Bacterial Communities and Antibiotic Resistance Communities in a Full-Scale Hospital Wastewater Treatment Plant by High-Throughput Pyrosequencing
Abstract
:1. Introduction
2. Materials and Methods
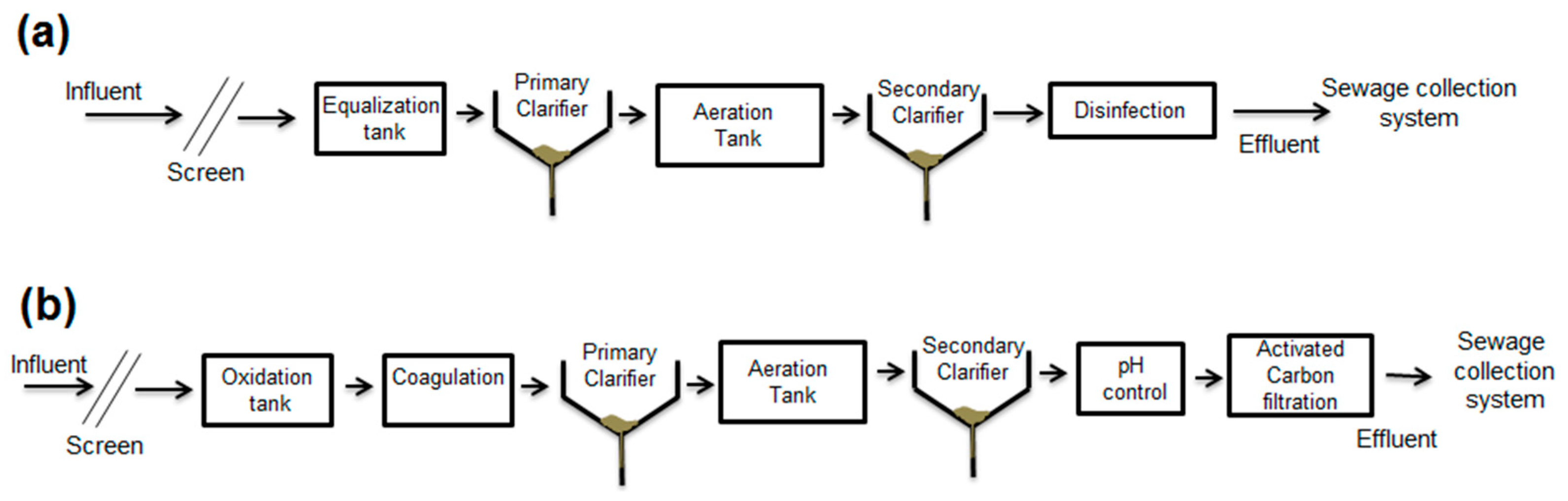
2.1. Description of Hospital Wastewater Treatment Plant
2.2. Water Quality Analysis
2.3. Microbial Analysis
2.3.1. DNA Extraction
2.3.2. 454 High-Throughput Pyrosequencing
2.3.3. Biodiversity Analysis and Antibiotic Resistance Gene Database
3. Results and Discussion
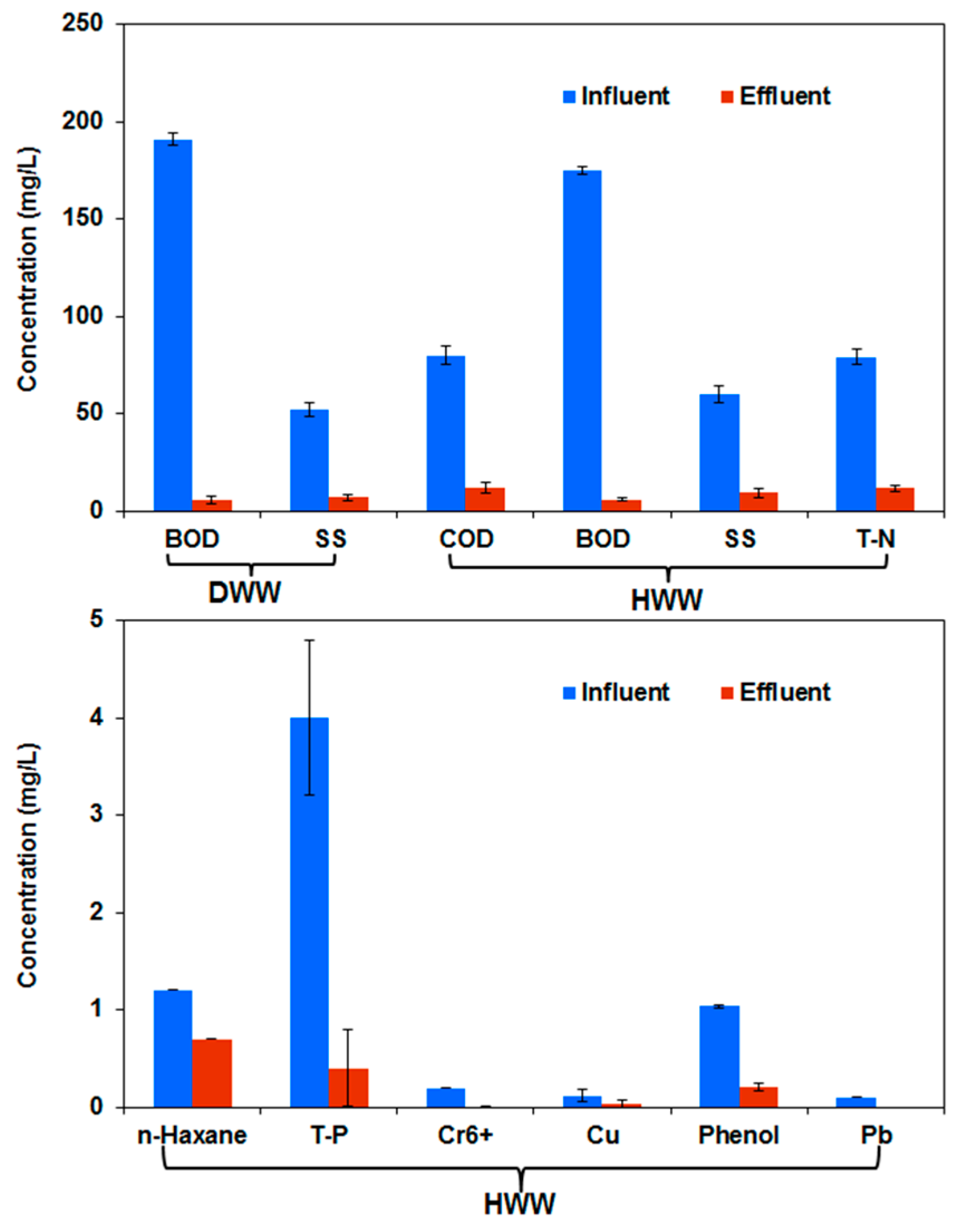
3.1. Water Quality in a Full-Scale Hospital Wastewater Treatment
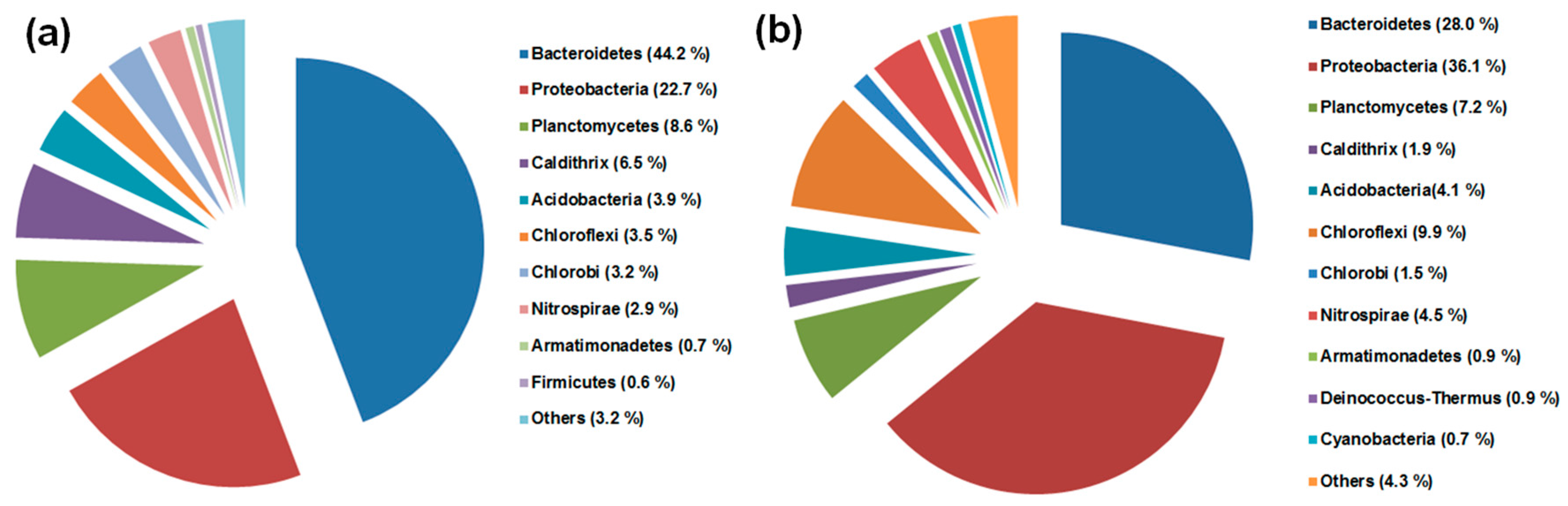
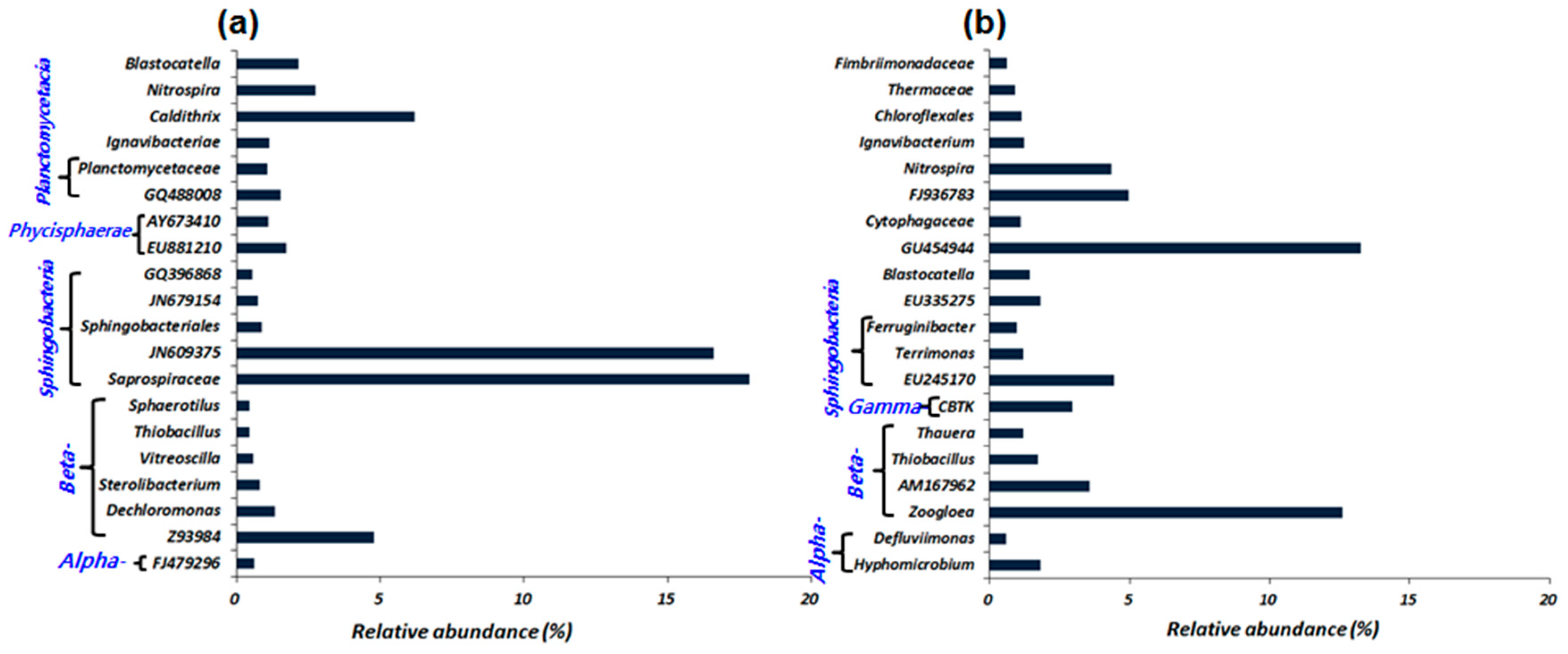
3.2. Microbial Diversity and Comparison in the DWW and the HWW
3.3. Correlation of Wastewater Type and Antibiotic Resistance Bacteria
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Abuzaid, A.; Hamouda, A.; Amyes, S.G.B. Klebsiella pneumoniae susceptibility to biocides and its association with cepA, qacΔE and qacE efflux pump genes and antibiotic resistance. J. Hosp. Infect. 2012, 81, 87–91. [Google Scholar] [CrossRef] [PubMed]
- Brooks, J.P.; Adeli, A.; McLaughlin, M.R. Microbial ecology, bacterial pathogens, and antibiotic resistant genes in swine manure wastewater as influenced by three swine management systems. Water Res. 2014, 57, 96–103. [Google Scholar] [CrossRef] [PubMed]
- Laxminarayan, R.; Duse, A.; Wattal, C.; Zaidi, A.K.M.; Wertheim, H.F.L.; Sumpradit, N.; Vlieghe, E.; Hara, G.L.; Gould, I.M.; Goossens, H.; et al. Antibiotic resistance—The need for global solutions. Lancet Infect. Dis. 2013, 13, 1057–1098. [Google Scholar] [CrossRef]
- Zhang, X.X.; Zhang, T.; Fang, H.H.P. Antibiotic resistance genes in water environment. Appl. Microbiol. Biotechnol. 2009, 82, 397–414. [Google Scholar] [CrossRef] [PubMed]
