, P.; Tahara, E.B.; Kowaltowski, A.J.; Elias, M.C.; Silber, A.M. Proline dehydrogenase regulates redox state and respiratory metabolism in Trypanosoma cruzi. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [Green Version]
- Lima, L.; Espinosa-Alvarez, O.; Pinto, C.M.; Cavazzana, M.J.; Pavan, A.C.; Carranza, J.C.; Lim, B.K.; Campaner, M.; Takata, C.S.; Camargo, E.P.; et al. New insights into the evolution of the Trypanosoma cruzi clade provided by a new trypanosome species tightly linked to neotropical Pteronotus bats and related to an Australian lineage of trypanosomes. Parasit. Vectors 2015, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vizcaino, J.A.; Csordas, A.; Del-Toro, N.; Dianes, J.A.; Griss, J.; Lavidas, I.; Mayer, G.; Perez-Riverol, Y.; Reisinger, F.; Ternent, T.; et al. 2016 update of the pride database and its related tools. Nucleic acids Res. 2016, 44, 11033. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef] [PubMed]
- Alves, M.J.; Kawahara, R.; Viner, R.; Colli, W.; Mattos, E.C.; Thaysen-Andersen, M.; Larsen, M.R.; Palmisano, G. Comprehensive glycoprofiling of the epimastigote and trypomastigote stages of Trypanosoma cruzi. J. Proteom. 2017, 151, 182–192. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, G.S.; Kawahara, R.; Rosa-Fernandes, L.; Mule, S.N.; Avila, C.C.; Teixeira, M.M.G.; Larsen, M.R.; Palmisano, G. Development of a Trypanosoma cruzi strain typing assay using MS2 peptide spectral libraries (Tc-STAMS2). PLoS Negl. Trop. Dis. 2018, 12. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, G.S.; Kawahara, R.; Rosa-Fernandes, L.; Avila, C.C.; Larsen, M.R.; Pereira Alves, J.M.; Palmisano, G. Novel DNA coding regions and protein arginylation reveal unexplored T. cruzi proteome and PTMs. Int. J. Mass Spectrom. 2017, 418, 51–66. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Sinitcyn, P. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Minning, T.A.; Weatherly, D.B.; Atwood, J., 3rd; Orlando, R.; Tarleton, R.L. The steady-state transcriptome of the four major life-cycle stages of Trypanosoma cruzi. BMC Genom. 2009, 10. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Shah-Simpson, S.; Okrah, K.; Belew, A.T.; Choi, J.; Caradonna, K.L.; Padmanabhan, P.; Ndegwa, D.M.; Temanni, M.R.; Corrada Bravo, H.; et al. Transcriptome remodeling in Trypanosoma cruzi and human cells during intracellular infection. PLoS Pathog. 2016, 12. [Google Scholar] [CrossRef]
- Smircich, P.; Eastman, G.; Bispo, S.; Duhagon, M.A.; Guerra-Slompo, E.P.; Garat, B.; Goldenberg, S.; Munroe, D.J.; Dallagiovanna, B.; Holetz, F.; et al. Ribosome profiling reveals translation control as a key mechanism generating differential gene expression in Trypanosoma cruzi. BMC Genom. 2015, 16. [Google Scholar] [CrossRef] [PubMed]
- Aron Marchler-Bauer, R.G. Cdd ftp-archive. Available online: ftp://ftp.ncbi.nih.gov/pub/mmdb/cdd (accessed on 7 June 2018).
- Marchler-Bauer, A.; Derbyshire, M.K.; Gonzales, N.R.; Lu, S.; Chitsaz, F.; Geer, L.Y.; Geer, R.C.; He, J.; Gwadz, M.; Hurwitz, D.I.; et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015, 43, D222–D226. [Google Scholar] [CrossRef] [PubMed]
- NCBI. Available online: https: //www.ncbi.nlm.nih.gov (accessed on 9 June 2018).
- Caballero, Z.C.; Costa-Martins, A.G.; Ferreira, R.C.; P Alves, J.M.; Serrano, M.G.; Camargo, E.P.; Buck, G.A.; Minoprio, P.; G Teixeira, M.M. Phylogenetic and syntenic data support a single horizontal transference to a Trypanosoma ancestor of a prokaryotic proline racemase implicated in parasite evasion from host defences. Parasit. Vectors 2015, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgar, R.C. Muscle: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galtier, N.; Gouy, M.; Gautier, C. SEAVIEW and PHYLO_WIN: Two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biosci. 1996, 12, 543–548. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef] [PubMed]
- Goloboff, P.A.; Farris, J.S.; Nixon, K.C. TNT, a free program for phylogenetic analysis. Cladistics 2008, 24, 774–786. [Google Scholar] [CrossRef]
- Rambaut, A. Molecular Evolution, Phylogenetics and Epidemiology. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 7 June 2018).
