Phosphorylation-Dependent Inhibition of Akt1
Abstract
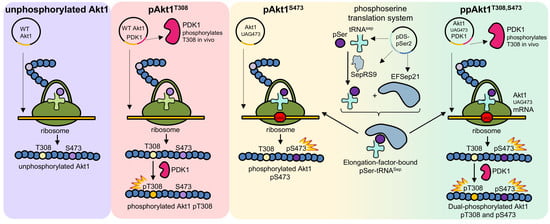
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Plasmids
2.2. Protein and Phosphoprotein Production
2.3. Parallel-Reaction Monitoring Mass Spectrometry of ppAkt1
2.4. MALDI-TOF/TOF Mass Spectrometry Analysis
2.5. Akt1 Kinase Activity Assay
2.6. Kinase Inhibition Assay
3. Results
3.1. Production of Recombinant Akt1 Variants
3.2. Recombinant Akt1 Produced in E. coli Versus Sf9 Cells
3.3. Impact of PH Domain Deletion on Differentially Phosphorylated Akt1
3.4. Chemical Inhibition of Phosphorylated Akt1 Variants
4. Discussion
4.1. Phosphorylation-Dependent Auto-Inhibition of Akt1
4.2. Phosphorylation-Dependent Chemical Inhibition of Akt1
4.3. Synthetic Biology Approach to Generate Active Akt1
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Manning, B.D.; Cantley, L.C. AKT/PKB signaling: Navigating downstream. Cell 2007, 129, 1261–1274. [Google Scholar] [CrossRef] [PubMed]
- Manning, B.D.; Toker, A. AKT/PKB Signaling: Navigating the Network. Cell 2017, 169, 381–405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agarwal, E.; Brattain, M.G.; Chowdhury, S. Cell survival and metastasis regulation by Akt signaling in colorectal cancer. Cell. Signal 2013, 25, 1711–1719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spencer, A.; Yoon, S.S.; Harrison, S.J.; Morris, S.R.; Smith, D.A.; Brigandi, R.A.; Gauvin, J.; Kumar, R.; Opalinska, J.B.; Chen, C. The novel AKT inhibitor afuresertib shows favorable safety, pharmacokinetics, and clinical activity in multiple myeloma. Blood 2014, 124, 2190–2195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antonelli, M.; Massimino, M.; Morra, I.; Garre, M.L.; Gardiman, M.P.; Buttarelli, F.R.; Arcella, A.; Giangaspero, F. Expression of pERK and pAKT in pediatric high grade astrocytomas: Correlation with YKL40 and prognostic significance. Neuropathology 2012, 32, 133–138. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, Y.; Shirai, K.; Oka, K.; Mobaraki, A.; Yoshida, Y.; Noda, S.E.; Okamoto, M.; Suzuki, Y.; Itoh, J.; Itoh, H.; et al. Higher pAkt expression predicts a significant worse prognosis in glioblastomas. J. Radiat. Res. 2010, 51, 343–348. [Google Scholar] [CrossRef] [PubMed]
- Blachly, J.S.; Baiocchi, R.A. Targeting PI3-kinase (PI3K), AKT and mTOR axis in lymphoma. Br. J. Haematol. 2014, 167, 19–32. [Google Scholar] [CrossRef] [PubMed]
- Westin, J.R. Status of PI3K/Akt/mTOR pathway inhibitors in lymphoma. Clin. Lymphoma Myeloma Leuk 2014, 14, 335–342. [Google Scholar] [CrossRef] [PubMed]
- Jung-Testas, I.; Hu, Z.Y.; Baulieu, E.E.; Robel, P. Neurosteroids: Biosynthesis of pregnenolone and progesterone in primary cultures of rat glial cells. Endocrinology 1989, 125, 2083–2091. [Google Scholar] [CrossRef] [PubMed]
