Evaluation of Continuous Membrane Chromatography Concepts with an Enhanced Process Simulation Approach
Abstract
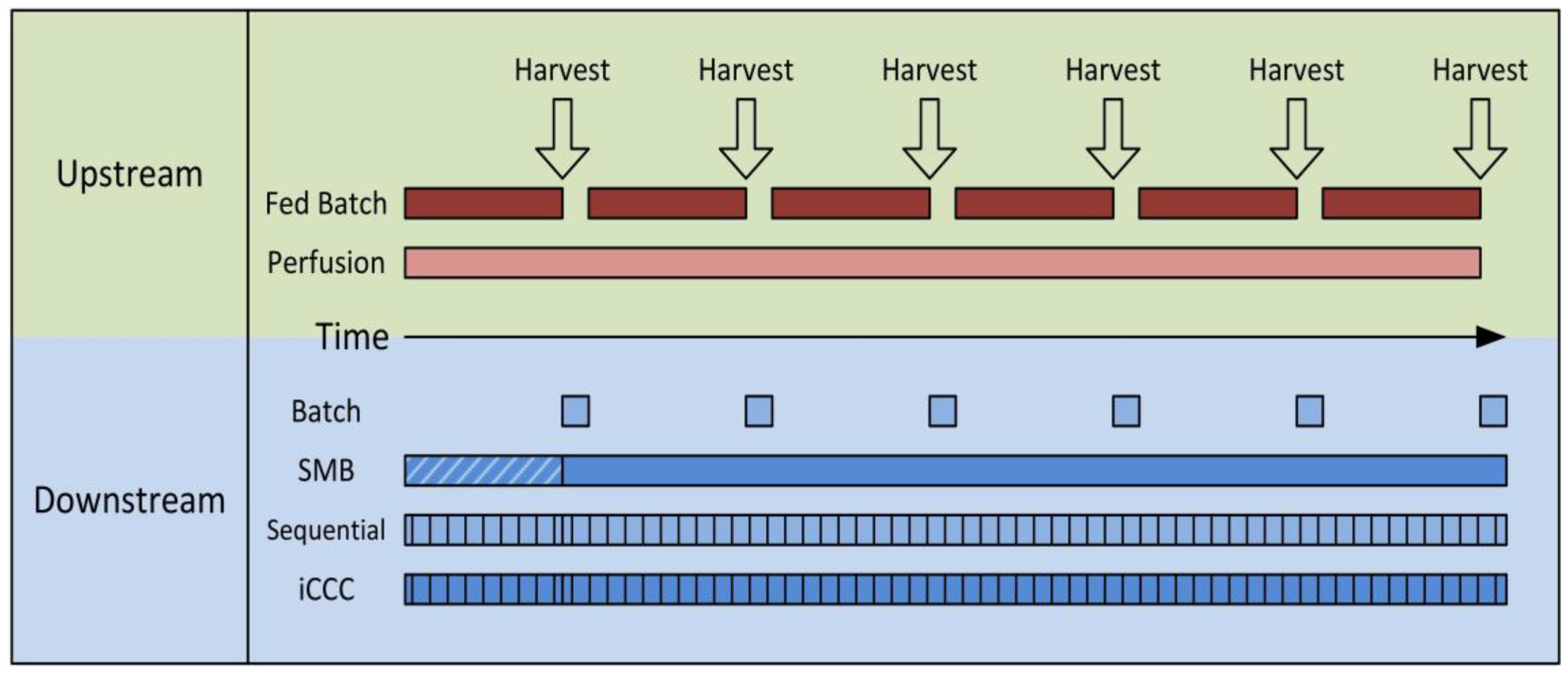
:1. Introduction
2. Materials and Methods
2.1. Proteins, Buffers, and Columns
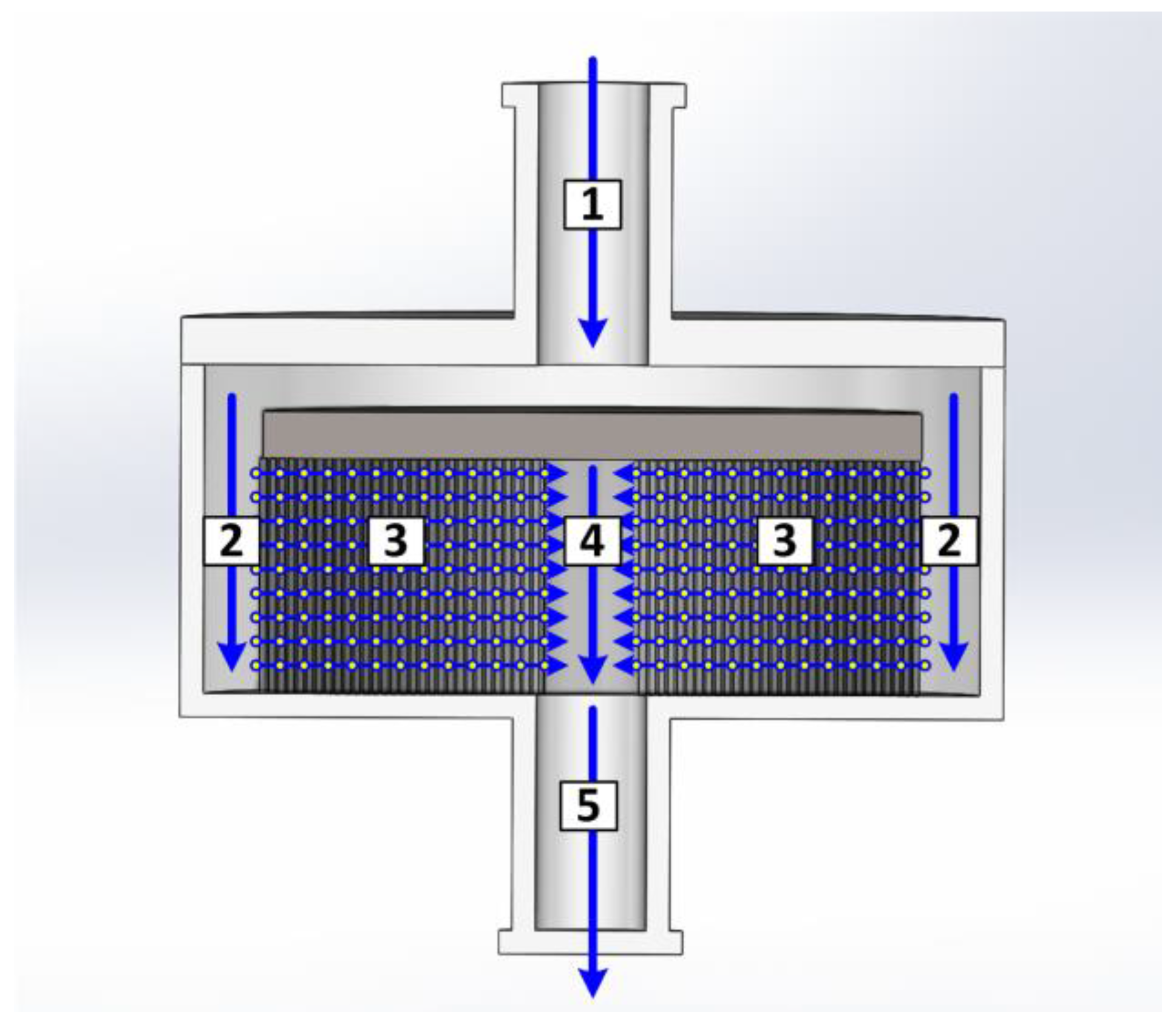
2.2. Devices and Instruments
2.3. Parameter Determination
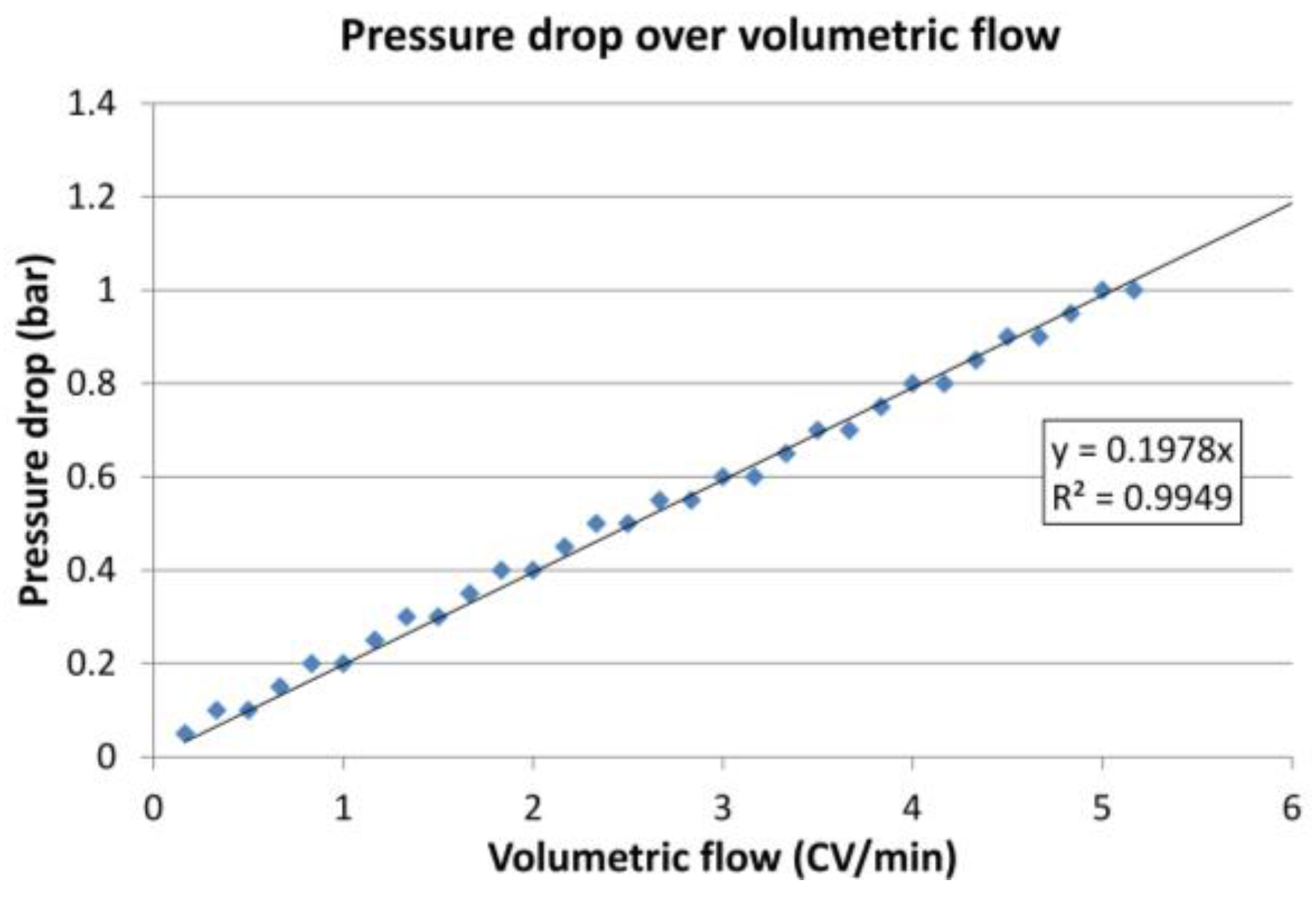
2.3.1. Fluid Dynamics
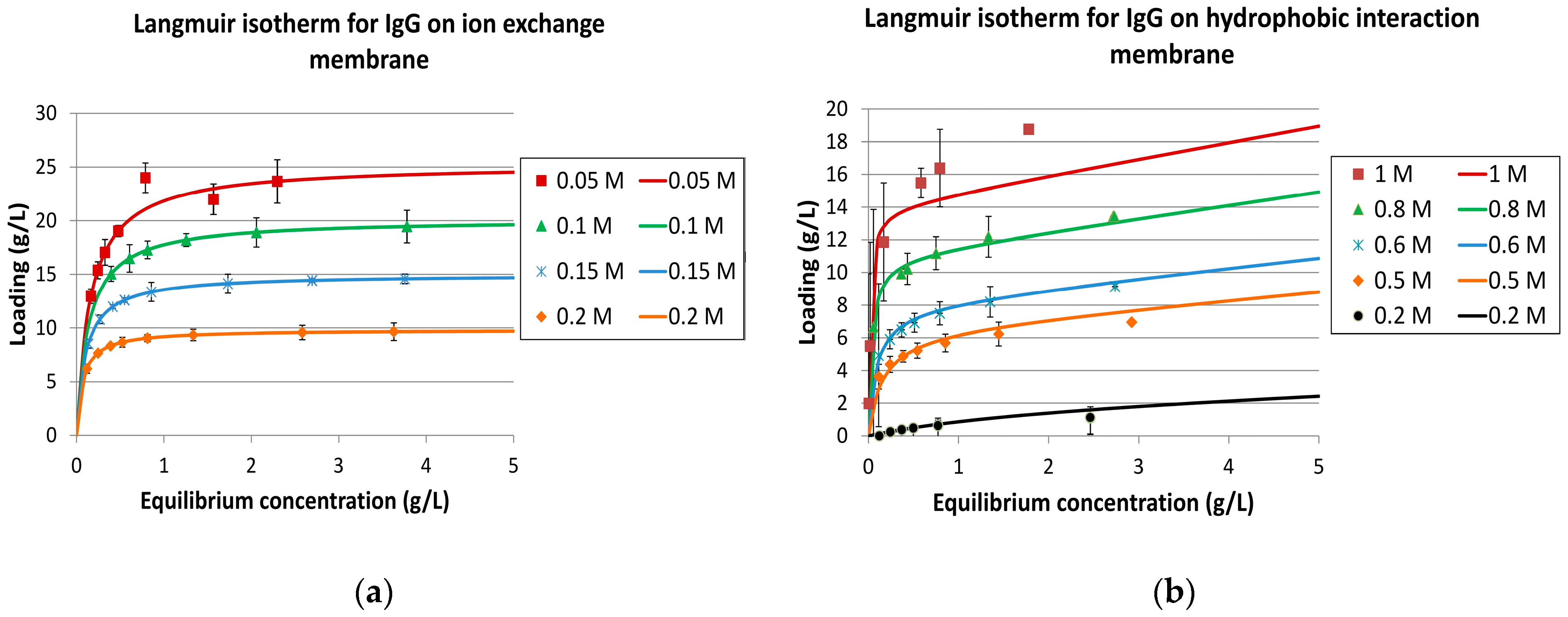
2.3.2. Isotherms
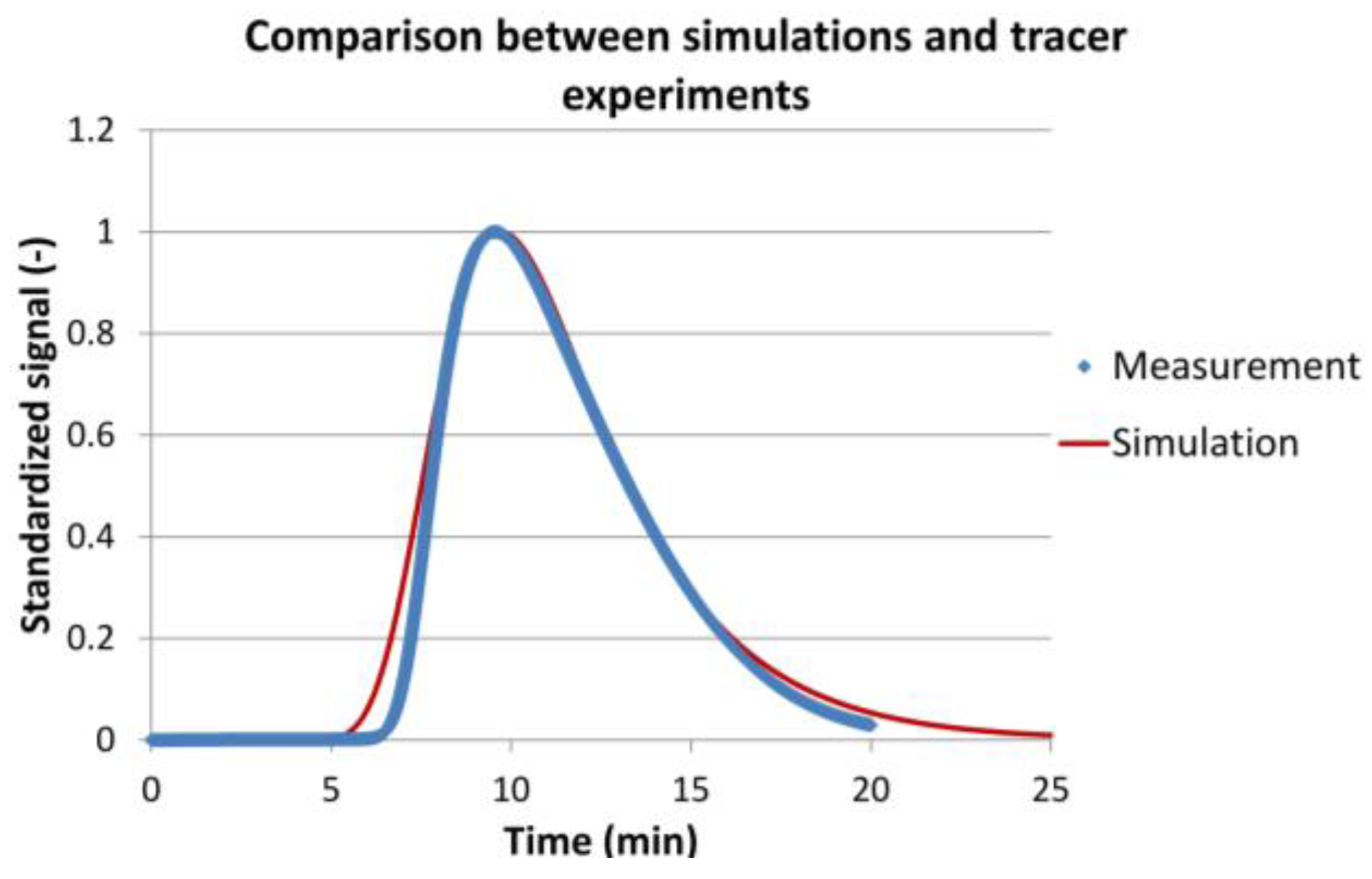
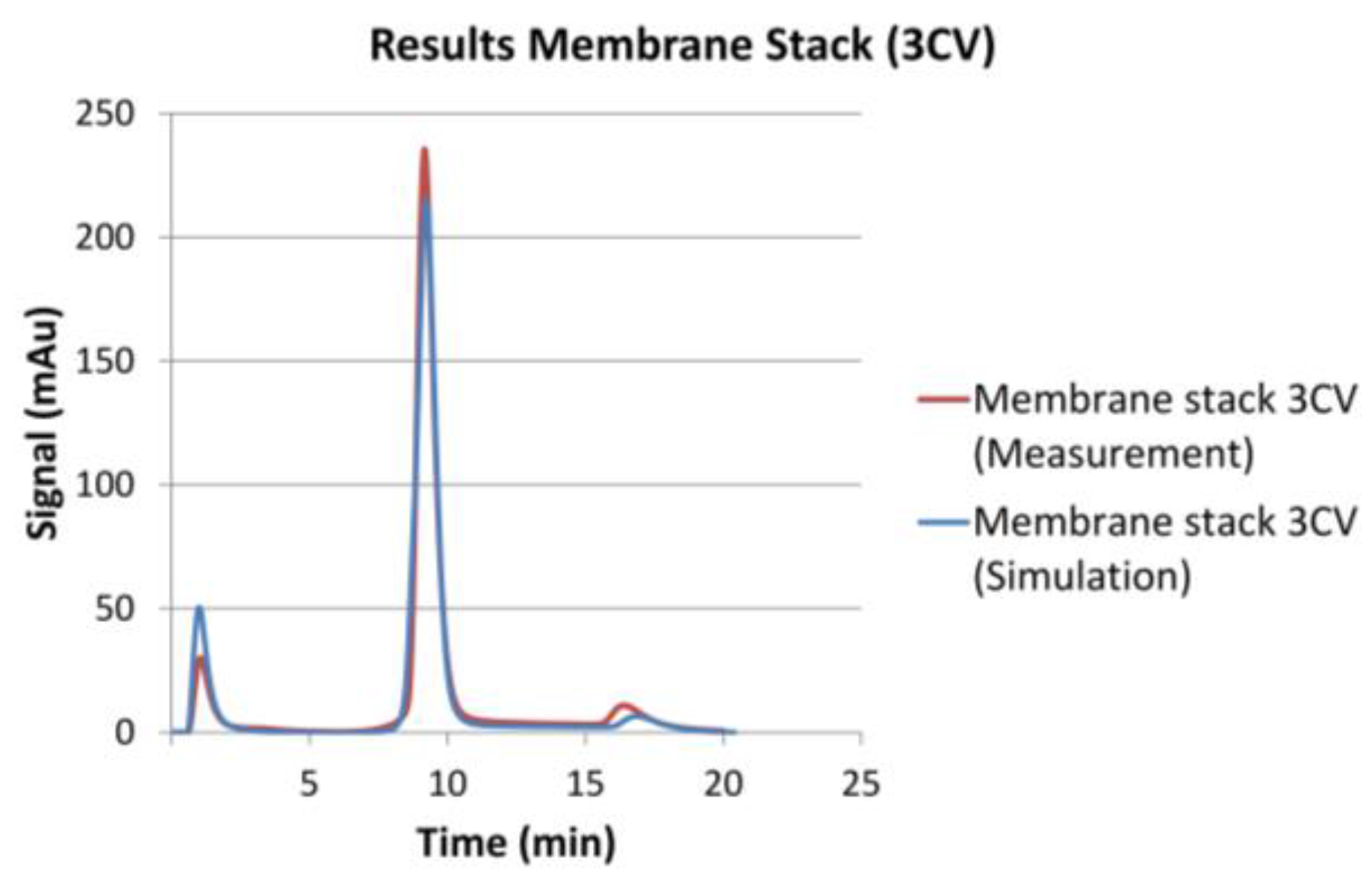
2.3.3. Model Validation
3. Process Modeling
3.1. Fluid Dynamics, Mass Transport, and Isotherms
3.2. Model Validation
4. Results and Discussion
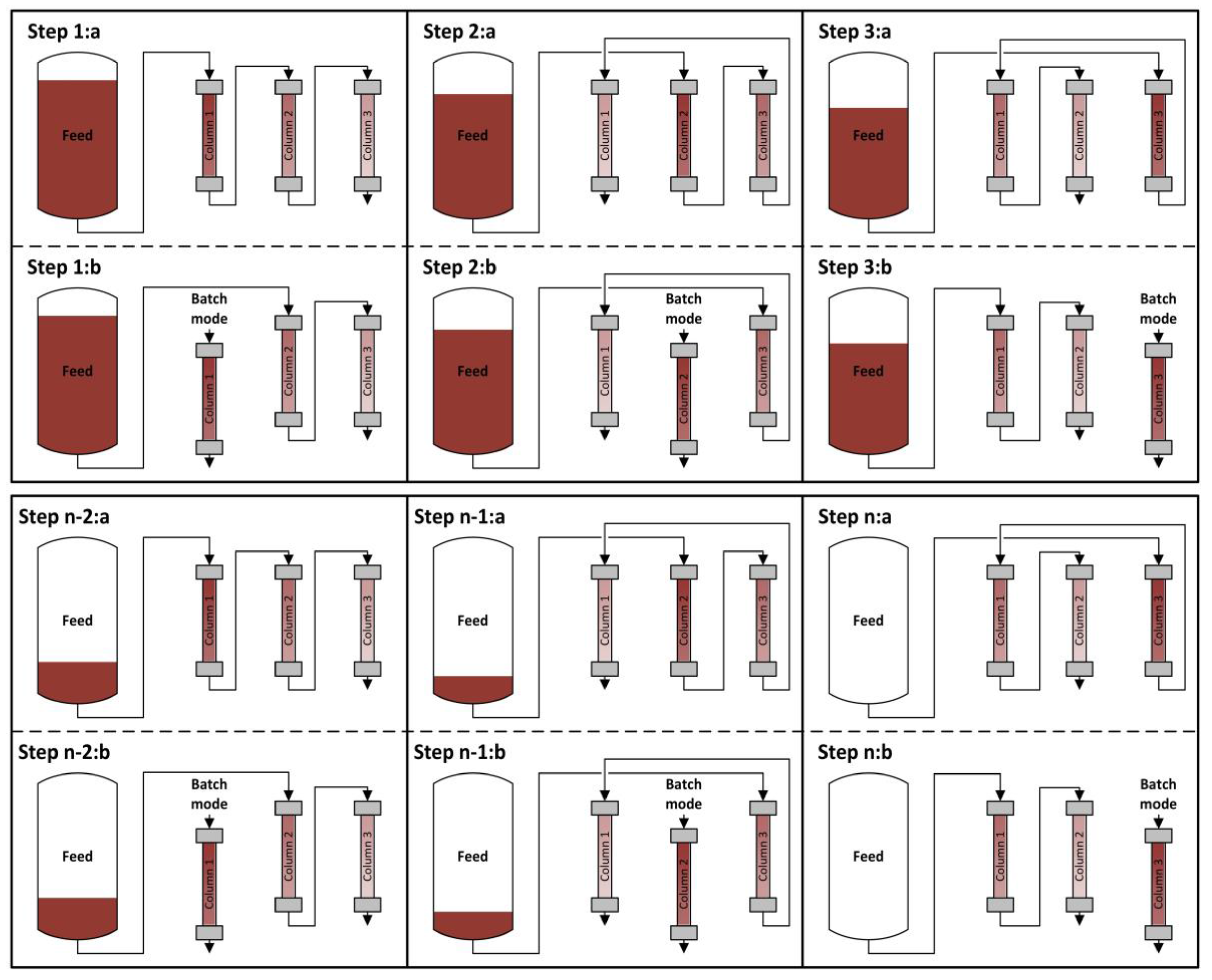
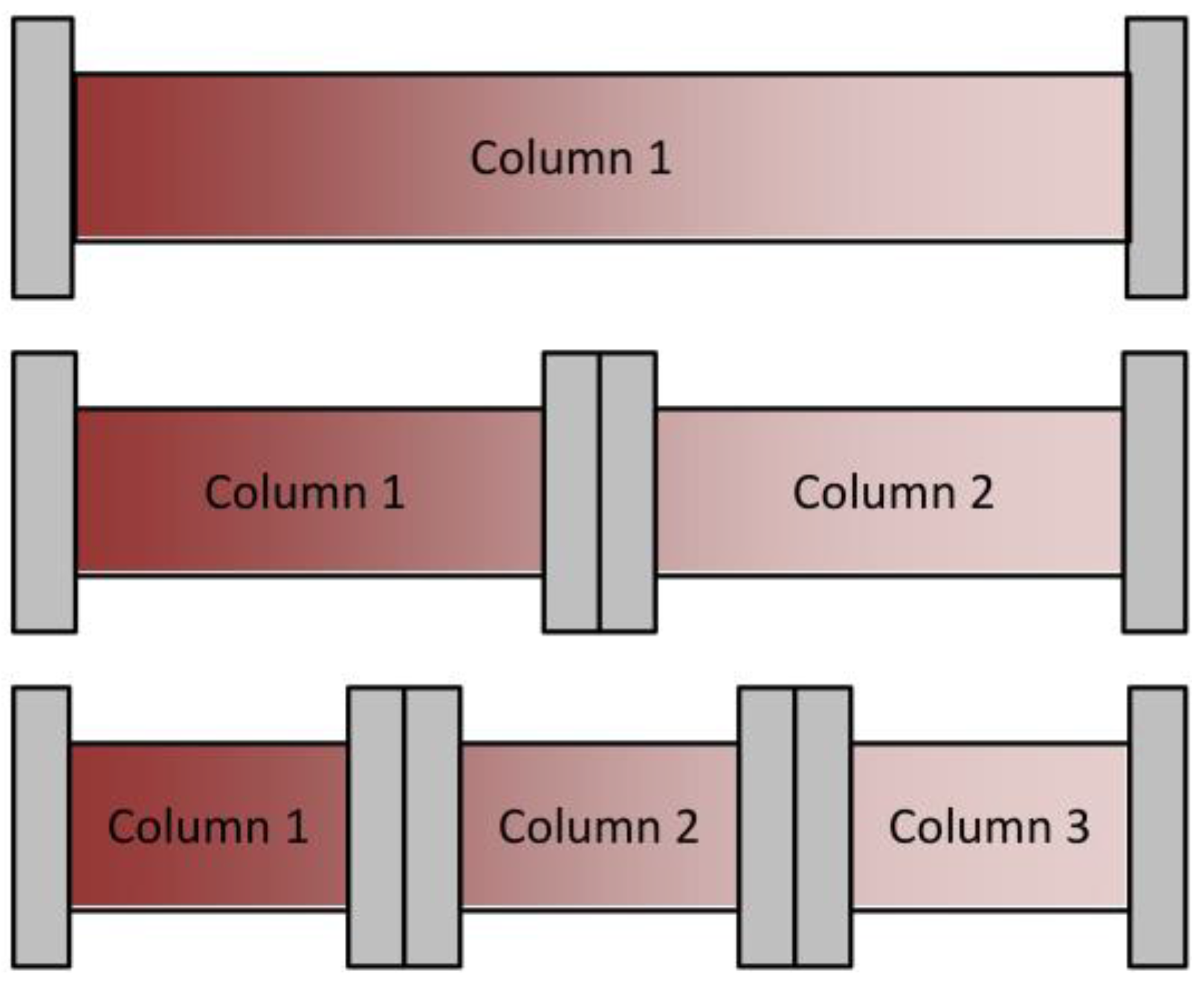
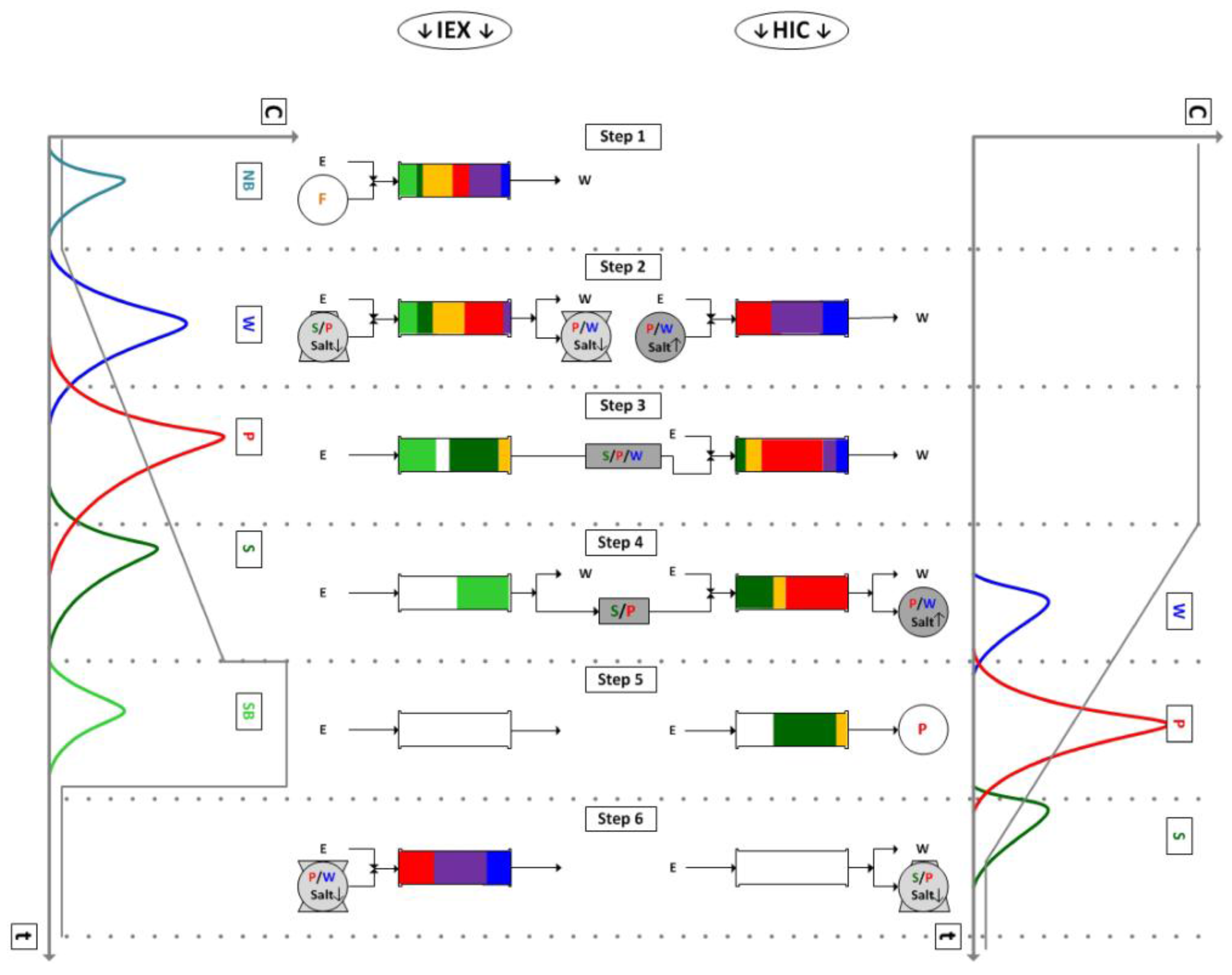
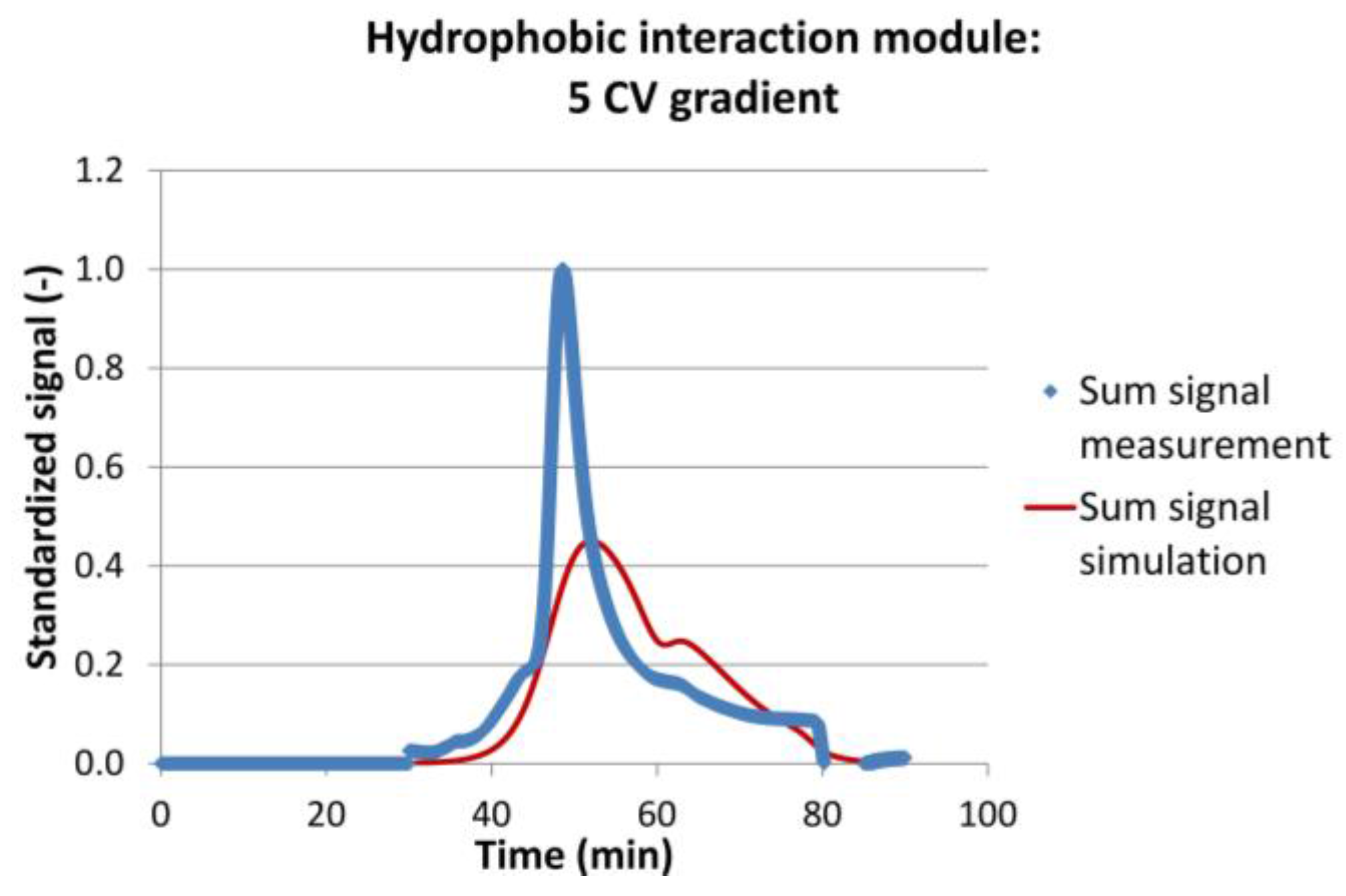
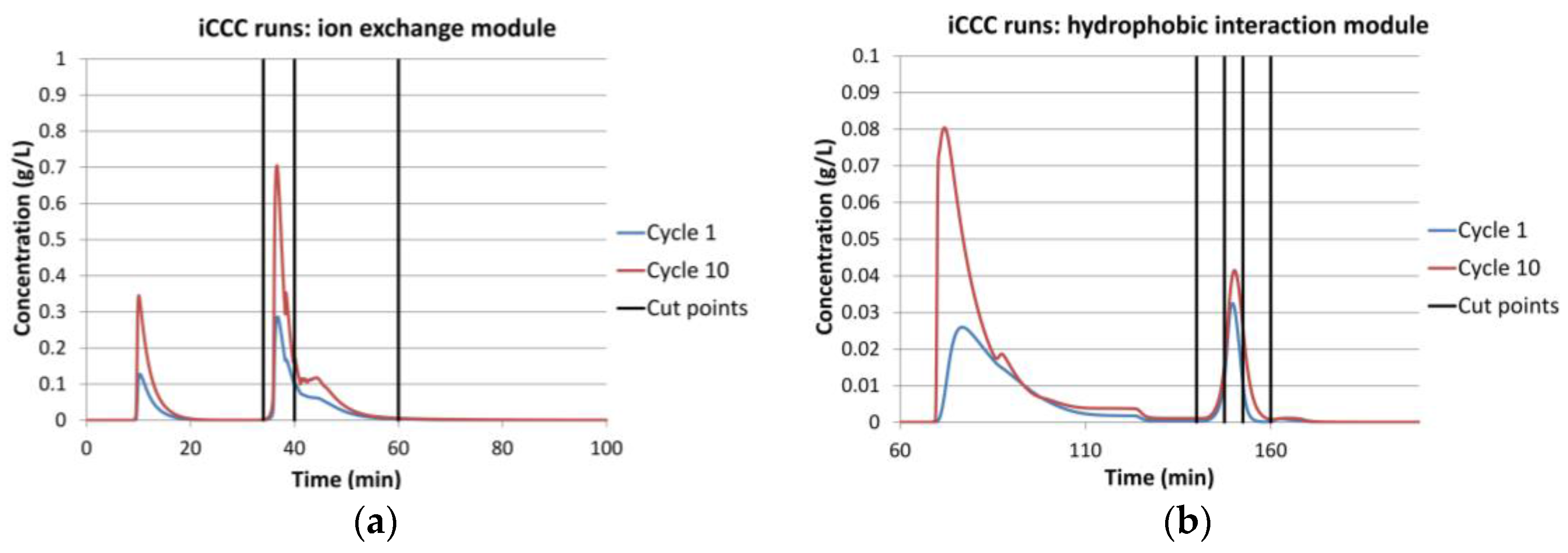
4.1. Integrated Counter Current Chromatography (iCCC)
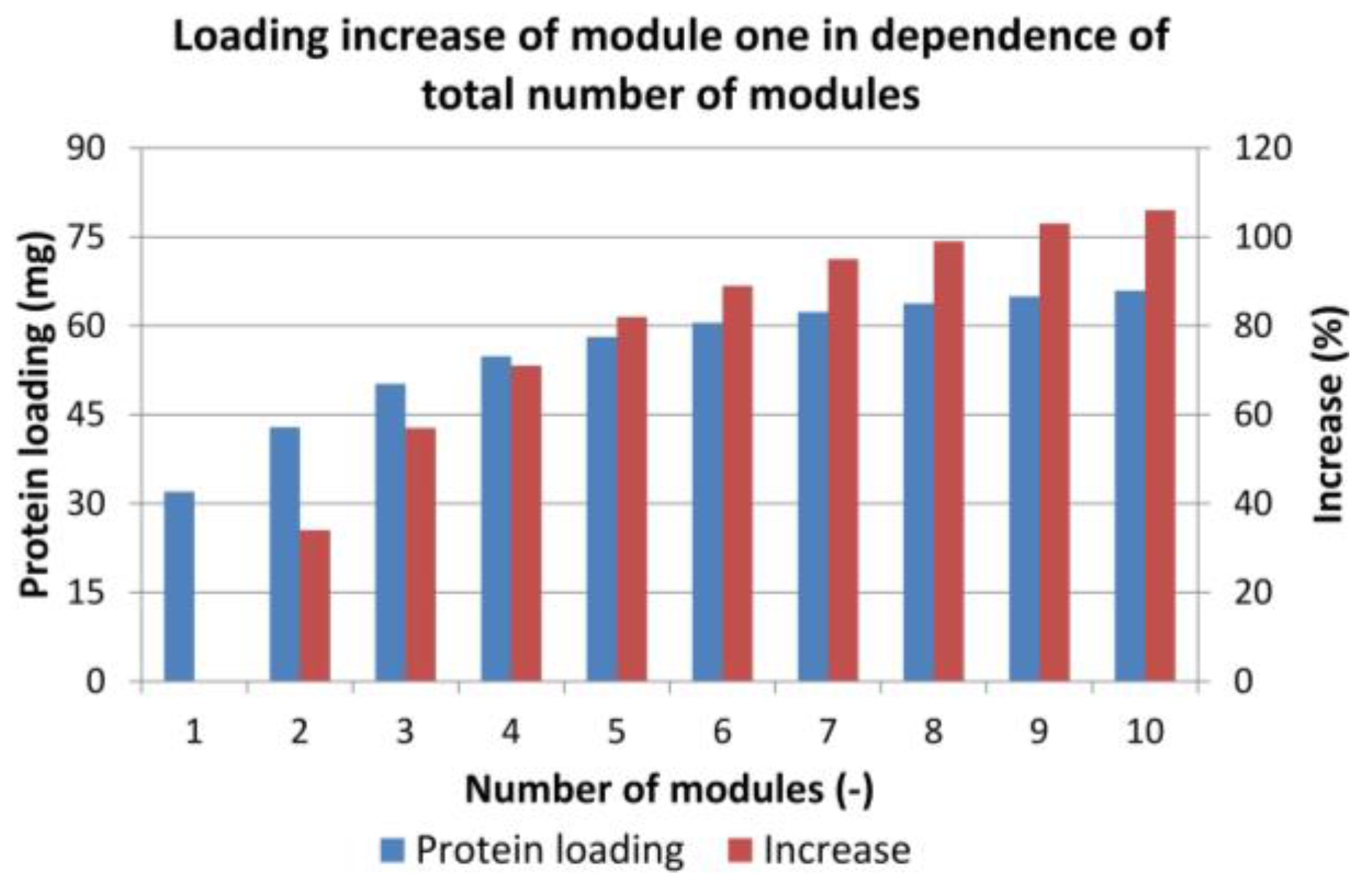
4.2. Sequential Chromatography
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| c (mg/mL) | Concentration |
| CV (mL) | Column Volume |
| Dax (cm2/s) | Axial Dispersion Coefficient |
| H (-) | Henry Coefficient |
| HIC | Hydrophobic Interaction Chromatography |
| iCCC | integrated Counter Current Chromatography |
| IEX | Ion-Exchange Chromatography |
| IgG | Immunoglobulin G |
| l (cm) | Length |
| kla (1/s) | Effective Mass Transfer Resistance and Area |
| MCSGP | Multicolumn Counter Current Solvent Gradient Purification |
| NaPi | Sodium Phosphate Buffer |
| PCC | Periodic Counter Current Chromatography |
| PFR | Plug Flow Reactor |
| q (mg/mL) | Loading |
| SEC | Size-Exclusion Chromatography |
| SMB | Simulated Moving Bed |
| SMCC | Sequential Multicolumn Chromatography |
| (s) | mean residence time |
| V (mL) | Volume |
| v (cm/s) | Velocity |
| (mL/min) | Volumetric Flow |
| ε (-) | Voidage |
| σ2 (s2) | Variance |
References
- Carroll, S. Goodbye blockbuster medicines; hello new pharmaceutical business models. Pharm. J. 2009, 282, 681–682. [Google Scholar]
- Hester, R.E.; Harrison, R.M. (Eds.) Pharmaceuticals in the Environment; The Royal Society of Chemistry: Cambridge, UK, 2015. [Google Scholar]
