Inactivation Effect of Antibiotic-Resistant Gene Using Chlorine Disinfection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Inactivation of Suspensions of Vancomycin-Resistant Enterococci (VRE)
2.2. Measurement of Enterococci Concentration
2.3. DNA Extraction and Resistance Gene Detection
2.4. Inactivation of VRE in Secondary Effluent Samples
3. Results and Discussion
3.1. Inactivation Ratio of VRE and Detection of vanA in Suspensions of VRE (Suspended in PBS Solution)
3.2. Inactivation Ratio of VRE and Detection of vanA in Secondary Effluent Water Sample
4. Conclusions
Acknowledgment
Author Contributions
Conflicts of Interest
References
- Sharma, V.K.; Johnson, N.; Cizmas, L.; McDonald, T.J.; Kim, H. A review of the influence of treatment strategies on antibiotic resistant bacteria and antibiotic resistance genes. Chemosphere 2016, 150, 702–714. [Google Scholar] [CrossRef] [PubMed]
- Ham, Y.S.; Kobori, H.; Kang, J.H.; Matsuzaki, T.; Iino, M.; Nomura, H. Distribution of antibiotic resistance in urban watershed in Japan. Environ. Pollut. 2012, 162, 98–103. [Google Scholar] [CrossRef] [PubMed]
- Sidrach-Cardona, R.; Bècares, E. Fecal indicator bacteria resistance to antibiotics in experimental constructed wetlands. Ecol. Eng. 2013, 50, 107–111. [Google Scholar] [CrossRef]
- Suzuki, Y.; Kajii, S.; Nishiyama, M.; Iguchi, A. Susceptibility of Pseudomonas aeruginosa isolates collected from river water in Japan to antipseudomonal agents. Sci. Total Environ. 2013, 450–451, 148–154. [Google Scholar] [CrossRef] [PubMed]
- Urase, T.; Sato, T. Quantitative monitoring of resistance in Escherichia coli to clinically important antimicrobials in urban watershed. J. Water Environ. Technol. 2016, 14, 341–349. [Google Scholar] [CrossRef]
- Ash, R.J.; Mauck, B.; Morgan, M. Antibiotic resistance of gram-negative bacteria in rivers, United States. Emerg. Infect. Dis. 2002, 8, 713–716. [Google Scholar] [CrossRef] [PubMed]
- Rosi-Marshall, E.J.; Kelly, J.J. Antibiotic stewardship should consider environmental fate of antibiotics. Environ. Sci. Technol. 2015, 49, 5257–5258. [Google Scholar] [CrossRef] [PubMed]
- Silbergeld, E.K.; Graham, J.; Price, L.B. Industrial food animal production, antimicrobial resistance, and human health. Annu. Rev. Public Health 2008, 29, 151–169. [Google Scholar] [CrossRef] [PubMed]
- Novo, A.; André, S.; Viana, P.; Nunes, O.C.; Manaia, C.M. Antibiotic resistance, antimicrobial residues and bacterial community composition in urban wastewater. Water Res. 2013, 47, 1875–1887. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, L.; Manaia, C.; Merlin, C.; Schwartz, T.; Dagot, C.; Ploy, M.C.; Michael, I.; Fatta-Kassinos, D. Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: A review. Sci. Total Environ. 2013, 447, 345–360. [Google Scholar] [CrossRef] [PubMed]