- Nitulescu, G.M.; Margina, D.; Juzenas, P.; Peng, Q.; Olaru, O.T.; Saloustros, E.; Fenga, C.; Spandidos, D.; Libra, M.; Tsatsakis, A.M. Akt inhibitors in cancer treatment: The long journey from drug discovery to clinical use (Review). Int. J. Oncol. 2016, 48, 869–885. [Google Scholar] [CrossRef] [PubMed]
- Altomare, D.A.; Testa, J.R. Perturbations of the AKT signaling pathway in human cancer. Oncogene 2005, 24, 7455–7464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balasuriya, N.; Kunkel, M.T.; Liu, X.; Biggar, K.K.; Li, S.S.; Newton, A.C.; O’Donoghue, P. Genetic code expansion and live cell imaging reveal that Thr308 phosphorylation is irreplaceable and sufficient for Akt1 activity. J. Biol. Chem. 2018, 293, 10744–10756. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.X.; Ng, Y.; Meoli, C.C.; Kumar, A.; Khoo, P.S.; Fazakerley, D.J.; Junutula, J.R.; Vali, S.; James, D.E.; Stockli, J. Amplification and demultiplexing in insulin-regulated Akt protein kinase pathway in adipocytes. J. Biol. Chem. 2012, 287, 6128–6138. [Google Scholar] [CrossRef] [PubMed]
- Parker, L.; Levinger, I.; Mousa, A.; Howlett, K.; de Courten, B. Plasma 25-hydroxyvitamin D is related to protein signaling involved in glucose homeostasis in a tissue-specific manner. Nutrients 2016, 8, 631. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Wu, Y.; Liu, M.; Qin, Y.; Wang, H.; Wang, L.; Li, S.; Zhu, H.; He, Z.; Luo, J.; et al. SEMA3B improves the survival of patients with esophageal squamous cell carcinoma by upregulating p53 and p21. Oncol. Rep. 2016, 36, 900–908. [Google Scholar] [CrossRef] [PubMed]
- Hart, J.R.; Vogt, P.K. Phosphorylation of AKT: A mutational analysis. Oncotarget 2011, 2, 467–476. [Google Scholar] [CrossRef] [PubMed]
- Kunkel, M.T.; Ni, Q.; Tsien, R.Y.; Zhang, J.; Newton, A.C. Spatio-temporal dynamics of protein kinase B/Akt signaling revealed by a genetically encoded fluorescent reporter. J. Biol. Chem. 2005, 280, 5581–5587. [Google Scholar] [CrossRef] [PubMed]
- Parikh, C.; Janakiraman, V.; Wu, W.I.; Foo, C.K.; Kljavin, N.M.; Chaudhuri, S.; Stawiski, E.; Lee, B.; Lin, J.; Li, H.; et al. Disruption of PH-kinase domain interactions leads to oncogenic activation of AKT in human cancers. Proc. Natl. Acad. Sci. USA 2012, 109, 19368–19373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- George, S.; Wang, S.M.; Bi, Y.; Treidlinger, M.; Barber, K.R.; Shaw, G.S.; O’Donoghue, P. Ubiquitin phosphorylated at Ser57 hyper-activates parkin. Biochim. Biophys. Acta 2017, 1861, 3038–3046. [Google Scholar] [CrossRef] [PubMed]
- George, S.; Aguirre, J.D.; Spratt, D.E.; Bi, Y.; Jeffery, M.; Shaw, G.S.; O’Donoghue, P. Generation of phospho-ubiquitin variants by orthogonal translation reveals codon skipping. FEBS Lett. 2016, 590, 1530–1542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aerni, H.R.; Shifman, M.A.; Rogulina, S.; O’Donoghue, P.; Rinehart, J. Revealing the amino acid composition of proteins within an expanded genetic code. Nucleic Acids Res. 2015, 43, e8. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Oh, S.; Yang, A.; Kim, J.; Soll, D.; Lee, D.; Park, H.S. A facile strategy for selective incorporation of phosphoserine into histones. Angew. Chem. Int. Ed. Engl. 2013, 52, 5771–5775. [Google Scholar] [CrossRef] [PubMed]