- Taylor, D. The Pharmaceutical Industry and the Future of Drug Development. In Pharmaceuticals in the Environment; Hester, R.E., Harrison, R.M., Eds.; The Royal Society of Chemistry: Cambridge, UK, 2015; pp. 1–33. [Google Scholar]
- Padmanabhan, S. Handbook of Pharmacogenomics and Stratified Medicine; Academic Press: London, UK, 2014. [Google Scholar]
- Subramanian, G. (Ed.) Continuous Biomanufacturing: Innovative Technologies and Methods; WILEY-VCH: Weinheim, Germany, 2017. [Google Scholar]
- Konstantinov, K. Continous bioprocessing: An interview with Konstantin Konstantinov from Genzyme. Interviewed by Prof. Alois Jungbauer and Dr. Judy Peng. Biotechnol. J. 2011, 6, 1431–1433. [Google Scholar] [CrossRef] [PubMed]
- Konstantinov, K.; Goudar, C.; Ng, M.; Meneses, R.; Thrift, J.; Chuppa, S.; Matanguihan, C.; Michaels, J.; Naveh, D. The “push-to-low” approach for optimization of high-density perfusion cultures of animal cells. Adv. Biochem. Eng./Biotechnol. 2006, 101, 75–98. [Google Scholar]
- Pollock, J.; Ho, S.V.; Farid, S.S. Fed-batch and perfusion culture processes: Economic, environmental, and operational feasibility under uncertainty. Biotechnol. Bioeng. 2013, 110, 206–219. [Google Scholar] [CrossRef] [PubMed]
- Warikoo, V.; Godawat, R.; Brower, K.; Jain, S.; Cummings, D.; Simons, E.; Johnson, T.; Walther, J.; Yu, M.; Wright, B.; et al. Integrated continuous production of recombinant therapeutic proteins. Biotechnol. Bioeng. 2012, 109, 3018–3029. [Google Scholar] [CrossRef] [PubMed]
- Zydney, A.L. Continuous downstream processing for high value biological products: A Review. Biotechnol. Bioeng. 2016, 113, 465–475. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, G. (Ed.) Continuous Processing in Pharmaceutical Manufacturing; WILEY-VCH: Weinheim, Germany, 2015. [Google Scholar]
- Schulte, M.; Britsch, L.; Strube, J. Continuous preparative liquid chromatography in the downstrem processing of biotechnological products. Acta Biotechnol. 2000, 20, 3–15. [Google Scholar] [CrossRef]
- Zobel, S.; Helling, C.; Ditz, R.; Strube, J. Design and Operation of Continuous Countercurrent Chromatography in Biotechnological Production. Ind. Eng. Chem. Res. 2014, 53, 9169–9185. [Google Scholar] [CrossRef]
- Belcheva, D. Theoretische und Experimentelle Studie der Gradienten-Gegenstromchromatographie unter Linearen Bedingungen. Ph.D. Thesis, Otto-von-Guericke-Universität Magdeburg, Fort Made, Germany, 2004. (In German). [Google Scholar]