- Lamour, N.; Riviere, L.; Coustou, V.; Coombs, G.H.; Barrett, M.P.; Bringaud, F. Proline metabolism in procyclic Trypanosoma Brucei is down-regulated in the presence of glucose. J. Biol. Chem. 2005, 280, 11902–11910. [Google Scholar] [CrossRef] [PubMed]
- Hunter, G.R.; Hellman, U.; Cazzulo, J.J.; Nowicki, C. Tetrameric and dimeric malate dehydrogenase isoenzymes in Trypanosoma cruzi epimastigotes. Mol. Biochem. Parasitol. 2000, 105, 203–214. [Google Scholar] [CrossRef]
- Tyler, K.M.; Engman, D.M. Flagellar elongation induced by glucose limitation is preadaptive for Trypanosoma cruzi differentiation. Cell Motil. Cytoskel. 2000, 46, 269–278. [Google Scholar] [CrossRef]
- Bonaldo, M.C.; Souto-Padron, T.; de Souza, W.; Goldenberg, S. Cell-substrate adhesion during Trypanosoma cruzi differentiation. J. Cell Biol. 1988, 106, 1349–1358. [Google Scholar] [CrossRef] [PubMed]
- Goncalves, C.S.; Avila, A.R.; de Souza, W.; Motta, M.C.M.; Cavalcanti, D.P. Revisiting the Trypanosoma cruzi metacyclogenesis: Morphological and ultrastructural analyses during cell differentiation. Parasit. Vectors 2018, 11. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, L.R.; Dossin Fde, M.; Ramos, T.C.; Freymuller, E.; Schenkman, S. Active transcription and ultrastructural changes during Trypanosoma cruzi metacyclogenesis. An. Acad. Bras. Cienc. 2008, 80, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Atwood, J.A., 3rd; Weatherly, D.B.; Minning, T.A.; Bundy, B.; Cavola, C.; Opperdoes, F.R.; Orlando, R.; Tarleton, R.L. The Trypanosoma cruzi proteome. Science 2005, 309, 473–476. [Google Scholar] [CrossRef] [PubMed]
- Schwanhausser, B.; Busse, D.; Li, N.; Dittmar, G.; Schuchhardt, J.; Wolf, J.; Chen, W.; Selbach, M. Global quantification of mammalian gene expression control. Nature 2011, 473, 337–342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tonelli, R.R.; Augusto Lda, S.; Castilho, B.A.; Schenkman, S. Protein synthesis attenuation by phosphorylation of eIF2α is required for the differentiation of Trypanosoma cruzi into infective forms. PLoS ONE 2011, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conte, I.; Labriola, C.; Cazzulo, J.J.; Docampo, R.; Parodi, A.J. The interplay between folding-facilitating mechanisms in Trypanosoma cruzi endoplasmic reticulum. Mol. Biol. Cell 2003, 14, 3529–3540. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, L.; Ramirez, G.; Valck, C.; Molina, M.C.; Rojas, A.; Schwaeble, W.; Ferreira, V.; Ferreira, A. F(ab’)2 antibody fragments against Trypanosoma cruzi calreticulin inhibit its interaction with the first component of human complement. Biol. Res. 2005, 38, 187–195. [Google Scholar] [CrossRef] [PubMed]
- De Paiva, R.M.; Grazielle-Silva, V.; Cardoso, M.S.; Nakagaki, B.N.; Mendonca-Neto, R.P.; Canavaci, A.M.; Souza Melo, N.; Martinelli, P.M.; Fernandes, A.P.; daRocha, W.D.; et al. Amastin knockdown in Leishmania braziliensis affects parasite-macrophage interaction and results in impaired viability of intracellular amastigotes. PLoS Pathog. 2015, 11. [Google Scholar] [CrossRef] [PubMed]
- Rochette, A.; McNicoll, F.; Girard, J.; Breton, M.; Leblanc, E.; Bergeron, M.G.; Papadopoulou, B. Characterization and developmental gene regulation of a large gene family encoding amastin surface proteins in Leishmania spp. Mol. Biochem. Parasitol. 2005, 140, 205–220. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, S.M.; Russell, D.G.; Kirchhoff, L.V.; Donelson, J.E. A differentially expressed gene family encoding “amastin,” a surface protein of Trypanosoma cruzi amastigotes. J. Biol. Chem. 1994, 269, 20509–20516. [Google Scholar] [PubMed]
