VNARs: An Ancient and Unique Repertoire of Molecules That Deliver Small, Soluble, Stable and High Affinity Binders of Proteins
Abstract
:1. Introduction: Their Basic Beginnings
1.1. Adapting to Attack
1.2. To be or Not to Be an Antibody

2. Exploiting the Benefits of VNAR
2.1. Biologics Are Leading the Way
2.2. VNAR as a Candidate Therapeutic Domain
2.2.1. Naturally Adapted for Drug Development
2.2.2. Small Size but Large Diversity

| Domain Type and Orientations | Mean KD Albumin (nM) | Mean t1/2 (days) | ||
| Mouse | Monkey | Mouse | Monkey | |
| Camel VHH | ||||
| ABVHH | 73 | N/A | 0.5 | N/A |
| VHH-ABVHH | 470 | N/A | 0.6 | 4.8 |
| VHH-ABVHH-VHH | 180 | N/A | 1.4 | N/A |
| Shark VNAR | ||||
| ABVNAR | 0.7 | 0.2 | N/A | N/A |
| VNAR-ABVNAR | 2.5 | 0.9 | 1.4 | 6.7 |
| VNAR-ABVNAR-VNAR | 4.0 | 1.0 | ||
2.2.3. Molecular Malleability
2.2.4. Remarkable Stability
3. VNARs in Development
3.1. NDure™—The First Clinical Candidate
3.1.1. E06—Fit for Purpose

3.1.2. Becoming Human—the Creation of NDure™
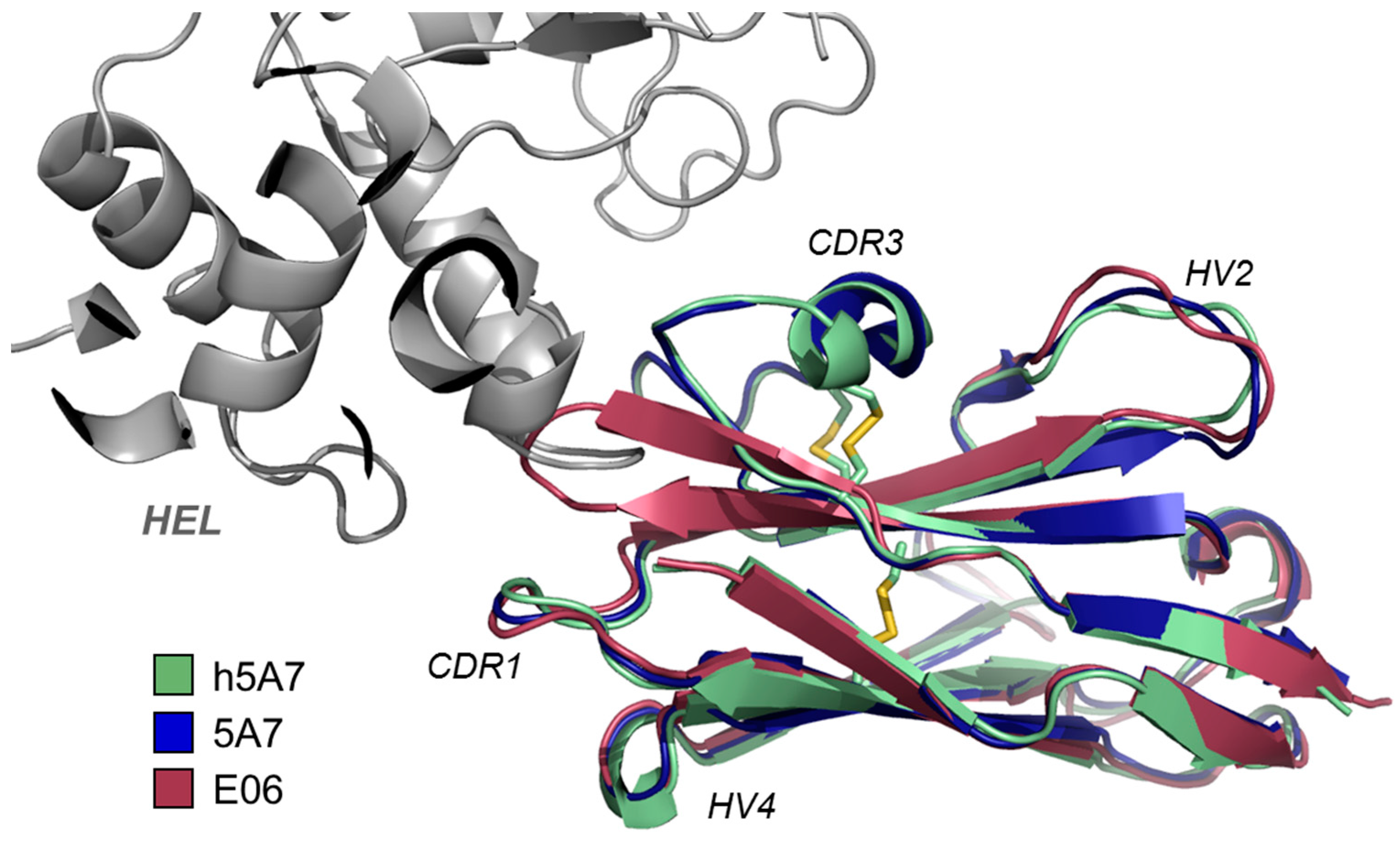
3.2. Other Reported VNAR Domains
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Papermaster, B.W.; Condie, R.M.; Finstad, J.; Good, R.A. Evolution of the Immune Response. I. the Phylogenetic Development of Adaptive Immunologic Responsiveness in Vertebrates. J. Exp. Med. 1964, 119, 105–130. [Google Scholar] [CrossRef] [PubMed]
- Parton, A.; Bayne, C.J.; Barnes, D.W. Analysis and functional annotation of expressed sequence tags from in vitro cell lines of elasmobranchs: Spiny dogfish shark (Squalus acanthias) and little skate (Leucoraja erinacea). Comp. Biochem. Physiol. Part D. Genomics Proteomics 2010, 5, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Marchalonis, J.J.; Schluter, S.F.; Bernstein, R.M.; Shen, S.; Edmundson, A.B. Phylogenetic emergence and molecular evolution of the immunoglobulin family. Adv. Immunol. 1998, 70, 417–506. [Google Scholar] [PubMed]
- Flajnik, M.F.; Rumfelt, L.L. The immune system of cartilaginous fish. Curr. Top. Microbiol. Immunol. 2000, 248, 249–270. [Google Scholar] [PubMed]
- Fange, R.; Pulsford, A. Structural studies on lymphomyeloid tissues of the dogfish, Scyliorhinus canicula L. Cell Tissue Res. 1983, 230, 337–351. [Google Scholar] [CrossRef] [PubMed]
- Rumfelt, L.L.; Avila, D.; Diaz, M.; Bartl, S.; McKinney, E.C.; Flajnik, M.F. A shark antibody heavy chain encoded by a nonsomatically rearranged VDJ is preferentially expressed in early development and is convergent with mammalian IgG. Proc. Natl. Acad. Sci. USA 2001, 98, 1775–1780. [Google Scholar] [CrossRef] [PubMed]
- Miracle, A.L.; Anderson, M.K.; Litman, R.T.; Walsh, C.J.; Luer, C.A.; Rothenberg, E.V.; Litman, G.W. Complex expression patterns of lymphocyte-specific genes during the development of cartilaginous fish implicate unique lymphoid tissues in generating an immune repertoire. Int. Immunol. 2001, 13, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Rumfelt, L.L.; McKinney, E.C.; Taylor, E.; Flajnik, M.F. The development of primary and secondary lymphoid tissues in the nurse shark Ginglymostoma cirratum: B-cell zones precede dendritic cell immigration and T-cell zone formation during ontogeny of the spleen. Scand. J. Immunol. 2002, 56, 130–148. [Google Scholar] [CrossRef] [PubMed]