- Holzer, M.; Osuna-Sanchez, H.; David, L. Multicolumn Chromatography: A New Approach to Relieving Capacity Bottlenecks for Downstream Processing Efficiency. BioProcess Int. 2008, 6, 74–82. [Google Scholar]
- Godawat, R.; Brower, K.; Jain, S.; Konstantinov, K.; Riske, F.; Warikoo, V. Periodic counter-current chromatography–design and operational considerations for integrated and continuous purification of proteins. Biotechnol. J. 2012, 7, 1496–1508. [Google Scholar] [CrossRef] [PubMed]
- Mahajan, E.; George, A.; Wolk, B. Improving affinity chromatography resin efficiency using semi-continuous chromatography. J. Chromatogr. A 2012, 1227, 154–162. [Google Scholar] [CrossRef] [PubMed]
- Angarita, M.; Müller-Späth, T.; Baur, D.; Lievrouw, R.; Lissens, G.; Morbidelli, M. Twin-column CaptureSMB: A novel cyclic process for protein A affinity chromatography. J. Chromatogr. A 2015, 1389, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Baur, D.; Angarita, M.; Muller-Spath, T.; Steinebach, F.; Morbidelli, M. Comparison of batch and continuous multi-column protein A capture processes by optimal design. Biotechnol. J. 2016, 11, 920–931. [Google Scholar] [CrossRef] [PubMed]
- Gjoka, X.; Rogler, K.; Martino, R.A.; Gantier, R.; Schofield, M. A straightforward methodology for designing continuous monoclonal antibody capture multi-column chromatography processes. J. Chromatogr. A 2015, 1416, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Whitford, W.G. Single-Use Systems As Principal Components in Bioproduction. BioProcess Int. 2010, 8, 34–44. [Google Scholar]
- Eggersgluess, J.K.; Richter, M.; Dieterle, M.; Strube, J. Multi-Stage Aqueous Two-Phase Extraction for the Purification of Monoclonal Antibodies. Chem. Eng. Technol. 2014, 37, 675–682. [Google Scholar] [CrossRef]
- Eggersglüss, J.; Both, S.; Strube, J. Process development for the extraction of biomolecules Application for downstream processing of proteins in aqueous two-phase systems. Chem. Today 2012, 30, 32–36. [Google Scholar]
- Muendges, J.; Stark, I.; Mohammad, S.; Górak, A.; Zeiner, T. Single stage aqueous two-phase extraction for monoclonal antibody purification from cell supernatant. Fluid Phase Equilibria 2015, 385, 227–236. [Google Scholar] [CrossRef]