- Kangussu-Marcolino, M.M.; de Paiva, R.M.; Araujo, P.R.; de Mendonca-Neto, R.P.; Lemos, L.; Bartholomeu, D.C.; Mortara, R.A.; daRocha, W.D.; Teixeira, S.M. Distinct genomic organization, mRNA expression and cellular localization of members of two amastin sub-families present in Trypanosoma cruzi. BMC Microbiol. 2013, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arco, A.D.; Satrustegui, J. New mitochondrial carriers: An overview. Cell. Mol. Life Sci. 2005, 62, 2204–2227. [Google Scholar] [CrossRef] [PubMed]
- Pena-Diaz, P.; Pelosi, L.; Ebikeme, C.; Colasante, C.; Gao, F.; Bringaud, F.; Voncken, F. Functional characterization of tbMCP5, a conserved and essential ADP/ATP carrier present in the mitochondrion of the human pathogen Trypanosoma brucei. J. Biol. Chem. 2012, 287, 41861–41874. [Google Scholar] [CrossRef] [PubMed]
- Gnipova, A.; Subrtova, K.; Panicucci, B.; Horvath, A.; Lukes, J.; Zikova, A. The ADP/ATP carrier and its relationship to oxidative phosphorylation in ancestral protist Trypanosoma brucei. Eukaryot. Cell 2015, 14, 297–310. [Google Scholar] [CrossRef] [PubMed]
- Van Der Heyden, N.; Docampo, R. Proton and sodium pumps regulate the plasma membrane potential of different stages of Trypanosoma cruzi. Mol. Biochem. Parasitol. 2002, 120, 127–139. [Google Scholar] [CrossRef]
- Martins, R.M.; Covarrubias, C.; Rojas, R.G.; Silber, A.M.; Yoshida, N. Use of l-Proline and ATP production by Trypanosoma cruzi metacyclic forms as requirements for host cell invasion. Infect. Immun. 2009, 77, 3023–3032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Baumgart, E.; Dong, G.X.; Morrell, J.C.; Jimenez-Sanchez, G.; Valle, D.; Smith, K.D.; Gould, S.J. PEX11α is required for peroxisome proliferation in response to 4-phenylbutyrate but is dispensable for peroxisome proliferator-activated receptor alpha-mediated peroxisome proliferation. Mol. Cell. Biol. 2002, 22, 8226–8240. [Google Scholar] [CrossRef] [PubMed]
- Subramani, S.; Koller, A.; Snyder, W.B. Import of peroxisomal matrix and membrane proteins. Annu. Rev. Biochem. 2000, 69, 399–418. [Google Scholar] [CrossRef] [PubMed]
- Parsons, M.; Furuya, T.; Pal, S.; Kessler, P. Biogenesis and function of peroxisomes and glycosomes. Mol. Biochem. Parasitol. 2001, 115, 19–28. [Google Scholar] [CrossRef]
- Erdmann, R.; Blobel, G. Giant peroxisomes in oleic acid-induced Saccharomyces cerevisiae lacking the peroxisomal membrane protein Pmp27p. J. Cell Biol. 1995, 128, 509–523. [Google Scholar] [CrossRef] [PubMed]
- Aman, R.A.; Wang, C.C. Identification of two integral glycosomal membrane proteins in Trypanosoma brucei. Mol. Biochem. Parasitol. 1987, 25, 83–92. [Google Scholar] [CrossRef]
- Lorenz, P.; Maier, A.G.; Baumgart, E.; Erdmann, R.; Clayton, C. Elongation and clustering of glycosomes in Trypanosoma brucei overexpressing the glycosomal Pex11p. EMBO J. 1998, 17, 3542–3555. [Google Scholar] [CrossRef] [PubMed]
- Maier, A.; Lorenz, P.; Voncken, F.; Clayton, C. An essential dimeric membrane protein of trypanosome glycosomes. Mol. Microbiol. 2001, 39, 1443–1451. [Google Scholar] [CrossRef] [Green Version]