- MacLean, B.; Tomazela, D.M.; Shulman, N.; Chambers, M.; Finney, G.L.; Frewen, B.; Kern, R.; Tabb, D.L.; Liebler, D.C.; MacCoss, M.J. Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 2010, 26, 966–968. [Google Scholar] [CrossRef] [PubMed]
- Turowec, J.P.; Duncan, J.S.; French, A.C.; Gyenis, L.; St Denis, N.A.; Vilk, G.; Litchfield, D.W. Protein kinase CK2 is a constitutively active enzyme that promotes cell survival: Strategies to identify CK2 substrates and manipulate its activity in mammalian cells. Method Enzymol. 2010, 484, 471–493. [Google Scholar]
- Lindsley, C.W.; Zhao, Z.; Leister, W.H.; Robinson, R.G.; Barnett, S.F.; Defeo-Jones, D.; Jones, R.E.; Hartman, G.D.; Huff, J.R.; Huber, H.E.; et al. Allosteric Akt (PKB) inhibitors: Discovery and SAR of isozyme selective inhibitors. Bioorg. Med. Chem. Lett. 2005, 15, 761–764. [Google Scholar] [CrossRef] [PubMed]
- Alessi, D.R.; James, S.R.; Downes, C.P.; Holmes, A.B.; Gaffney, P.R.; Reese, C.B.; Cohen, P. Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Bα. Curr. Biol. 1997, 7, 261–269. [Google Scholar] [CrossRef]
- Klein, S.; Geiger, T.; Linchevski, I.; Lebendiker, M.; Itkin, A.; Assayag, K.; Levitzki, A. Expression and purification of active PKB kinase from Escherichia coli. Protein Expr. Purif. 2005, 41, 162–169. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, S.; Yamane, H.; Wahl, R.; Ali, A.; Lofgren, J.A.; Kendall, R.L. Kinetic mechanism of AKT/PKB enzyme family. J. Biol. Chem. 2006, 281, 13949–13956. [Google Scholar] [CrossRef] [PubMed]
- Fabbro, D.; Batt, D.; Rose, P.; Schacher, B.; Roberts, T.M.; Ferrari, S. Homogeneous purification of human recombinant GST-Akt/PKB from Sf9 cells. Protein Expr. Purif. 1999, 17, 83–88. [Google Scholar] [CrossRef] [PubMed]
- Alessi, D.R.; Caudwell, F.B.; Andjelkovic, M.; Hemmings, B.A.; Cohen, P. Molecular basis for the substrate specificity of protein kinase B; comparison with MAPKAP kinase-1 and p70 S6 kinase. FEBS Lett. 1996, 399, 333–338. [Google Scholar] [CrossRef] [Green Version]
- Reuveni, H.; Livnah, N.; Geiger, T.; Klein, S.; Ohne, O.; Cohen, I.; Benhar, M.; Gellerman, G.; Levitzki, A. Toward a PKB inhibitor: Modification of a selective PKA inhibitor by rational design. Biochemistry 2002, 41, 10304–10314. [Google Scholar] [CrossRef] [PubMed]
- Yap, T.A.; Walton, M.I.; Grimshaw, K.M.; Te Poele, R.H.; Eve, P.D.; Valenti, M.R.; de Haven Brandon, A.K.; Martins, V.; Zetterlund, A.; Heaton, S.P.; et al. AT13148 is a novel, oral multi-AGC kinase inhibitor with potent pharmacodynamic and antitumor activity. Clin. Cancer Res. 2012, 18, 3912–3923. [Google Scholar] [CrossRef] [PubMed]