- Muendges, J.; Zalesko, A.; Górak, A.; Zeiner, T. Multistage aqueous two-phase extraction of a monoclonal antibody from cell supernatant. Biotechnol. Prog. 2015, 31, 925–936. [Google Scholar] [CrossRef] [PubMed]
- Gronemeyer, P.; Strube, J. Purification of Antibodies and their Fragments by ATPE and Precipitation: One Step towards a Chromatography-Free Manufacturing Process. Chem. Ing. Tech. 2015, 87, 1057–1058. [Google Scholar] [CrossRef]
- Hammerschmidt, N.; Tscheliessnig, A.; Sommer, R.; Helk, B.; Jungbauer, A. Economics of recombinant antibody production processes at various scales: Industry-standard compared to continuous precipitation. Biotechnol. J. 2014, 9, 766–775. [Google Scholar] [CrossRef] [PubMed]
- Schwellenbach, J.; Zobel, S.; Taft, F.; Villain, L.; Strube, J. Purification of Monoclonal Antibodies Using a Fiber Based Cation-Exchange Stationary Phase: Parameter Determination and Modeling. Bioengineering 2016, 3, 24. [Google Scholar] [CrossRef] [PubMed]
- Fröhlich, H.; Villian, L.; Melzner, D.; Strube, J. Membrane Technology in Bioprocess Science. Chem. Ing. Tech. 2012, 297, 905–917. [Google Scholar] [CrossRef]
- Ghosh, R. Protein separation using membrane chromatography: Opportunities and challenges. J. Chromatogr. A 2002, 952, 13–27. [Google Scholar] [CrossRef]
- Saxena, A.; Tripathi, B.P.; Kumar, M.; Shahi, V.K. Membrane-based techniques for the separation and purification of proteins: An overview. Adv. Colloid Interface Sci. 2009, 145, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Thoemmes, J.; Kula, M.-R. Membrane chromatography—An integrative concept in the downstream processing of proteins. Biotechnol. Prog. 1995, 11, 357–367. [Google Scholar] [CrossRef]
- Van Reis, R.; Zydney, A. Membrane separations in biotechnology. Curr. Opin. Biotechnol. 2001, 12, 208–211. [Google Scholar] [CrossRef]
- Charcosset, C. Membrane processes in biotechnology: An overview. Biotechnol. Adv. 2006, 24, 482–492. [Google Scholar] [CrossRef] [PubMed]
- Fornstedt, T.; Forssén, P.; Samuelsson, J. Modeling of preparative liquid chromatography. In Liquid Chromatography: Fundamentals and Instrumentation; Fanali, S., Haddad, P.R., Poole, C., Riekkola, M.-L., Eds.; Elsevier Science: Saint Louis, MO, USA, 2017; pp. 573–592. [Google Scholar]