- Voncken, F.; van Hellemond, J.J.; Pfisterer, I.; Maier, A.; Hillmer, S.; Clayton, C. Depletion of GIM5 causes cellular fragility, a decreased glycosome number, and reduced levels of ether-linked phospholipids in trypanosomes. J. Biol. Chem. 2003, 278, 35299–35310. [Google Scholar] [CrossRef] [PubMed]
- Cevallos, A.M.; Segura-Kato, Y.X.; Merchant-Larios, H.; Manning-Cela, R.; Alberto Hernandez-Osorio, L.; Marquez-Duenas, C.; Ambrosio, J.R.; Reynoso-Ducoing, O.; Hernandez, R. Trypanosoma cruzi: Multiple actin isovariants are observed along different developmental stages. Exp. Parasitol. 2011, 127, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Cevallos, A.M.; Perez-Escobar, M.; Espinosa, N.; Herrera, J.; Lopez-Villasenor, I.; Hernandez, R. The stabilization of housekeeping transcripts in Trypanosoma cruzi epimastigotes evidences a global regulation of RNA decay during stationary phase. FEMS Microbiol. Lett. 2005, 246, 259–264. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Kirisako, T.; Kamada, Y.; Mizushima, N.; Noda, T.; Ohsumi, Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001, 20, 5971–5981. [Google Scholar] [CrossRef] [PubMed]
- Sou, Y.S.; Waguri, S.; Iwata, J.; Ueno, T.; Fujimura, T.; Hara, T.; Sawada, N.; Yamada, A.; Mizushima, N.; Uchiyama, Y.; et al. The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol. Biol. Cell 2008, 19, 4762–4775. [Google Scholar] [CrossRef] [PubMed]
- Kuma, A.; Mizushima, N.; Ishihara, N.; Ohsumi, Y. Formation of the approximately 350-kda Apg12-Apg5·Apg16 multimeric complex, mediated by Apg16 oligomerization, is essential for autophagy in yeast. J. Biol. Chem. 2002, 277, 18619–18625. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, V.E.; Kosec, G.; Sant’Anna, C.; Turk, V.; Cazzulo, J.J.; Turk, B. Autophagy is involved in nutritional stress response and differentiation in Trypanosoma cruzi. J. Biol. Chem. 2008, 283, 3454–3464. [Google Scholar] [CrossRef] [PubMed]
- Vanrell, M.C.; Losinno, A.D.; Cueto, J.A.; Balcazar, D.; Fraccaroli, L.V.; Carrillo, C.; Romano, P.S. The regulation of autophagy differentially affects Trypanosoma cruzi metacyclogenesis. PLoS Negl. Trop. Dis. 2017, 11. [Google Scholar] [CrossRef] [PubMed]
- Romano, P.S.; Arboit, M.A.; Vazquez, C.L.; Colombo, M.I. The autophagic pathway is a key component in the lysosomal dependent entry of Trypanosoma cruzi into the host cell. Autophagy 2009, 5, 6–18. [Google Scholar] [CrossRef] [PubMed]
- Neer, E.J.; Schmidt, C.J.; Nambudripad, R.; Smith, T.F. The ancient regulatory-protein family of WD-repeat proteins. Nature 1994, 371, 297–300. [Google Scholar] [CrossRef] [PubMed]
- Smith, T.F.; Gaitatzes, C.; Saxena, K.; Neer, E.J. The WD repeat: A common architecture for diverse functions. Trends Biochem. Sci. 1999, 24, 181–185. [Google Scholar] [CrossRef]
- Zingales, B.; Miles, M.A.; Campbell, D.A.; Tibayrenc, M.; Macedo, A.M.; Teixeira, M.M.; Schijman, A.G.; Llewellyn, M.S.; Lages-Silva, E.; Machado, C.R.; et al. The revised Trypanosoma cruzi subspecific nomenclature: Rationale, epidemiological relevance and research applications. Infect. Genet. Evol. 2012, 12, 240–253. [Google Scholar] [CrossRef] [PubMed]