- Pruden, A.; Pei, R.; Storteboom, H.; Carlson, K.H. Antibiotic resistance genes as emerging contaminants: Studies in Northern Colorado. Environ. Sci. Technol. 2006, 40, 7445–7450. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Li, B.; Ju, F.; Zhang, T. Exploring variation of antibiotic resistance genes in activated sludge over a four-year period through a metagenomic approach. Environ. Sci. Technol. 2013, 47, 10197–10205. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, L.; Manaia, C.; Merlin, C.; Schwartz, T.; Dagot, C.; Ploy, M.C.; Michael, I.; Fatta-Kassinos, D. Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: A review. Sci. Total Environ. 2013, 447, 345–360. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Zhang, T. Mass flows and removal of antibiotics in two municipal wastewater treatment plants. Chemosphere 2011, 83, 1284–1289. [Google Scholar] [CrossRef] [PubMed]
- Philippe, H.; Douady, C.J. Horizontal gene transfer and phylogenetics. Curr. Opin. Microbiol. 2003, 6, 498–505. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Zhang, X.X.; Ye, L. Plasmid metagenome reveals high levels of antibiotic resistance genes and mobile genetic elements in activated sludge. PLoS ONE 2011, 6, e26041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heuer, H.; Schmitt, H.; Smalla, K. Antibiotic resistance gene spread due to manure application on agricultural fields. Curr. Opin. Microbiol. 2011, 14, 236–243. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Ding, R.; Zhang, Y.; Gao, Y.; Tian, Z.; Zhang, T.; Yang, M. Abundance and distribution of Macrolide-Lincosamide-Streptogramin resistance genes in an anaerobic-aerobic system treating spiramycin production wastewater. Water Res. 2014, 63, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Shi, P.; Jia, S.; Zhang, X.X.; Zhang, T.; Cheng, S.; Li, A. Metagenomic