- Forrer, N. Antibody Purification with Ion-Exchange Chromatography. Bachelor’s Thesis, Eidgenössische Technische Hochschule, Zürich, Switzerland, 2008. [Google Scholar]
- Aumann, L.; Stroehlein, G.; Morbidelli, M. Parametric study of a 6-column countercurrent solvent gradient purification (MCSGP) unit. Biotechnol. Bioeng. 2007, 98, 1029–1042. [Google Scholar] [CrossRef] [PubMed]
- Aumann, L.; Morbidelli, M. A continuous multicolumn countercurrent solvent gradient purification (MCSGP) process. Biotechnol. Bioeng. 2007, 98, 1043–1055. [Google Scholar] [CrossRef] [PubMed]
- Strube, J. Simulation und Optimierung kontinuierlicher Simulated-Moving-Bed (SMB)-Chromatographie-Prozesse; Dortmund University: Dortmund, Germany, 1996. [Google Scholar]
- Rodrigues, A. Simulated Moving Bed Technology. Principles, Design and Process Applications; Elsevier Science: Burlington, ON, Canada, 2015. [Google Scholar]
- Müller-Späth, T.; Aumann, L.; Melter, L.; Ströhlein, G.; Morbidelli, M. Chromatographic separation of three monoclonal antibody variants using multicolumn countercurrent solvent gradient purification (MCSGP). Biotechnol. Bioeng. 2008, 100, 1166–1177. [Google Scholar] [CrossRef] [PubMed]
- Steinebach, F.; Muller-Spath, T.; Morbidelli, M. Continuous counter-current chromatography for capture and polishing steps in biopharmaceutical production. Biotechnol. J. 2016, 11, 1126–1141. [Google Scholar] [CrossRef] [PubMed]
- Helling, C.; Borrmann, C.; Strube, J. Optimal Integration of Directly Combined Hydrophobic Interaction and Ion Exchange Chromatography Purification Processes. Chem. Eng. Technol. 2012, 35, 1786–1796. [Google Scholar] [CrossRef]
- Zobel-Roos, S.; Thiess, H.; Gronemeyer, P.; Ditz, R.; Strube, J. Chapter 13: Continuous Chromatography as a Fully Integrated Process in Continuous Biomanufacturing. In Continuous Biomanufacturing: Innovative Technologies and Methods; Subramanian, G., Ed.; WILEY-VCH: Weinheim, Germany, 2017; pp. 369–393. [Google Scholar]
- Levenspiel, O. Chemical Reaction Engineering, 3rd ed.; Wiley: New York, NY, USA, 1999. [Google Scholar]
- Danckwerts, P.V. Continuous flow systems. Distribution of residence times. Chem. Eng. Sci. 1953, 2, 1–13. [Google Scholar] [CrossRef]
- Carta, G.; Jungbauer, A. Protein Chromatography. Process Development and Scale-Up; WILEY-VCH: Weinheim, Germany, 2010. [Google Scholar]