- Levantesi, C.; La Mantia, R.; Masciopinto, C.; Böckelmann, U.; Ayuso-Gabella, M.N.; Salgot, M.; Tandoi, V.; Van Houtte, E.; Wintgens, T.; Grohmann, E. Quantification of pathogenic microorganisms and microbial indicators in three wastewater reclamation and managed aquifer recharge facilities in Europe. Sci. Total Environ. 2010, 408, 4923–4930. [Google Scholar] [CrossRef] [PubMed]
- Nagulapally, S.R.; Ahmad, A.; Henry, A.; Marchin, G.L.; Zurek, L.; Bhandari, A. Occurrence of ciprofloxacin-, trimethoprim-sulfamethoxazole-, and vancomycin-resistant bacteria in a municipal wastewater treatment plant. Water Environ. Res. 2009, 81, 82–90. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg Goldstein, R.E.; Micallef, S.A.; Gibbs, S.G.; Davis, J.A.; He, X.; George, A.; Kleinfelter, L.M.; Schreiber, N.A.; Mukherjee, S.; Sapkota, A.; et al. Methicillin-resistant Staphylococcus aureus (MRSA) Detected at Four U.S. wastewater treatment plants. Environ. Health Perspect. 2012, 120, 1551–1558. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg Goldstein, R.E.; Micallef, S.A.; Gibbs, S.G.; George, A.; Claye, E.; Sapkota, A.; Joseph, S.W.; Sapkota, A.R. Detection of vancomycin-resistant enterococci (VRE) at four U.S. wastewater treatment plants that provide effluent for reuse. Sci. Total Environ. 2014, 466–467, 404–411. [Google Scholar] [CrossRef] [PubMed]
- Garcia, S.; Wade, B.; Bauer, C.; Craig, C.; Nakaoka, K.; Lorowitz, W. The effect of wastewater treatment on antibiotic resistance in Escherichia coli and Enterococcus sp. Water Environ. Res. 2007, 79, 2387–2395. [Google Scholar] [CrossRef] [PubMed]
- Carey, S.A.; Goldstein, R.E.R.; Gibbs, S.G.; Claye, E.; He, X.; Sapkota, A.R. Occurrence of vancomycin-resistant and -susceptible Enterococcus spp. in reclaimed water used for spray irrigation. Environ. Res. 2016, 147, 350–355. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, T.; Hashimoto, R.; Mekata, T. Quantification of vancomycin-resistant enterococci and corresponding resistance genes in a sewage treatment plant. J. Environ. Sci. Health A Tox. Hazard. Subst. Environ. Eng. 2015, 50, 989–995. [Google Scholar] [CrossRef] [PubMed]
- World Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. Available online: http://www.who.int/medicines/publications/WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf?ua=1 (accessed on 19 June 2017).
- Gold, H.S.; Moellering, R.C., Jr. Antimicrobial-drug resistance. N. Eng. J. Med. 1996, 335, 1445–1453. [Google Scholar] [CrossRef]
- U.S. Environmental Protection Agency. Wastewater Technology Fact Sheet: Chlorine Disinfection, EPA 832-F-99-062; U.S. EPA: Washington, DC, USA, 1999.
- Yuan, Q.; Guo, M.; Yang, J. Fate of antibiotic resistant bacteria and genes during wastewater chlorination: Implication for antibiotic resistance control. PLoS ONE 2015, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Dutka-Malen, S.; Evers, S.; Courvalin, P. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J. Clin. Microbiol. 1995, 33, 24–27, PMCID:PMC227872. [Google Scholar] [PubMed]