insights into chlorination effects on microbial antibiotic resistance in drinking water. Water Res. 2013, 47, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Varela, A.R.; Andre, S.; Nunes, O.C.; Manaia, C.M. Insights into the relationship between antimicrobial residues and bacterial populations in a hospital-urban wastewater treatment plant system. Water Res. 2014, 54, 327–336. [Google Scholar] [CrossRef] [PubMed]
- Michael, I.; Rizzo, L.; McArdell, C.S.; Manaia, C.M.; Merlin, C.; Schwartz, T.; Dagot, C.; Fatta-Kassinos, D. Urban wastewater treatment plants as hotspots for the release of antibiotics in the environment: A review. Water Res. 2013, 47, 957–995. [Google Scholar] [CrossRef] [PubMed]
- Xia, S.Q.; Dual, L.; Song, Y.; Li, J.; Piceno, Y.M.; Andersen, G.L.; Alvarez-Cohen, L.; Moreno-Andrade, I.; Huang, C.; Hermanowicz, S.W. Bacterial community structure in geographically distributed biological wastewater treatment reactors. Environ. Sci. Technol. 2010, 44, 7391–7396. [Google Scholar] [CrossRef] [PubMed]
- Schmieder, R.; Edwards, R. Insights into antibiotic resistance through metagenomic approaches. Future Microbiol. 2012, 7, 73–89. [Google Scholar] [CrossRef] [PubMed]
- Sogin, M.L.; Morrison, H.G.; Huber, J.A.; Weich, D.M.; Huse, S.M.; Neal, P.R.; Arrieta, J.M.; Herndl, G.J. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc. Natl. Acad. Sci. USA 2006, 103, 12115–12120. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Liu, Y. Biodegradation of oil sands process affected water in sequencing batch reactors and microbial community analysis by high-throughput pyrosequencing. Int. Biodeter. Biodegrad. 2014, 92, 79–85. [Google Scholar] [CrossRef]