- Green, C.J.; Goransson, O.; Kular, G.S.; Leslie, N.R.; Gray, A.; Alessi, D.R.; Sakamoto, K.; Hundal, H.S. Use of Akt inhibitor and a drug-resistant mutant validates a critical role for protein kinase B/Akt in the insulin-dependent regulation of glucose and system A amino acid uptake. J. Biol. Chem. 2008, 283, 27653–27667. [Google Scholar] [CrossRef] [PubMed]
- Barnett, S.F.; Defeo-Jones, D.; Fu, S.; Hancock, P.J.; Haskell, K.M.; Jones, R.E.; Kahana, J.A.; Kral, A.M.; Leander, K.; Lee, L.L.; et al. Identification and characterization of pleckstrin-homology-domain-dependent and isoenzyme-specific Akt inhibitors. Biochem. J. 2005, 385, 399–408. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Cron, P.; Good, V.M.; Thompson, V.; Hemmings, B.A.; Barford, D. Crystal structure of an activated Akt/protein kinase B ternary complex with GSK3-peptide and AMP-PNP. Nat. Struct. Biol. 2002, 9, 940–944. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.I.; Voegtli, W.C.; Sturgis, H.L.; Dizon, F.P.; Vigers, G.P.; Brandhuber, B.J. Crystal structure of human AKT1 with an allosteric inhibitor reveals a new mode of kinase inhibition. PLoS ONE 2010, 5, e12913. [Google Scholar] [CrossRef] [PubMed]
- Massihnia, D.; Avan, A.; Funel, N.; Maftouh, M.; van Krieken, A.; Granchi, C.; Raktoe, R.; Boggi, U.; Aicher, B.; Minutolo, F.; et al. Phospho-Akt overexpression is prognostic and can be used to tailor the synergistic interaction of Akt inhibitors with gemcitabine in pancreatic cancer. J. Hematol. Oncol. 2017, 10, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Slipicevic, A.; Holm, R.; Nguyen, M.T.; Bohler, P.J.; Davidson, B.; Florenes, V.A. Expression of activated Akt and PTEN in malignant melanomas: Relationship with clinical outcome. Am. J. Clin. Pathol. 2005, 124, 528–536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; Chen, X.; Hay, N. Akt as a target for cancer therapy: More is not always better (lessons from studies in mice). Br. J. Cancer 2017, 117, 159–163. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Wan, M.; Leavens, K.F.; Chu, Q.; Monks, B.R.; Fernandez, S.; Ahima, R.S.; Ueki, K.; Kahn, C.R.; Birnbaum, M.J. Insulin regulates liver metabolism in vivo in the absence of hepatic Akt and Foxo1. Nat. Med. 2012, 18, 388–395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crouthamel, M.C.; Kahana, J.A.; Korenchuk, S.; Zhang, S.Y.; Sundaresan, G.; Eberwein, D.J.; Brown, K.K.; Kumar, R. Mechanism and management of AKT inhibitor-induced hyperglycemia. Clin. Cancer Res. 2009, 15, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.; Mu, J.; Kim, J.K.; Thorvaldsen, J.L.; Chu, Q.; Crenshaw, E.B.; Kaestner, K.H.; Bartolomei, M.S.; Shulman, G.I.; Birnbaum, M.J. Insulin resistance and a diabetes mellitus-like syndrome in mice lacking the protein kinase Akt2 (PKBβ). Science 2001, 292, 1728–1731. [Google Scholar] [CrossRef] [PubMed]