- Clem, L.W.; De Boutard, F.; Sigel, M.M. Phylogeny of immunoglobulin structure and function - II. Immunoglobulins of the nurse shark. J. Immunol. 1967, 99, 1226–1235. [Google Scholar]
- Clem, L.W.; Small, P.A., Jr. Phylogeny of immunoglobulin structure and function. I. Immunoglobulins of the lemon shark. J. Exp. Med. 1967, 125, 893–920. [Google Scholar] [CrossRef]
- Marchalonis, J.; Edelman, G.M. Phylogenetic origins of antibody structure. I. Multichain structure of immunoglobulins in the smooth dogfish (Mustelus canis). J. Exp. Med. 1965, 122, 601–618. [Google Scholar] [CrossRef] [PubMed]
- Marchalonis, J.; Edelman, G.M. Polypeptide chains of immunoglobulins from the smooth dogfish (Mustelus canis). Science 1966, 154, 1567–1568. [Google Scholar] [CrossRef] [PubMed]
- Morrow, W.; Harris, J.E.; Pulsford, A. Immunological responses of the dogfish (Scyliorhinus canicula L.) to cellular antigens. Acta Zoologica (Stockh.) 1982, 63, 153–159. [Google Scholar] [CrossRef]
- Morrow, W.J.W.; Harris, J.E.; Davies, D.; Pulsford, A.A. Isolation and partial characterization of dogfish (Scyliorhinus canicula L) antibody. J. Mar. Biol. Ass. UK 1983, 63, 409–418. [Google Scholar] [CrossRef]
- Rumfelt, L.L.; Diaz, M.; Lohr, R.L.; Mochon, E.; Flajnik, M.F. Unprecedented multiplicity of Ig transmembrane and secretory mRNA forms in the cartilaginous fish. J. Immunol. 2004, 173, 1129–1139. [Google Scholar] [CrossRef]
- Rumfelt, L.L.; Lohr, R.L.; Dooley, H.; Flajnik, M.F. Diversity and repertoire of IgW and IgM VH families in the newborn nurse shark. BMC Immunol. 2004, 5, 8–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greenberg, A.S.; Avila, D.; Hughes, M.; Hughes, A.; McKinney, E.C.; Flajnik, M.F. A new antigen receptor gene family that undergoes rearrangement and extensive somatic diversification in sharks. Nature 1995, 374, 168–173. [Google Scholar] [CrossRef] [PubMed]
- Dooley, H.; Stanfield, R.L.; Brady, R.A.; Flajnik, M.F. First molecular and biochemical analysis of in vivo affinity maturation in an ectothermic vertebrate. Proc. Natl. Acad. Sci. USA 2006, 103, 1846–1851. [Google Scholar] [CrossRef] [PubMed]
- Feige, M.J.; Grawert, M.A.; Marcinowski, M.; Hennig, J.; Behnke, J.; Auslander, D.; Herold, E.M.; Peschek, J.; Castro, C.D.; Flajnik, M.; et al. The structural analysis of shark IgNAR antibodies reveals evolutionary principles of immunoglobulins. Proc. Natl. Acad. Sci. USA 2014, 111, 8155–8160. [Google Scholar] [CrossRef] [PubMed]
- Dooley, H.; Flajnik, M.F.; Porter, A.J. Selection and characterization of naturally occurring single-domain (IgNAR) antibody fragments from immunized sharks by phage display. Mol. Immunol. 2003, 40, 25–33. [Google Scholar] [CrossRef]