- Lima, L.; Espinosa-Alvarez, O.; Ortiz, P.A.; Trejo-Varon, J.A.; Carranza, J.C.; Pinto, C.M.; Serrano, M.G.; Buck, G.A.; Camargo, E.P.; Teixeira, M.M. Genetic diversity of Trypanosoma cruzi in bats, and multilocus phylogenetic and phylogeographical analyses supporting tcbat as an independent DTU (discrete typing unit). Acta Trop. 2015, 151, 166–177. [Google Scholar] [CrossRef] [PubMed]
- Drazic, A.; Myklebust, L.M.; Ree, R.; Arnesen, T. The world of protein acetylation. Biochim. Biophys. Acta 2016, 1864, 1372–1401. [Google Scholar] [CrossRef] [PubMed]
- Aksnes, H.; Hole, K.; Arnesen, T. Molecular, cellular, and physiological significance of N-terminal acetylation. Int. Rev. Cell Mol. Biol. 2015, 316, 267–305. [Google Scholar] [PubMed]
- Brownell, J.E.; Zhou, J.; Ranalli, T.; Kobayashi, R.; Edmondson, D.G.; Roth, S.Y.; Allis, C.D. Tetrahymena histone acetyltransferase A: A homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell 1996, 84, 843–851. [Google Scholar] [CrossRef]
- Carrozza, M.J.; Utley, R.T.; Workman, J.L.; Cote, J. The diverse functions of histone acetyltransferase complexes. Trends Genet. 2003, 19, 321–329. [Google Scholar] [CrossRef]
- Boitz, J.M.; Ullman, B. Leishmania donovani singly deficient in HGPRT, APRT or XPRT are viable in vitro and within mammalian macrophages. Mol. Biochem. Parasitol. 2006, 148, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Luscher, A.; Lamprea-Burgunder, E.; Graf, F.E.; de Koning, H.P.; Maser, P. Trypanosoma brucei adenine-phosphoribosyltransferases mediate adenine salvage and aminopurinol susceptibility but not adenine toxicity. Int. J. Parasitol. Drugs Drug Resist. 2014, 4, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Wallace, L.J.; Candlish, D.; Hagos, A.; Seley, K.L.; de Koning, H.P. Selective transport of a new class of purine antimetabolites by the protozoan parasite Trypanosoma brucei. Nucleosides Nucleotides Nucleic Acids 2004, 23, 1441–1444. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Huertas, P.; Mejia-Jaramillo, A.M.; Gonzalez, L.; Triana-Chavez, O. Transcriptome and functional genomics reveal the participation of adenine phosphoribosyltransferase in Trypanosoma cruzi resistance to benznidazole. J. Cell Biochem. 2017, 118, 1936–1945. [Google Scholar] [CrossRef] [PubMed]
- Tzfati, Y.; Abeliovich, H.; Avrahami, D.; Shlomai, J. Universal minicircle sequence binding protein, a CCHC-type zinc finger protein that binds the universal minicircle sequence of trypanosomatids. Purification and characterization. J. Biol. Chem. 1995, 270, 21339–21345. [Google Scholar] [CrossRef] [PubMed]
- Milman, N.; Motyka, S.A.; Englund, P.T.; Robinson, D.; Shlomai, J. Mitochondrial origin-binding protein UMSBP mediates DNA replication and segregation in trypanosomes. Proc. Natl. Acad. Sci. USA 2007, 104, 19250–19255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, S.; Dubey, V.K. Quantitative proteome analysis of Leishmania donovani under spermidine starvation. PLoS ONE 2016, 11. [Google Scholar] [CrossRef] [PubMed]
- Starheim, K.K.; Gevaert, K.; Arnesen, T. Protein N-terminal acetyltransferases: When the start matters. Trends Biochem. Sci. 2012, 37, 152–161. [Google Scholar] [CrossRef] [PubMed]
- Whiteway, M.; Szostak, J.W. The ARD1 gene of yeast functions in the switch between the mitotic cell cycle and alternative developmental pathways. Cell 1985, 43, 483–492. [Google Scholar] [CrossRef]
- Park, E.C.; Szostak, J.W. ARD1 and NAT1 proteins form a complex that has N-terminal acetyltransferase activity. EMBO J. 1992, 11, 2087–2093. [Google Scholar] [PubMed]