- Calleja, V.; Alcor, D.; Laguerre, M.; Park, J.; Vojnovic, B.; Hemmings, B.A.; Downward, J.; Parker, P.J.; Larijani, B. Intramolecular and intermolecular interactions of protein kinase B define its activation in vivo. PLoS Biol. 2007, 5, e95. [Google Scholar] [CrossRef] [PubMed]
- Lucic, I.; Rathinaswamy, M.K.; Truebestein, L.; Hamelin, D.J.; Burke, J.E.; Leonard, T.A. Conformational sampling of membranes by Akt controls its activation and inactivation. Proc. Natl. Acad. Sci. USA 2018, 115, E3940–E3949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agarwal, A.K. How to explain the AKT phosphorylation of downstream targets in the wake of recent findings. Proc. Natl. Acad. Sci. USA 2018, 115, E6099–E6100. [Google Scholar] [CrossRef] [PubMed]
- Ebner, M.; Lucic, I.; Leonard, T.A.; Yudushkin, I. PI(3,4,5)P3 engagement restricts akt activity to cellular membranes. Mol. Cell 2017, 65, 416–431. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, S.J.; Azimifar, S.B.; Mann, M. High-throughput phosphoproteomics reveals in vivo insulin signaling dynamics. Nat. Biotechnol. 2015, 33, 990–995. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Hu, C.; Li, J.; Xue, P.; He, X.; Ge, C.; Qin, W.; Yao, G.; Gu, J. Real-time imaging nuclear translocation of Akt1 in HCC cells. Biochem. Biophys. Res. Commun. 2007, 356, 1038–1043. [Google Scholar] [CrossRef] [PubMed]
- Hixon, M.L.; Boekelheide, K. Expression and localization of total Akt1 and phosphorylated Akt1 in the rat seminiferous epithelium. J. Androl. 2003, 24, 891–898. [Google Scholar] [CrossRef] [PubMed]
- Freudlsperger, C.; Horn, D.; Weissfuss, S.; Weichert, W.; Weber, K.J.; Saure, D.; Sharma, S.; Dyckhoff, G.; Grabe, N.; Plinkert, P.; et al. Phosphorylation of AKT(Ser473) serves as an independent prognostic marker for radiosensitivity in advanced head and neck squamous cell carcinoma. Int. J. Cancer 2015, 136, 2775–2785. [Google Scholar] [CrossRef] [PubMed]
- Okuzumi, T.; Fiedler, D.; Zhang, C.; Gray, D.C.; Aizenstein, B.; Hoffman, R.; Shokat, K.M. Inhibitor hijacking of Akt activation. Nat. Chem. Biol. 2009, 5, 484–493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wisinski, K.B.; Tevaarwerk, A.J.; Burkard, M.E.; Rampurwala, M.; Eickhoff, J.; Bell, M.C.; Kolesar, J.M.; Flynn, C.; Liu, G. Phase I study of an AKT inhibitor (MK-2206) combined with lapatinib in adult solid tumors followed by dose expansion in advanced HER2+ breast cancer. Clin. Cancer Res. 2016, 22, 2659–2667. [Google Scholar] [CrossRef] [PubMed]
- Winder, A.; Unno, K.; Yu, Y.; Lurain, J.; Kim, J.J. The allosteric AKT inhibitor, MK2206, decreases tumor growth and invasion in patient derived xenografts of endometrial cancer. Cancer Biol. Ther. 2017, 18, 958–964. [Google Scholar] [CrossRef] [PubMed]
- Chandarlapaty, S.; Sawai, A.; Scaltriti, M.; Rodrik-Outmezguine, V.; Grbovic-Huezo, O.; Serra, V.; Majumder, P.K.; Baselga, J.; Rosen, N. AKT inhibition relieves feedback suppression of receptor tyrosine kinase expression and activity. Cancer Cell 2011, 19, 58–71. [Google Scholar] [CrossRef] [PubMed]