- Dooley, H.; Flajnik, M.F. Shark immunity bites back: affinity maturation and memory response in the nurse shark, Ginglymostoma cirratum. Eur. J. Immunol. 2005, 35, 936–945. [Google Scholar] [CrossRef] [PubMed]
- Diaz, M.; Velez, J.; Singh, M.; Cerny, J.; Flajnik, M.F. Mutational pattern of the nurse shark antigen receptor gene (NAR) is similar to that of mammalian Ig genes and to spontaneous mutations in evolution: The translesion synthesis model of somatic hypermutation. Int. Immunol. 1999, 11, 825–833. [Google Scholar] [CrossRef] [PubMed]
- Nuttall, S.D.; Krishnan, U.V.; Hattarki, M.; De Gori, R.; Irving, R.A.; Hudson, P.J. Isolation of the new antigen receptor from wobbegong sharks, and use as a scaffold for the display of protein loop libraries. Mol. Immunol. 2001, 38, 313–326. [Google Scholar] [CrossRef]
- Nuttall, S.D.; Krishnan, U.V.; Doughty, L.; Nathanielsz, A.; Ally, N.; Pike, R.N.; Hudson, P.J.; Kortt, A.A.; Irving, R.A. A naturally occurring NAR variable domain binds the Kgp protease from Porphyromonas gingivalis. FEBS Lett. 2002, 516, 80–86. [Google Scholar] [CrossRef]
- Liu, J.L.; Anderson, G.P.; Delehanty, J.B.; Baumann, R.; Hayhurst, A.; Goldman, E.R. Selection of cholera toxin specific IgNAR single-domain antibodies from a naive shark library. Mol. Immunol. 2007, 44, 1775–1783. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.L.; Anderson, G.P.; Goldman, E.R. Isolation of anti-toxin single domain antibodies from a semi-synthetic spiny dogfish shark display library. BMC Biotechnol. 2007, 7, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Crouch, K.; Smith, L.E.; Williams, R.; Cao, W.; Lee, M.; Jensen, A.; Dooley, H. Humoral immune response of the small-spotted catshark, Scyliorhinus canicula. Fish Shellfish Immunol. 2013, 34, 1158–1169. [Google Scholar] [CrossRef] [PubMed]
- Ohtani, M.; Hikima, J.; Jung, T.S.; Kondo, H.; Hirono, I.; Aoki, T. Construction of an artificially randomized IgNAR phage display library: Screening of variable regions that bind to hen egg white lysozyme. Mar. Biotechnol. (NY) 2013, 15, 56–62. [Google Scholar] [CrossRef] [PubMed]
- Ohtani, M.; Hikima, J.; Jung, T.S.; Kondo, H.; Hirono, I.; Takeyama, H.; Aoki, T. Variable domain antibodies specific for viral hemorrhagic septicemia virus (VHSV) selected from a randomized IgNAR phage display library. Fish Shellfish Immunol. 2013, 34, 724–728. [Google Scholar] [CrossRef] [PubMed]
- Zielonka, S.; Weber, N.; Becker, S.; Doerner, A.; Christmann, A.; Christmann, C.; Uth, C.; Fritz, J.; Schafer, E.; Steinmann, B.; et al. Shark Attack: high affinity binding proteins derived from shark vNAR domains by stepwise in vitro affinity maturation. J. Biotechnol. 2014, 191, 236–245. [Google Scholar] [CrossRef] [PubMed]
- Müller, M.R.; Saunders, K.; Grace, C.; Jin, M.; Piche-Nicholas, N.; Steven, J.; O'Dwyer, R.; Wu, L.; Khetemenee, L.; Vugmeyster, Y.; et al. Improving the pharmacokinetic properties of biologics by fusion to an anti-HSA shark VNAR domain. MAbs 2012, 4, 673–685. [Google Scholar] [CrossRef] [PubMed]