- Moretti, N.S.; Cestari, I.; Anupama, A.; Stuart, K.; Schenkman, S. Comparative proteomic analysis of lysine acetylation in trypanosomes. J. Proteome Res. 2018, 17, 374–385. [Google Scholar] [CrossRef] [PubMed]
- Marcili, A.; Lima, L.; Cavazzana, M.; Junqueira, A.C.; Veludo, H.H.; Maia Da Silva, F.; Campaner, M.; Paiva, F.; Nunes, V.L.; Teixeira, M.M. A new genotype of Trypanosoma cruzi associated with bats evidenced by phylogenetic analyses using SSU rDNA, cytochrome b and histone H2B genes and genotyping based on ITS1 rDNA. Parasitology 2009, 136, 641–655. [Google Scholar] [CrossRef] [PubMed]
- Nogueira, N.P.; Saraiva, F.M.; Sultano, P.E.; Cunha, P.R.; Laranja, G.A.; Justo, G.A.; Sabino, K.C.; Coelho, M.G.; Rossini, A.; Atella, G.C.; et al. Proliferation and differentiation of Trypanosoma cruzi inside its vector have a new trigger: Redox status. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- Nozaki, T.; Shigeta, Y.; Saito-Nakano, Y.; Imada, M.; Kruger, W.D. Characterization of transsulfuration and cysteine biosynthetic pathways in the protozoan hemoflagellate, Trypanosoma cruzi. Isolation and molecular characterization of cystathionine β-synthase and serine acetyltransferase from trypanosoma. J. Biol. Chem. 2001, 276, 6516–6523. [Google Scholar] [CrossRef] [PubMed]
- Ohmori, S.; Nawata, Y.; Kiyono, K.; Murata, H.; Tsuboi, S.; Ikeda, M.; Akagi, R.; Morohashi, K.I.; Ono, B. Saccharomyces cerevisiae cultured under aerobic and anaerobic conditions: Air-level oxygen stress and protection against stress. Biochim. Biophys. Acta 1999, 1472, 587–594. [Google Scholar] [CrossRef]
- Liu, H.; Ponniah, G.; Neill, A.; Patel, R.; Andrien, B. Accurate determination of protein methionine oxidation by stable isotope labeling and LC-MS analysis. Anal. Chem. 2013, 85, 11705–11709. [Google Scholar] [CrossRef] [PubMed]
- Marchese, L.; Nascimento, J.F.; Damasceno, F.S.; Bringaud, F.; Michels, P.A.M. The uptake and metabolism of amino acids, and their unique role in the biology of pathogenic trypanosomatids. Pathogens 2018, 7, 36. [Google Scholar] [CrossRef]
- Tonelli, R.R.; Silber, A.M.; Almeida-de-Faria, M.; Hirata, I.Y.; Colli, W.; Alves, M.J. L-proline is essential for the intracellular differentiation of Trypanosoma cruzi. Cell. Microbiol. 2004, 6, 733–741. [Google Scholar] [CrossRef] [PubMed]
- Magdaleno, A.; Ahn, I.Y.; Paes, L.S.; Silber, A.M. Actions of a proline analogue, l-thiazolidine-4-carboxylic acid (T4C), on Trypanosoma cruzi. PLoS ONE 2009, 4. [Google Scholar] [CrossRef] [PubMed]
- Saye, M.; Miranda, M.R.; di Girolamo, F.; de los Milagros Camara, M.; Pereira, C.A. Proline modulates the Trypanosoma cruzi resistance to reactive oxygen species and drugs through a novel D, L-proline transporter. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Mantilla, B.S.; Paes, L.S.; Pral, E.M.; Martil, D.E.; Thiemann, O.H.; Fernandez-Silva, P.; Bastos, E.L.; Silber, A.M. Role of Δ1-pyrroline-5-carboxylate dehydrogenase supports mitochondrial metabolism and host-cell invasion of Trypanosoma cruzi. J. Biol. Chem. 2015, 290, 7767–7790. [Google Scholar] [CrossRef] [PubMed]









| Proteins | Name | Exp/St |
|---|---|---|
| Q4E088 | 40S ribosomal protein S10 | 10.09 |
| Q4E078 | Ribosomal protein L24 | 6.33 |
| Q4E3M0 | 40S ribosomal protein S21 | 4.69 |
| Q4DZ31 | 60S ribosomal protein L18a | 4.75 |