- Luo, J.; Yan, H.L.L.; Ma, J.; Yang, Y.; Li, G. Microbial community structures in a closed raw distribution system biofilm as revealed by 454-pyrosequencing analysis and the effect of microbial biofilm communities on raw water quality. Bioresour. Technol. 2013, 148, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.; Zhang, T. Bacterial communities in different sections of a municipal wastewater treatment plant revealed by 16S rDNA 454 pyrosequencing. Appl. Microbiol. Biotechnol. 2013, 97, 2681–2690. [Google Scholar] [CrossRef] [PubMed]
- American Public Health Association (APHA). Standard Methods for the Examination of Water and Wastewater, 21st ed.; APHA: Washington, DC, USA, 2005. [Google Scholar]
- Resource Central. Available online: http://oklbb.ezbiocloud.net/content/1001 (accessed on 28 August 2015).
- Jeon, Y.S.; Chun, J.; Kim, B.S. Identification of household bacterial community and analysis of species shared with human microbiome. Curr. Microbiol. 2013, 67, 557–563. [Google Scholar] [CrossRef] [PubMed]
- Eddy, S.R. Accelerated profile HMM searches. PLoS Comput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef] [PubMed]
- Kim, O.S.; Cho, Y.J.; Lee, K.; Yoon, S.H.; Kim, M.; Na, H.; Park, S.C.; Jeon, Y.S.; Lee, J.H.; Yi, H. Introducing EzTaxon-e: A prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Microbiol. 2012, 62, 716–721. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Pop, M. ARDB-Antibiotic resistance genes database. Nucleic Acids Res. 2009, 37, 443–447. [Google Scholar] [CrossRef] [PubMed]
- Monser, L.; Adhoum, N. Modified activated carbon for the removal of copper, zinc, chromium and cyanide from wastewater. Sep. Purif. Technol. 2002, 26, 137–146. [Google Scholar] [CrossRef]
- Lofmark, S.; Jernberg, C.; Jansson, J.K.; Edlund, C. Clindamycin-induced enrichment and long-term persistence of resistant Bacteroides spp. and resistance genes. J. Antimicrob. Chemother. 2006, 58, 1160–1167. [Google Scholar] [CrossRef] [PubMed]