- Tree, J.A.; Adams, M.R.; Lees, D.N. Chlorination of indicator bacteria and viruses in primary sewage effluent. Appl. Environ. Microbiol. 2003, 69, 2038–2043. [Google Scholar] [CrossRef] [PubMed]
- Pang, Y.; Huang, J.; Xi, J.; Hu, H.; Zhu, Y. Effect of ultraviolet irradiation and chlorination on ampicillin-resistant Escherichia coli and its ampicillin resistance gene. Front. Environ. Sci. Eng. 2016, 10, 522–530. [Google Scholar] [CrossRef]
- Al-Jassim, N.; Ansari, M.; Harb, M.; Hong, P.Y. Removal of bacterial contaminants and antibiotic resistance genes by conventional wastewater treatment processes in Saudi Arabia: Is the treated wastewater safe to reuse for agricultural irrigation? Water Res. 2015, 73, 277–290. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Hu, H.; Wu, Y.; Wei, B.; Lu, Y. Effect of chlorination and ultraviolet disinfection on tetA-mediated tetracycline resistance of Escherichia coli. Chemosphere 2013, 90, 2247–2253. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhuang, Y.; Geng, J.; Ren, H.; Zhang, Y.; Ding, L.; Xu, K. Inactivation of antibiotic resistance genes in municipal wastewater effluent by chlorination and sequential UV/chlorination disinfection. Sci. Total Environ. 2015, 512–513, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, J.L.; Shigeno, D.S.; Calomiris, J.J.; Seidler, R.J. Antibiotic-resistant bacteria in drinking water. Appl. Environ. Microbiol. 1981, 42, 277–283, PMCID:PMC244002. [Google Scholar]
- Jia, S.; Shi, P.; Hu, Q.; Li, B.; Zhang, T.; Zhang, X.X. Bacterial community shift drives antibiotic resistance promotion during drinking water chlorination. Environ. Sci. Technol. 2015, 49, 12271–12279. [Google Scholar] [CrossRef] [PubMed]
- Oliver, J.D. The viable but nonculturable state in bacteria. J. Microbiol. 2005, 43, 93–100. [Google Scholar] [PubMed]
- Hashimoto, H.; Murayama, S. Emergence and evolution of microbial drug-resistance. J. Jpn. Soc. Clin. Microbiol. 2013, 23, 1–11. [Google Scholar]
- Lederberg, J. Genetic recombination in bacteria: A discovery account. Annu. Rev. Genet. 1987, 21, 23–46. [Google Scholar] [CrossRef] [PubMed]
- Lederberg, J.; Tatum, E.L. Gene recombination in Escherichia coli. Nature 1946, 158, 558, PMCID:PMC518375. [Google Scholar] [CrossRef]
- Zinder, N.D.; Lederberg, J. Genetic exchange in Salmonella. J. Bacteriol. 1952, 64, 679–699, PMCID:PMC169409. [Google Scholar]
- Dell’anno, A.; Faviano, M.; Mei, M.L.; Danovaro, R. Pelagic-benthic coupling of nucleic acids in an abyssal location of the northeastern Atlantic Ocean. Appl. Environ. Microbiol. 1999, 65, 4451–4457, PMCID:PMC91592. [Google Scholar]
- Lorenz, M.G.; Wackernagel, W. Bacterial gene transfer by natural genetic transformation in the environment. Microbiol. Rev. 1994, 58, 563–602, PMCID:PMC372978. [Google Scholar]