- Müller, M.R.; O'Dwyer, R.; Kovaleva, M.; Rudkin, F.; Dooley, H.; Barelle, C.J. Generation and isolation of target-specific single-domain antibodies from shark immune repertoires. Methods Mol. Biol. 2012, 907, 177–194. [Google Scholar]
- Camacho-Villegas, T.; Mata-Gonzalez, T.; Paniagua-Solis, J.; Sanchez, E.; Licea, A. Human TNF cytokine neutralization with a vNAR from Heterodontus francisci shark: a potential therapeutic use. MAbs 2013, 5, 80–85. [Google Scholar] [CrossRef] [PubMed]
- Kovalenko, O.V.; Olland, A.; Piche-Nicholas, N.; Godbole, A.; King, D.; Svenson, K.; Calabro, V.; Muller, M.R.; Barelle, C.J.; Somers, W.; et al. Atypical Antigen Recognition Mode of a Shark IgNAR Variable Domain Characterized by Humanization and Structural Analysis. J. Biol. Chem. 2013, 288, 17408–17419. [Google Scholar] [CrossRef] [PubMed]
- Dooley, H.; Flajnik, M.F. Antibody repertoire development in cartilaginous fish. Dev. Comp. Immunol. 2006, 30, 43–56. [Google Scholar] [CrossRef] [PubMed]
- Flajnik, M.F. Immunogenetics: Alternative strategies in adaptive immunity and the rise of comparative immunogenomics. Curr. Opin. Immunol. 2007, 19, 522–525. [Google Scholar] [CrossRef] [PubMed]
- Criscitiello, M.F.; Saltis, M.; Flajnik, M.F. An evolutionarily mobile antigen receptor variable region gene: Doubly rearranging NAR-TcR genes in sharks. Proc. Natl. Acad. Sci. USA 2006, 103, 5036–5041. [Google Scholar] [CrossRef] [PubMed]
- Hinds, K.R.; Litman, G.W. Major reorganization of immunoglobulin VH segmental elements during vertebrate evolution. Nature 1986, 320, 546–549. [Google Scholar] [CrossRef] [PubMed]
- Litman, G.W.; Anderson, M.K.; Rast, J.P. Evolution of antigen binding receptors. Annu. Rev. Immunol. 1999, 17, 109–147. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, A.S.; Hughes, A.L.; Guo, J.; Avila, D.; McKinney, E.C.; Flajnik, M.F. A novel "chimeric" antibody class in cartilaginous fish: IgM may not be the primordial immunoglobulin. Eur. J. Immunol. 1996, 26, 1123–1129. [Google Scholar] [CrossRef] [PubMed]
- Venkatesh, B.; Lee, A.P.; Ravi, V.; Maurya, A.K.; Lian, M.M.; Swann, J.B.; Ohta, Y.; Flajnik, M.F.; Sutoh, Y.; Kasahara, M.; et al. Elephant shark genome provides unique insights into gnathostome evolution. Nature 2014, 505, 174–179. [Google Scholar] [CrossRef] [PubMed]
- Muyldermans, S. Nanobodies: Natural Single-Domain Antibodies. Annu. Rev. Biochem. 2013, 82, 775–797. [Google Scholar] [CrossRef] [PubMed]
- Flajnik, M.F.; Deschacht, N.; Muyldermans, S. A case of convergence: Why did a simple alternative to canonical antibodies arise in sharks and camels? PLoS Biol. 2011, 9, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Kovaleva, M.; Ferguson, L.; Steven, J.; Porter, A.; Barelle, C. Shark variable new antigen receptor biologics - a novel technology platform for therapeutic drug development. Expert Opin. Biol. Ther. 2014, 14, 1527–1539. [Google Scholar] [CrossRef] [PubMed]