- Cayrou, C.; Raoult, D.; Drancourt, M. Broad-spectrum antibiotic resistance of Planctomycetes organisms determined by Etest. J. Antimicrob. Chemother. 2010, 65, 2119–2122. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Qi, R.; Yang, M.; Zhang, Y.; Yu, T. Bacterial community characteristics under long-term antibiotic selection pressures. Water Res. 2011, 45, 6063–6073. [Google Scholar] [CrossRef] [PubMed]
- Kraigher, B.; Kosjek, T.; Heath, E.; Kompare, B.; Mandic-Mulec, I. Influence of pharmaceutical residues on the structure of activated sludge bacterial communities in wastewater treatment bioreactors. Water Res. 2008, 42, 4578–4588. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Sadowsky, M.J.; Roberts, M.C.; Gralnick, J.A.; LaPara, T.M. Sphingobacterium sp. strain PM2-P1-29 harbours a functional tet (X) gene encoding for the degradation of tetracycline. J. Appl. Microbiol. 2009, 106, 1336–1342. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Zhang, Y.; Yang, M.; Tian, Z.; Ren, L.; Zhang, S. Abundance and distribution of tetracycline resistance genes and mobile elements in an oxytetracycline production wastewater treatment system. Environ. Sci. Technol. 2012, 46, 7551–7557. [Google Scholar] [CrossRef] [PubMed]
- Egli, K.; Bosshard, F.; Werlen, C.; Lais, P.; Siegrist, H.; Zehnder, A.J.B.; van der Meer, J.R. Microbial composition and structure of a rotating biological contactor biofilm treating ammonium-rich wastewater without organic carbon. Microb. Ecol. 2003, 45, 419–432. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Zhang, Y.; Ding, R.; Gao, Y.; Yang, M. Response of activated sludge to the treatment of oxytetracycline production waste stream. Appl. Microbiol. Biotechnol. 2013, 97, 8805–8812. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Hu, M.; Xia, Y.; Wen, X.; Ding, K. Pyrosequencing analysis of bacterial diversity in 14 wastewater treatment systems in China. Appl. Environ. Microbiol. 2012, 78, 7042–7047. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, P.H.; Saunders, A.M.; Hansen, A.A.; Larsen, P.; Nielsen, J.L. Microbial communities involved in enhanced biological phosphorus removal from wastewater—A model system in environmental biotechnology. Curr. Opin. Biotechnol. 2012, 23, 452–459. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Holzem, R.M.; Gunsch, C.K. Effects of pharmaceutically active compounds on a mixed microbial community originating from a municipal wastewater treatment plant. Environ. Sci. Technol. 2008, 42, 1091–1095. [Google Scholar] [CrossRef] [PubMed]
- Tomasz, A. Antibiotic resistance in Streptococcus pneumonia. Clin. Infect. Dis. 1997, 24, S85–S88. [Google Scholar] [CrossRef] [PubMed]
- Gu, C.; Karthikeyan, K.G. Interaction of tetracycline with aluminum and iron hydrous oxides. Environ. Sci. Technol. 2005, 39, 2660–2667. [Google Scholar] [CrossRef] [PubMed]
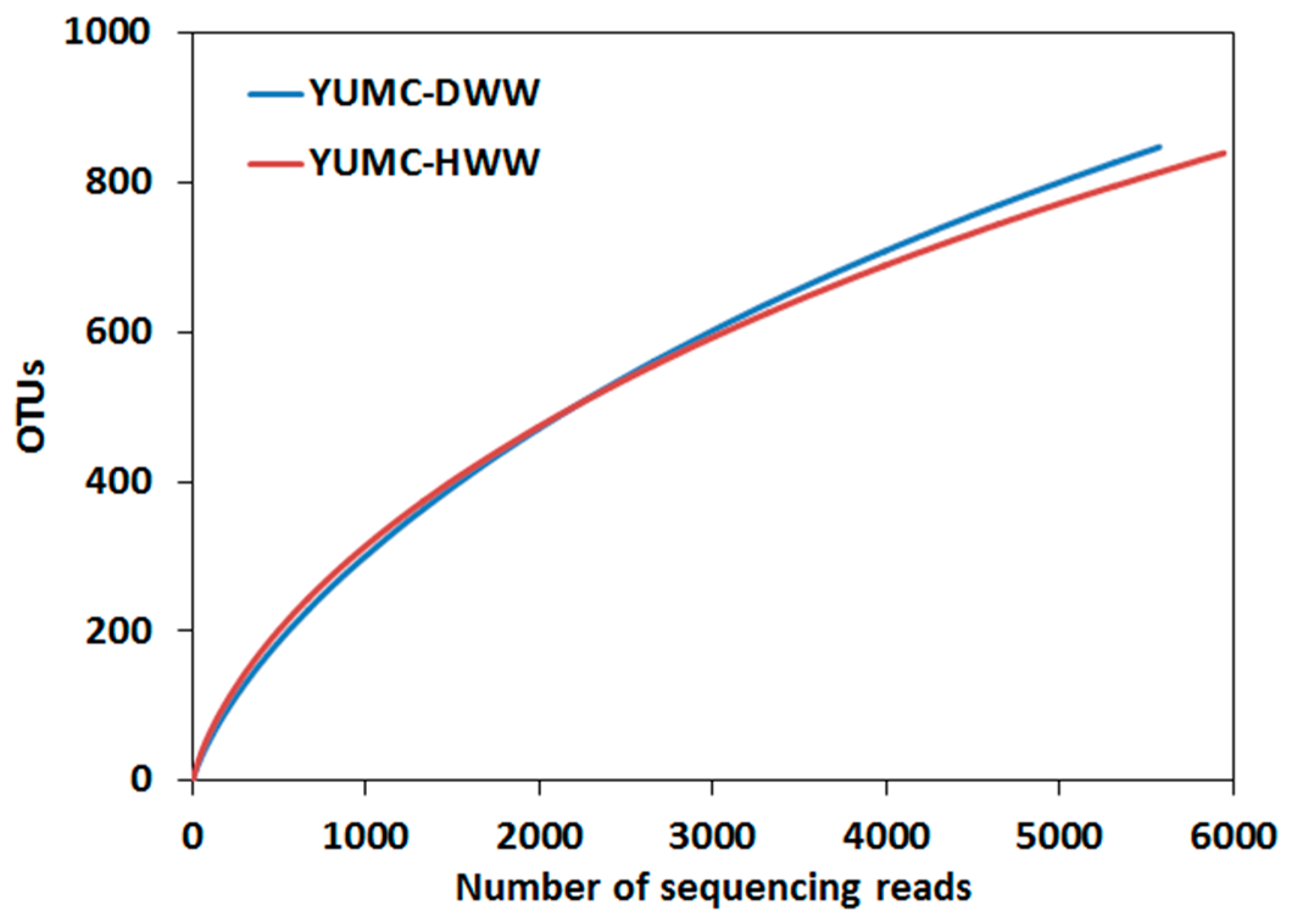

| Samples | Total Reads | Analyzed Reads | Read Length (bp) | Observed OTUs | Chao1 Estimation | Shannon Index | Goods Coverage | |
|---|---|---|---|---|---|---|---|---|
| Mean Length | Maximum Length | |||||||
| DWW | 9128 | 5572 | 478 | 642 | 848 | 1493 | 4.71 | 0.921 |
| HWW | 10,990 | 5944 | 472 | 673 | 840 | 1414 | 5.24 | 0.932 |
| Samples | Genus | Relative Abundance (%) | Antibiotic Resistance Type |
|---|---|---|---|
| DWW | Aeromonas | 0.084 | Tetracycline, chloramphenicol, penicillin, cephalosporin, streptomycin, dibekacin |
| Azoarcus | 0.017 | Bacitracin, chloramphenicol | |
| Enterobacter | 0.017 | Dibekacin, fluoroquinolone, sulfonamide, tetracycline, trimethoprim | |