- Hashimoto, R.; Furukawa, T.; Suzuki, Y. Fate of vancomycin-resistant bacteria and corresponding resistance genes in a sewage treatment plant. In Proceedings of the Water and Environment Technology Conference 2016, Tokyo, Japan, 27–28 August 2016. [Google Scholar]
- Öncü, N.B.; Menceloğlu, Y.Z.; Balcıoğlu, I.A. Comparison of the effectiveness of chlorine, ozone, and photocatalytic disinfection in reducing the risk of antibiotic resistance pollution. J. Adv. Oxid. Technol. 2011, 14, 196–203. [Google Scholar] [CrossRef]
- Dunlop, P.S.M.; Ciavola, M.; Rizzo, L.; McDowell, D.A.; Byrne, J.A. Effect of photocatalysis on the transfer of antibiotic resistance genes in urban wastewater. Catal. Today 2015, 240, 55–60. [Google Scholar] [CrossRef]

| Time (min) | Log Reduction (log C/C0) | ||||
|---|---|---|---|---|---|
| 0.1 mg Cl2/L | 0.3 mg Cl2/L | 0.5 mg Cl2/L | 1.0 mg Cl2/L | 3.0 mg Cl2/L | |
| 3 | 0.00 | 0.33 | 1.59 | 5.48 | >7.85 |
| 5 | 0.08 | 0.66 | 1.73 | 6.27 | >7.85 |
| 10 | 0.10 | 0.83 | 1.78 | 7.76 | >7.85 |
| 20 | 0.09 | 0.91 | 2.49 | >7.77 | >7.85 |
| 40 | 0.09 | 1.14 | 7.01 | >7.77 | >7.85 |
| 60 | 0.10 | 1.46 | >7.93 | >7.77 | >7.85 |
| Time (min) | vanA Detection (-) | ||||
|---|---|---|---|---|---|
| 0.1 mg Cl2/L | 0.3 mg Cl2/L | 0.5 mg Cl2/L | 1.0 mg Cl2/L | 3.0 mg Cl2/L | |
| 0 | + | + | + | + | + |
| 3 | + | + | + | + | + |
| 5 | + | + | + | + | + |
| 10 | + | + | + | + | + |
| 20 | + | + | + | + | + |
| 40 | + | + | + | + | + |
| 60 | + | + | + | + | + |
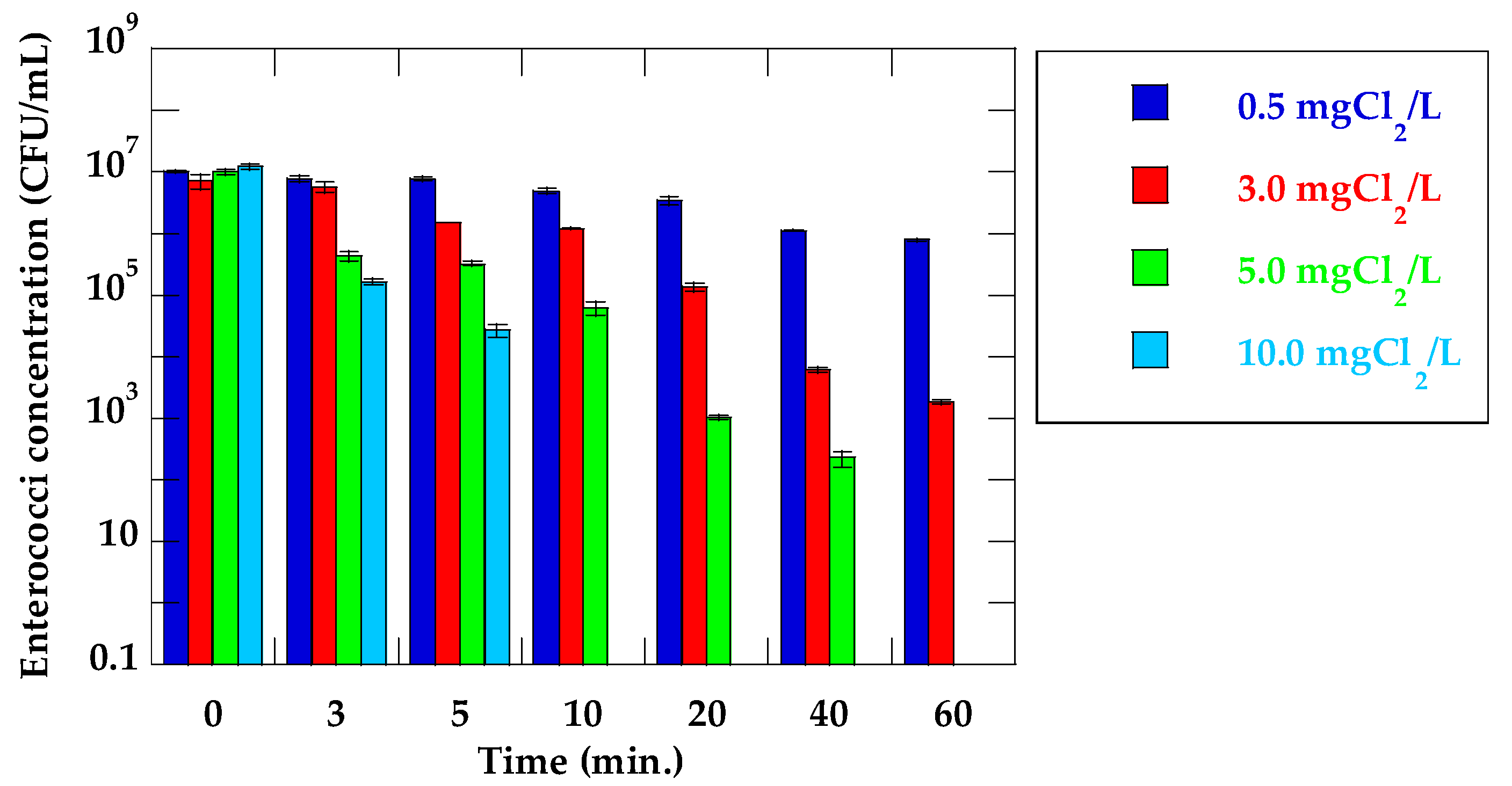
| Time (min) | Log Reduction (log C/C0) | |||
|---|---|---|---|---|
| 0.5 mg Cl2/L | 3.0 mg Cl2/L | 5.0 mg Cl2/L | 10 mg Cl2/L | |
| 3 | 0.12 | 0.33 | 1.37 | 1.86 |
| 5 | 0.11 | 0.90 | 1.50 | 2.65 |
| 10 | 0.32 | 0.77 | 2.21 | >7.09 |
| 20 | 0.47 | 1.73 | 3.99 | >7.09 |
| 40 | 0.96 | 3.07 | 4.65 | >7.09 |
| 60 | 1.12 | 3.59 | >7.01 | >7.09 |
| Time (min) | vanA Detection (-) | |||
|---|---|---|---|---|
| 0.5 mg Cl2/L | 3.0 mg Cl2/L | 5.0 mg Cl2/L | 10 mg Cl2/L | |
| 0 | + | + | + | + |
| 3 | + | + | + | + |
| 5 | + | + | + | + |
| 10 | + | + | + | + |
| 20 | + | + | + | + |
| 40 | + | + | + | + |
| 60 | + | + | + | + |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Furukawa, T.; Jikumaru, A.; Ueno, T.; Sei, K. Inactivation Effect of Antibiotic-Resistant Gene Using Chlorine Disinfection. Water 2017, 9, 547. https://doi.org/10.3390/w9070547
Furukawa T, Jikumaru A, Ueno T, Sei K. Inactivation Effect of Antibiotic-Resistant Gene Using Chlorine Disinfection. Water. 2017; 9(7):547. https://doi.org/10.3390/w9070547
Chicago/Turabian StyleFurukawa, Takashi, Atsushi Jikumaru, Takahisa Ueno, and Kazunari Sei. 2017. "Inactivation Effect of Antibiotic-Resistant Gene Using Chlorine Disinfection" Water 9, no. 7: 547. https://doi.org/10.3390/w9070547