| Q4D4L4 | 40S ribosomal protein S11 | 3.53 |
| Q4DEK4 | 40S ribosomal protein S16 | 2.87 |
| Q4D1T6 | Ubiquitin/ribosomal protein S27a | 2.91 |
| Q4DJZ6 | Ribosomal protein L15 OS | 2.78 |
| Q4DVM4 | 60S ribosomal protein L7 | 2.43 |
| Q4DG45 | 60S ribosomal protein L26 | 2.28 |
| Q4DVM5 | 60S ribosomal protein L7 | 2.21 |
| Q4D5P4 | 40S ribosomal protein S4 | 2.25 |
| Q4DI49 | 60S ribosomal protein L23a | 2.22 |
| Q4DTQ1 | 40S ribosomal protein S23 | 2.02 |
| Q4DC23 | 60S ribosomal protein L17 | 1.93 |
| Q4DPQ8 | 60S ribosomal protein L2 | 1.93 |
| Q4CLU9 | 40S ribosomal protein S8 | 1.91 |
| Q4CVQ4 | 60S acidic ribosomal protein P2 beta (H6.4) | 1.81 |
| Q4CQ63 | 40S ribosomal protein SA | 1.81 |
| Q4DW38 | 40S ribosomal protein S24 | 1.74 |
| Q4E4R1 | 60S ribosomal protein L23 | 1.73 |
| Q4DJY1 | Ribosomal protein S19 | 1.68 |
| Q4E3R2 | 60S ribosomal protein L7a | 1.64 |
| Q4DJX1 | Ribosomal protein L3 | 1.66 |
| Q4DZ41 | 40S ribosomal protein S3a-2 | 1.62 |
| Q4DD50 | 60S ribosomal protein L10 | 1.62 |
| Q4D6H7 | Ribosomal protein S20 | 1.56 |
| Q4E0N6 | 40S ribosomal protein S15a | 1.50 |
| Q4CTR5 | 40S ribosomal protein L14 | 1.50 |
| Q4DGZ5 | 40S ribosomal protein S15 | 1.47 |
| Q4DIZ9 | 40S ribosomal protein S2 | 1.41 |
| Q4DIV9 | 60S ribosomal protein L6 | 1.40 |
| Q4CU61 | 60S ribosomal protein L5 | 1.39 |
| Q4D991 | 60S acidic ribosomal protein | Exp |
| Q4DTK4 | 50S ribosomal protein L7Ae | Exp |
| Q4DN72 | 60S ribosomal protein L34 | Exp |
| Q4DMS9 | Ribosomal protein L27 | Exp |
| Q4CQG0 | 40S ribosomal protein S12 | St |
| Peptide | Proteins | Name | Nac_St | Nac_exp | Prot_St * | Prot_expo * |
|---|---|---|---|---|---|---|
| AVVQTHCFNWMDHDGTR | Q4DMP7 | Uncharacterized protein | 0 | 340,000,000 | 1,100,000,000 | 1,480,000,000 |
| MMEGFYGVEVASGQQVKPK | Q4D244 | Nucleolar RNA-binding protein, putative | 0 | 130,000,000 | 559,000,000 | 758,000,000 |
| MENLTVEEAR | Q4DQY7 | Acetyl-coenzyme A synthetase | 0 | 107,000,000 | 1,710,000,000 | 2,450,000,000 |
| ASVFYILDSK | Q4DX10 | Mu-adaptin 1, putative | 0 | 14,581,000 | 277,000,000 | 194,000,000 |
| ATAAVALAPTADAAGSVLEPLLDK | Q4E342 | Proteasome regulatory non-ATPase subunit 6, putative | 0 | 56,185,000 | 340,000,000 | 599,000,000 |
| STVEDFVLQALASTDAVESDK | Q4DY45 | Phenylalanyl-tRNA synthetase alpha chain, putative | 0 | 58,198,000 | 320,000,000 | 290,000,000 |
| SDPDKSNTAASQEDASGNVASK | Q4CNC1 | Aspartyl-tRNA synthetase, putative | 0 | 58,968,500 | 745,000,000 | 688,000,000 |
| SALTESLLDLHK | Q4E3B2 | Uncharacterized protein | 0 | 51,456,500 | 71,460,000 | 151,000,000 |
| TAFLDNKLEYLQK | Q4E1S6 | Adenylate kinase, putative | 0 | 47,744,500 | 590,000,000 | 347,000,000 |
| SLTLQSEQFQHIVR | Q4E093 | 40S ribomal protein S18, putative | 0 | 60,961,500 | 3,230,000,000 | 3,550,000,000 |
| SVFGVDFGNLNSTVAITR | Q4D673 | Heat shock protein, putative (Fragment) | 0 | 41,490,000 | 29,524,000 | 180,000,000 |
| AALVHLPDPFVTLPFR | Q4D509 | Uncharacterized protein | 0 | 26,870,850 | 217,000,000 | 206,000,000 |
| MLELPPVASLK | Q4D3W3 | Aspartate carbamoyltransferase, putative | 0 | 22,928,000 | 1,140,000,000 | 1,120,000,000 |
| MIVLNGISEEQKK | Q4DDB3 | Uncharacterized protein | 0 | 32,419,000 | 598,000,000 | 408,000,000 |
| TDKKEEQQNTEEYDYDR | Q4DTK4 | 50S ribomal protein L7Ae, putative | 0 | 46,915,500 | 0 | 75,524,000 |