- Lin, A.; Piao, H.L.; Zhuang, L.; Sarbassov dos, D.; Ma, L.; Gan, B. FoxO transcription factors promote AKT Ser473 phosphorylation and renal tumor growth in response to pharmacologic inhibition of the PI3K-AKT pathway. Cancer Res. 2014, 74, 1682–1693. [Google Scholar] [CrossRef] [PubMed]







| Protein Yields (μg/L Escherichia coli Culture) | ||
|---|---|---|
| Akt1 Variant | Full Length | ΔPH Akt1 |
| Akt1 (unphosphorylated) | 46 | 330 |
| pAkt1S473 | 100 | 150 |
| pAkt1T308 | 37 | 235 |
| ppAkt1T308,S473 | 45 | 224 |
| Akt1 Variant | Akt1 Amount (pmol) | Initial Velocity vo (fmol/min) | Apparent Catalytic Rate kapp (fmol/min/pmol Akt1) | Activation (Fold Increase) | Reference |
|---|---|---|---|---|---|
| Akt1 (unphosphorylated) | 18 | 0.6 ± 0.2 | 0.03 ± 0.01 | 1.0 ± 0.3 | [12] |
| pAkt1S473 | 18 | 46 ± 5 | 2.6 ± 0.3 | 85 ± 9 | [12] |
| pAkt1T308 | 1.8 | 22 ± 4 | 12 ± 2 | 400 ± 70 | [12] |
| ppAkt1T308,S473 | 1.8 | 79 ± 11 | 44 ± 6 | 1500 ± 200 | [12] |
| ΔPHAkt1 (unphosphorylated) | 18 | 1.4 ± 0.1 | 0.079 ± 0.006 | 2.7 ± 0.2 | this study |
| ΔPH pAkt1S473 | 18 | 210 ± 20 | 12 ± 1 | 390 ± 40 | this study |
| ΔPH pAkt1T308 | 0.18 | 370 ± 100 | 2100 ± 600 | 6900 ± 1800 | this study |
| ΔPH ppAkt1T308,S473 | 0.18 | 2300 ± 600 | (13 ± 3) × 103 | (4 ± 1) × 105 | this study |
| Commercial Akt1 (Abcam) | |||||
| Lot 1 | 18 | 100 ± 20 | 6 ± 1 | 200 ± 30 | this study |
| Lot 2 | 18 | 2200 ± 700 | 120 ± 40 | 4000 ± 1000 | this study |
| Akt1 Variant | Full-Length Akt1 | ΔPH-Akt1 |
|---|---|---|
| Akt1 (unphosphorylated) | 1.0 ± 0.3 | 2.7 ± 0.2 |
| pAkt1S473 | 1.0 ± 0.1 | 4.6 ± 0.4 |
| pAkt1T308 | 1.0 ± 0.2 | 175 ± 50 |
| ppAkt1T308,S473 | 1.0 ± 0.1 | 295 ± 68 |
| Akt1 Variant | Inhibitor Type | IC50 (μM) | Expression System | Reference |
|---|---|---|---|---|
| ppAkt1T308,S473 | Akti-1/2 inhibitor VIII | 1.2 ± 0.3 | E. coli | this study |
| pAkt1T308 | Akti-1/2 inhibitor VIII | 0.3 ± 0.1 | E. coli | this study |
| pAkt1S473 | Akti-1/2 inhibitor VIII | 1.3 ± 0.1 | E. coli | this study |
| active | Akti-1/2 inhibitor VIII | 0.058 | Drosophila S2 cells | [25] |
| active | Akti-1/2 inhibitor VIII | 0.1 | HEK 293 cells | [33] |
| active | Akti-1/2 inhibitor | 2.7 | Drosophila S2 cells | [34] |
| active | Akti-1 inhibitor | 4.6 | Drosophila S2 cells | [34] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Balasuriya, N.; McKenna, M.; Liu, X.; Li, S.S.C.; O’Donoghue, P. Phosphorylation-Dependent Inhibition of Akt1. Genes 2018, 9, 450. https://doi.org/10.3390/genes9090450
Balasuriya N, McKenna M, Liu X, Li SSC, O’Donoghue P. Phosphorylation-Dependent Inhibition of Akt1. Genes. 2018; 9(9):450. https://doi.org/10.3390/genes9090450
Chicago/Turabian StyleBalasuriya, Nileeka, McShane McKenna, Xuguang Liu, Shawn S. C. Li, and Patrick O’Donoghue. 2018. "Phosphorylation-Dependent Inhibition of Akt1" Genes 9, no. 9: 450. https://doi.org/10.3390/genes9090450