- Zielonka, S.; Empting, M.; Grzeschik, J.; Konning, D.; Barelle, C.J.; Kolmar, H. Structural insights and biomedical potential of IgNAR scaffolds from sharks. MAbs 2015, 7, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Stanfield, R.L.; Dooley, H.; Flajnik, M.F.; Wilson, I.A. Crystal structure of a shark single-domain antibody V region in complex with lysozyme. Science 2004, 305, 1770–1773. [Google Scholar] [CrossRef] [PubMed]
- Streltsov, V.A.; Carmichael, J.A.; Nuttall, S.D. Structure of a shark IgNAR antibody variable domain and modeling of an early-developmental isotype. Protein Sci. 2005, 14, 2901–2909. [Google Scholar] [CrossRef] [PubMed]
- Diaz, M.; Greenberg, A.S.; Flajnik, M.F. Somatic hypermutation of the new antigen receptor gene (NAR) in the nurse shark does not generate the repertoire: possible role in antigen-driven reactions in the absence of germinal centers. Proc. Natl. Acad. Sci. USA 1998, 95, 14343–14348. [Google Scholar] [CrossRef] [PubMed]
- Diaz, M.; Stanfield, R.L.; Greenberg, A.S.; Flajnik, M.F. Structural analysis, selection, and ontogeny of the shark new antigen receptor (IgNAR): Identification of a new locus preferentially expressed in early development. Immunogenetics 2002, 54, 501–512. [Google Scholar] [CrossRef] [PubMed]
- Shao, C.Y.; Secombes, C.J.; Porter, A.J. Rapid isolation of IgNAR variable single-domain antibody fragments from a shark synthetic library. Mol. Immunol. 2007, 44, 656–665. [Google Scholar] [CrossRef] [PubMed]
- Nuttall, S.D.; Krishnan, U.V.; Doughty, L.; Pearson, K.; Ryan, M.T.; Hoogenraad, N.J.; Hattarki, M.; Carmichael, J.A.; Irving, R.A.; Hudson, P.J. Isolation and characterization of an IgNAR variable domain specific for the human mitochondrial translocase receptor Tom70. Eur. J. Biochem. 2003, 270, 3543–3554. [Google Scholar] [CrossRef] [PubMed]
- Nuttall, S.D.; Humberstone, K.S.; Krishnan, U.V.; Carmichael, J.A.; Doughty, L.; Hattarki, M.; Coley, A.M.; Casey, J.L.; Anders, R.F.; Foley, M.; et al. Selection and affinity maturation of IgNAR variable domains targeting Plasmodium falciparum AMA1. Proteins 2004, 55, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Streltsov, V.A.; Varghese, J.N.; Carmichael, J.A.; Irving, R.A.; Hudson, P.J.; Nuttall, S.D. Structural evidence for evolution of shark Ig new antigen receptor variable domain antibodies from a cell-surface receptor. Proc. Natl. Acad. Sci. USA 2004, 101, 12444–12449. [Google Scholar] [CrossRef] [PubMed]
- Kopsidas, G.; Roberts, A.S.; Coia, G.; Streltsov, V.A.; Nuttall, S.D. In vitro improvement of a shark IgNAR antibody by Qbeta replicase mutation and ribosome display mimics in vivo affinity maturation. Immunol. Lett. 2006, 107, 163–168. [Google Scholar] [CrossRef] [PubMed]
- Henderson, K.A.; Streltsov, V.A.; Coley, A.M.; Dolezal, O.; Hudson, P.J.; Batchelor, A.H.; Gupta, A.; Bai, T.; Murphy, V.J.; Anders, R.F.; et al. Structure of an IgNAR-AMA1 complex: targeting a conserved hydrophobic cleft broadens malarial strain recognition. Structure 2007, 15, 1452–1466. [Google Scholar] [CrossRef] [PubMed]