| Lautropia | 0.168 | Bacitracin, chloramphenicol | |
| Paracaedibacter | 0.034 | Cephalosporin, tetracycline | |
| HWW | Acetobacteraceae | 0.018 | Tetracycline |
| Acidovorax | 0.269 | Spectinomycin, streptomycin, bacitracin | |
| Bacillus | 0.054 | Kanamycin, tobramycin | |
| Blautia | 0.018 | Tetracycline | |
| Clostridiales | 0.018 | Tetracycline | |
| Coprococcus | 0.018 | Bacitracin, tetracycline | |
| Enterobacter | 0.054 | Dibekacin, fluoroquinolone, sulfonamide, tetracycline, trimethoprim | |
| Escherichia | 0.072 | Dibekacin, amikacim, cephalosporin, cloxacillin, tetracycline | |
| Lactobacillus | 0.036 | Bacitracin, chloramphenicol | |
| Parabacteroides | 0.036 | Cephalosporin, tetracycline | |
| Polynucleobacter | 0.018 | Bacitracin | |
| Proteus | 0.018 | Astromicin, gentamicin, sisomicin | |
| Ralstonia | 0.126 | Bacitracin | |
| Streptococcus | 0.036 | Amikacin, butirosin, gentamincin, isepamicin, kanamycin, lividomycin, neomycin, paromomycin, ribostamycin, streptomycin | |
| Thauera | 0.413 | Bacitracin |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahn, Y.; Choi, J. Bacterial Communities and Antibiotic Resistance Communities in a Full-Scale Hospital Wastewater Treatment Plant by High-Throughput Pyrosequencing. Water 2016, 8, 580. https://doi.org/10.3390/w8120580
Ahn Y, Choi J. Bacterial Communities and Antibiotic Resistance Communities in a Full-Scale Hospital Wastewater Treatment Plant by High-Throughput Pyrosequencing. Water. 2016; 8(12):580. https://doi.org/10.3390/w8120580
Chicago/Turabian StyleAhn, Youngho, and Jeongdong Choi. 2016. "Bacterial Communities and Antibiotic Resistance Communities in a Full-Scale Hospital Wastewater Treatment Plant by High-Throughput Pyrosequencing" Water 8, no. 12: 580. https://doi.org/10.3390/w8120580