| AEHLLEQLR | Q4D4P8 | Uncharacterized protein | 0 | 127,000,000 | 0 | 123,000,000 |
| MDLDAFLNKK | Q4DUJ9 | Uncharacterized protein | 0 | 73,499,500 | 0 | 423,000,000 |
| MFHGFPDVQIAPR | Q4E3S5 | Uncharacterized protein | 0 | 59,240,000 | 0 | 128,000,000 |
| GDVEQIVEKEETDIQANVLAIPTFEAMGLK | Q4CMK2 | ATP-dependent DEAD/H RNA helicase, putative (Fragment) | 0 | 13,256,500 | 0 | 53,465,000 |
| AELLTPK | Q4E1W2 | Uncharacterized protein | 0 | 13,818,000 | 0 | 70,917,500 |
| MFFEGACAK | Q4CTR7 | Malate dehydrogenase | 0 | 26,409,500 | 0 | 193,000,000 |
| MLNNELANLVDQQK | Q4DM75 | Protein translation factor SUI1 homolog, putative | 0 | 121,000,000 | ||
| SVAEGFLSHGEPCTR | Q4DV00 | Uncharacterized protein | 0 | 81,500,500 | ||
| MLHEIPLHDASASAAER | Q4CX95 | Uncharacterized protein | 0 | 74,033,000 | ||
| TAPSTTAASTAVPFLEVK | Q4CQ45 | Glutaminyl-tRNA synthetase, putative (Fragment) | 0 | 50,153,500 | ||
| SLEEVEPNFFTLSPDSPLR | Q4DVM8 | Adenine phphoribyltransferase, putative | 0 | 48,383,000 | ||
| AILPHGFIEAIHASPLR | Q4DS12 | Uncharacterized protein | 0 | 39,947,500 | ||
| METAPHNIQTER | Q4DS85 | Uncharacterized protein | 0 | 16,433,000 | ||
| SSDIIEHSFFFTPLER | Q4DZY5 | Cholinephphate cytidylyltransferase A, putative | 0 | 25,039,000 | ||
| SAFNPDAPAFIPTFLR | Q4DNJ8 | RNA guanylyltransferase, putative | 0 | 45,679,500 | ||
| MDFLHHVQGIVVLNELGGR | Q4DEQ7 | Coatomer zeta subunit, putative | 0 | 51,783,000 | ||
| MFDALSALVETASAK | Q4D4V0 | Uncharacterized protein | 0 | 40,160,500 | ||
| SGRPESVQGGNVESQSSNVSQGGR | Q4E0M5 | Retrotranspon hot spot (RHS) protein, putative (Fragment) | 0 | 50,721,500 | ||
| SHAGNVTNVYDEK | Q4DXE3 | Uncharacterized protein | 0 | 30,738,000 | ||
| AYNTPAWNEEFGVLK | Q4DJ63 | Uncharacterized protein | 0 | 28,215,500 | ||
| AALVHLPDQFVTLPFR | Q4DP85 | Uncharacterized protein | 0 | 33,409,000 | ||
| SDSTEQSVSKPETSK | Q4CMM3 | Uncharacterized protein | 0 | 28,615,500 | ||
| ADEVPMTKR | Q4D6T8 | Universal minicircle sequence binding protein (UMSBP), putative | 37,170,000 | 0 | 4,410,000,000 | 4,790,000,000 |
| SLEEVEPNFFILSPDSPLR | Q4DNZ4 | Adenine phphoribyltransferase, putative | 117,000,000 | 0 | 186,000,000 | 0 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Avila, C.C.; Mule, S.N.; Rosa-Fernandes, L.; Viner, R.; Barisón, M.J.; Costa-Martins, A.G.; De Oliveira, G.S.; Teixeira, M.M.G.; Marinho, C.R.F.; Silber, A.M.; et al. Proteome-Wide Analysis of Trypanosoma cruzi Exponential and Stationary Growth Phases Reveals a Subcellular Compartment-Specific Regulation. Genes 2018, 9, 413. https://doi.org/10.3390/genes9080413
Avila CC, Mule SN, Rosa-Fernandes L, Viner R, Barisón MJ, Costa-Martins AG, De Oliveira GS, Teixeira MMG, Marinho CRF, Silber AM, et al. Proteome-Wide Analysis of Trypanosoma cruzi Exponential and Stationary Growth Phases Reveals a Subcellular Compartment-Specific Regulation. Genes. 2018; 9(8):413. https://doi.org/10.3390/genes9080413
Chicago/Turabian StyleAvila, Carla Cristi, Simon Ngao Mule, Livia Rosa-Fernandes, Rosa Viner, María Julia Barisón, André Guillherme Costa-Martins, Gilberto Santos De Oliveira, Marta Maria Geraldes Teixeira, Claudio Romero Farias Marinho, Ariel Mariano Silber, and et al. 2018. "Proteome-Wide Analysis of Trypanosoma cruzi Exponential and Stationary Growth Phases Reveals a Subcellular Compartment-Specific Regulation" Genes 9, no. 8: 413. https://doi.org/10.3390/genes9080413