- Zielonka, S.; Empting, M.; Konning, D.; Grzeschik, J.; Krah, S.; Becker, S.; Dickgiesser, S.; Kolmar, H. The Shark Strikes Twice: Hypervariable Loop 2 of Shark IgNAR Antibody Variable Domains and Its Potential to Function as an Autonomous Paratope. Mar. Biotechnol. (NY) 2015, 17, 386–392. [Google Scholar] [CrossRef] [PubMed]
- Fennell, B.J.; Darmanin-Sheehan, A.; Hufton, S.E.; Calabro, V.; Wu, L.; Muller, M.R.; Cao, W.; Gill, D.; Cunningham, O.; Finlay, W.J. Dissection of the IgNAR V domain: molecular scanning and orthologue database mining define novel IgNAR hallmarks and affinity maturation mechanisms. J. Mol. Biol. 2010, 400, 155–170. [Google Scholar] [CrossRef] [PubMed]
- Simmons, D.P.; Abregu, F.A.; Krishnan, U.V.; Proll, D.F.; Streltsov, V.A.; Doughty, L.; Hattarki, M.K.; Nuttall, S.D. Dimerisation strategies for shark IgNAR single domain antibody fragments. J. Immunol. Methods 2006, 315, 171–184. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.L.; Zabetakis, D.; Brown, J.C.; Anderson, G.P.; Goldman, E.R. Thermal stability and refolding capability of shark derived single domain antibodies. Mol. Immunol. 2014, 59, 194–199. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, K.; Dolezal, O.; Parisi, K.; Angerosa, J.; Dogovski, C.; Barraclough, M.; Sanalla, A.; Casey, J.; Gonzalez, I.; Perugini, M.; et al. Shark variable new antigen receptor (VNAR) single domain antibody fragments: Stability and diagnostic applications. Antibodies 2013, 2, 66–88. [Google Scholar] [CrossRef]
- Markussen, J.; Havelund, S.; Kurtzhals, P.; Andersen, A.S.; Halstrom, J.; Hasselager, E.; Larsen, U.D.; Ribel, U.; Schaffer, L.; Vad, K.; et al. Soluble, fatty acid acylated insulins bind to albumin and show protracted action in pigs. Diabetologia 1996, 39, 281–288. [Google Scholar] [CrossRef] [PubMed]
- Makrides, S.C.; Nygren, P.A.; Andrews, B.; Ford, P.J.; Evans, K.S.; Hayman, E.G.; Adari, H.; Uhlen, M.; Toth, C.A. Extended in vivo half-life of human soluble complement receptor type 1 fused to a serum albumin-binding receptor. J. Pharmacol. Exp. Ther. 1996, 277, 534–542. [Google Scholar] [PubMed]
- Smith, B.J.; Popplewell, A.; Athwal, D.; Chapman, A.P.; Heywood, S.; West, S.M.; Carrington, B.; Nesbitt, A.; Lawson, A.D.; Antoniw, P.; et al. Prolonged in vivo residence times of antibody fragments associated with albumin. Bioconjug. Chem. 2001, 12, 750–756. [Google Scholar] [CrossRef] [PubMed]
- Stork, R.; Muller, D.; Kontermann, R.E. A novel tri-functional antibody fusion protein with improved pharmacokinetic properties generated by fusing a bispecific single-chain diabody with an albumin-binding domain from streptococcal protein G. Protein Eng. Des. Sel. 2007, 20, 569–576. [Google Scholar] [CrossRef] [PubMed]
- Van Roy, M.; Ververken, C.; Beirnaert, E.; Hoefman, S.; Kolkman, J.; Vierboom, M.; Breedveld, E.; 't Hart, B.; Poelmans, S.; Bontinck, L.; et al. The preclinical pharmacology of the high affinity anti-IL-6R Nanobody(R) ALX-0061 supports its clinical development in rheumatoid arthritis. Arthritis Res. Ther. 2015, 17, 135–150. [Google Scholar] [CrossRef] [PubMed]