- Gronemeyer, P.; Ditz, R.; Strube, J. DoE based integration approach of upstream and downstream processing regarding HCP and ATPE as harvest operation. Biochem. Eng. J. 2016, 113, 158–166. [Google Scholar] [CrossRef]
- Gronemeyer, P.; Strube, J. Entwicklung einer Methode zur Integration von Upstream und Downstream Processing am Beispiel der Herstellung monoklonaler Antikörper. Bachelor’s Thesis, Technische Universität Clausthal, Clausthal, Germany, Shaker Verlag GmbH, Herzogenrath, Deutschland, 2017. [Google Scholar]
- Kornecki, M.; Mestmäcker, F.; Zobel-Roos, S.; Heikaus de Figueiredo, L.; Schlüter, H.; Strube, J. Host Cell Proteins in Biologics Manufacturing: The Good, the Bad, and the Ugly. Antibodies 2017, 6, 13. [Google Scholar] [CrossRef]
- Altenhöner, U.; Meurer, M.; Strube, J.; Schmidt-Traub, H. Parameter estimation for the simulation of liquid chromatography. J. Chromatogr. A 1997, 769, 59–69. [Google Scholar] [CrossRef]
- Strube, J. Technische Chromatographie: Auslegung, Optimierung, Betrieb und Wirtschaftlichkeit; Shaker: Aachen, Germany, 2000. [Google Scholar]
- Staby, A.; Rathore, A.S.; Ahuja, S. Preparative Chromatography for Separation of Proteins; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2017. [Google Scholar]
- Meurer, M.; Altenhöner, U.; Strube, J.; Schmidt-Traub, H. Dynamic simulation of simulated moving bed chromatographic reactors. J. Chromatogr. A 1997, 769, 71–79. [Google Scholar] [CrossRef]
- Meurer, M.; Altenhöner, U.; Strube, J.; Untiedt, A.; Schmidt-Traub, H. Dynamic Simulation of a Simulated-Moving-Bed Chromatographic Reactor for the Inversion of Sucrose. Starch/Stärke 1996, 48, 452–457. [Google Scholar] [CrossRef]
- Strube, J.; Altenhöner, U.; Meurer, M.; Schmidt-Traub, H.; Schulte, M. Dynamic simulation of simulated moving-bed chromatographic processes for the optimization of chiral separations. J. Chromatogr. A 1997, 769, 81–92. [Google Scholar] [CrossRef]
- Boi, C.; Dimartino, S.; Sarti, G.C. Modelling and simulation of affinity membrane adsorption. J. Chromatogr. A 2007, 1162, 24–33. [Google Scholar] [CrossRef] [PubMed]
- Ladd Effio, C.; Hahn, T.; Seiler, J.; Oelmeier, S.A.; Asen, I.; Silberer, C.; Villain, L.; Hubbuch, J. Modeling and simulation of anion-exchange membrane chromatography for purification of Sf9 insect cell-derived virus-like particles. J. Chromatogr. A 2016, 1429, 142–154. [Google Scholar] [CrossRef] [PubMed]