- Padlan, E.A. A possible procedure for reducing the immunogenicity of antibody variable domains while preserving their ligand-binding properties. Mol. Immunol. 1991, 28, 489–498. [Google Scholar] [CrossRef]
- Pelat, T.; Bedouelle, H.; Rees, A.R.; Crennell, S.J.; Lefranc, M.P.; Thullier, P. Germline humanization of a non-human primate antibody that neutralizes the anthrax toxin, by in vitro and in silico engineering. J. Mol. Biol. 2008, 384, 1400–1407. [Google Scholar] [CrossRef] [PubMed]
- Robert, R.; Streltsov, V.A.; Newman, J.; Pearce, L.A.; Wark, K.L.; Dolezal, O. Germline humanization of a murine Abeta antibody and crystal structure of the humanized recombinant Fab fragment. Protein Sci. 2010, 19, 299–308. [Google Scholar] [CrossRef] [PubMed]
- Cheung, N.K.; Guo, H.; Hu, J.; Tassev, D.V.; Cheung, I.Y. Humanizing murine IgG3 anti-GD2 antibody m3F8 substantially improves antibody-dependent cell-mediated cytotoxicity while retaining targeting in vivo. Oncoimmunology 2012, 1, 477–486. [Google Scholar] [CrossRef] [PubMed]
- Reichert, J.M. Marketed therapeutic antibodies compendium. MAbs 2012, 4, 413–415. [Google Scholar] [CrossRef] [PubMed]
- Goodchild, S.A.; Dooley, H.; Schoepp, R.J.; Flajnik, M.; Lonsdale, S.G. Isolation and characterisation of Ebolavirus-specific recombinant antibody fragments from murine and shark immune libraries. Mol. Immunol. 2011, 48, 2027–2037. [Google Scholar] [CrossRef] [PubMed]
- Walsh, R.; Nuttall, S.; Revill, P.; Colledge, D.; Cabuang, L.; Soppe, S.; Dolezal, O.; Griffiths, K.; Bartholomeusz, A.; Locarnini, S. Targeting the hepatitis B virus precore antigen with a novel IgNAR single variable domain intrabody. Virology 2011, 411, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Bojalil, R.; Mata-Gonzalez, M.T.; Sanchez-Munoz, F.; Yee, Y.; Argueta, I.; Bolanos, L.; Amezcua-Guerra, L.M.; Camacho-Villegas, T.A.; Sanchez-Castrejon, E.; Garcia-Ubbelohde, W.J.; et al. Anti-tumor necrosis factor VNAR single domains reduce lethality and regulate underlying inflammatory response in a murine model of endotoxic shock. BMC Immunol. 2013, 14, 17. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barelle, C.; Porter, A. VNARs: An Ancient and Unique Repertoire of Molecules That Deliver Small, Soluble, Stable and High Affinity Binders of Proteins. Antibodies 2015, 4, 240-258. https://doi.org/10.3390/antib4030240
Barelle C, Porter A. VNARs: An Ancient and Unique Repertoire of Molecules That Deliver Small, Soluble, Stable and High Affinity Binders of Proteins. Antibodies. 2015; 4(3):240-258. https://doi.org/10.3390/antib4030240
Chicago/Turabian StyleBarelle, Caroline, and Andy Porter. 2015. "VNARs: An Ancient and Unique Repertoire of Molecules That Deliver Small, Soluble, Stable and High Affinity Binders of Proteins" Antibodies 4, no. 3: 240-258. https://doi.org/10.3390/antib4030240