- Francis, P.; von Lieres, E.; Haynes, C. Zonal rate model for stacked membrane chromatography part II: Characterizing ion-exchange membrane chromatography under protein retention conditions. Biotechnol. Bioeng. 2012, 109, 615–629. [Google Scholar] [CrossRef] [PubMed]
- Francis, P.; von Lieres, E.; Haynes, C.A. Zonal rate model for stacked membrane chromatography. I: Characterizing solute dispersion under flow-through conditions. J. Chromatogr. A 2011, 1218, 5071–5078. [Google Scholar] [CrossRef] [PubMed]
- Kaczmarski, K.; Cavazzini, A.; Szabelski, P.; Zhou, D.; Liu, X.; Guiochon, G. Application of the general rate model and the generalized Maxwell–Stefan equation to the study of the mass transfer kinetics of a pair of enantiomers. J. Chromatogr. A 2002, 962, 57–67. [Google Scholar] [CrossRef]
- Kaczmarski, K.; Gubernak, M.; Zhou, D.; Guiochon, G. Application of the general rate model with the Maxwell–Stefan equations for the prediction of the band profiles of the 1-indanol enantiomers. Chem. Eng. Sci. 2003, 58, 2325–2338. [Google Scholar] [CrossRef]
- Guiochon, G.; Felinger, A.; Shirazi, D.G.; Katti, A.M. Fundamentals of Preparative and Nonlinear Chromatography, 2nd ed.; Elsevier Academic Press: San Diego, CA, USA, 2006. [Google Scholar]
- Roper, D.K.; Lightfoot, E.N. Estimating plate heights in stacked-membrane chromatography by flow reversal. J. Chromatogr. A 1995, 702, 69–80. [Google Scholar] [CrossRef]
- Ghosh, P.; Lin, M.; Vogel, J.H.; Choy, D.; Haynes, C.; von Lieres, E. Zonal rate model for axial and radial flow membrane chromatography, part II: Model-based scale-up. Biotechnol. Bioeng. 2014, 111, 1587–1594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghosh, P.; Vahedipour, K.; Lin, M.; Vogel, J.H.; Haynes, C.A.; von Lieres, E. Zonal rate model for axial and radial flow membrane chromatography. Part I: Knowledge transfer across operating conditions and scales. Biotechnol. Bioeng. 2013, 110, 1129–1141. [Google Scholar] [CrossRef] [PubMed]
- Morbidelli, M.; Servida, A.; Storti, G.; Carra, S. Simulation of multicomponent adsorption beds. Model analysis and numerical solution. Ind. Eng. Chem. Fund. 1982, 21, 123–131. [Google Scholar] [CrossRef]
- Sommerfeld, S.; Strube, J. Challenges in biotechnology production—generic processes and process optimization for monoclonal antibodies. Chem. Eng. Process Process Intensif. 2005, 44, 1123–1137. [Google Scholar] [CrossRef]

| Module | Volumetric Flow | Gradient | Mean Residence Time (IgG) | Coefficient of Determination | ||
|---|---|---|---|---|---|---|
| Experiment | Simulation | Deviation | R² | |||
| (mL/min) | (CV) | (min) | (min) | (%) | (-) | |
| Ion exchange | 0.5 | 1 | 38.73 | 38.09 | 1.66 | 0.864 |
| 3 | 47.20 | 47.18 | 0.04 | 0.777 | ||
| 5 | 53.60 | 52.28 | 2.46 | 0.770 | ||
| Hydrophobic interaction | 0.5 | 1 | 40.26 | 39.42 | 2.09 | 0.945 |
| 3 | 46.54 | 47.18 | −1.38 | 0.848 | ||
| 5 | 53.51 | 57.36 | −7.19 | 0.647 | ||
| Mean value | 2.468 | 0.809 | ||||
| 20 L Fermenter | 200 L Fermenter | 2000 L Fermenter | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Batch | Sequential | Batch | Sequential | Batch | Sequential | ||||
| Total membrane volume | (L) | 15 | 9 | 150 | 88 | 1500 | 877 | ||
| Module size | (L) | 5 | 0.15 | 0.003 | 5 | 5 | 0.15 | 5 | 5 |
| Number of modules per cycle | (-) | 1 | 4 | 4 | 1 | 4 | 4 | 1 | 4 |
| Runs/cycles | (-) | 3 | 15 | 731 | 30 | 4 | 4 | 300 | 44 |
| Runtime | (h) | 0.16 | 3.66 | 183.24 | 1.63 | 1.10 | 36.65 | 16.33 | 10.99 |
| Productivity | |||||||||
| Run/Cycle | (g/L/day) | 2939 | 4366 | 2939 | 4366 | 2939 | 4366 | ||
| Batch to batch | (g/L/day) | 1.11 | 1.90 | 1.11 | 1.90 | 1.11 | 1.90 | ||
| Eluents consumption | (L/g) | 2.45 | 1.52 | 2.45 | 1.52 | 2.45 | 1.52 | ||
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zobel-Roos, S.; Stein, D.; Strube, J. Evaluation of Continuous Membrane Chromatography Concepts with an Enhanced Process Simulation Approach. Antibodies 2018, 7, 13. https://doi.org/10.3390/antib7010013
Zobel-Roos S, Stein D, Strube J. Evaluation of Continuous Membrane Chromatography Concepts with an Enhanced Process Simulation Approach. Antibodies. 2018; 7(1):13. https://doi.org/10.3390/antib7010013
Chicago/Turabian StyleZobel-Roos, Steffen, Dominik Stein, and Jochen Strube. 2018. "Evaluation of Continuous Membrane Chromatography Concepts with an Enhanced Process Simulation Approach" Antibodies 7, no. 1: 13. https://doi.org/10.3390/antib7010013





