Antibody Screening by Microarray Technology—Direct Identification of Selective High-Affinity Clones
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents, Buffers, Materials, and Equipment
2.2. Preparation of Epoxy Slides
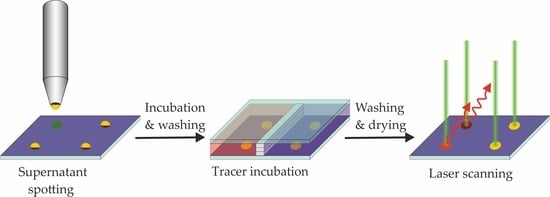
2.3. Sample Printing and Incubation
2.4. Design and Synthesis of Hapten–Fluorophore Conjugates
2.5. Competition Experiments
3. Results
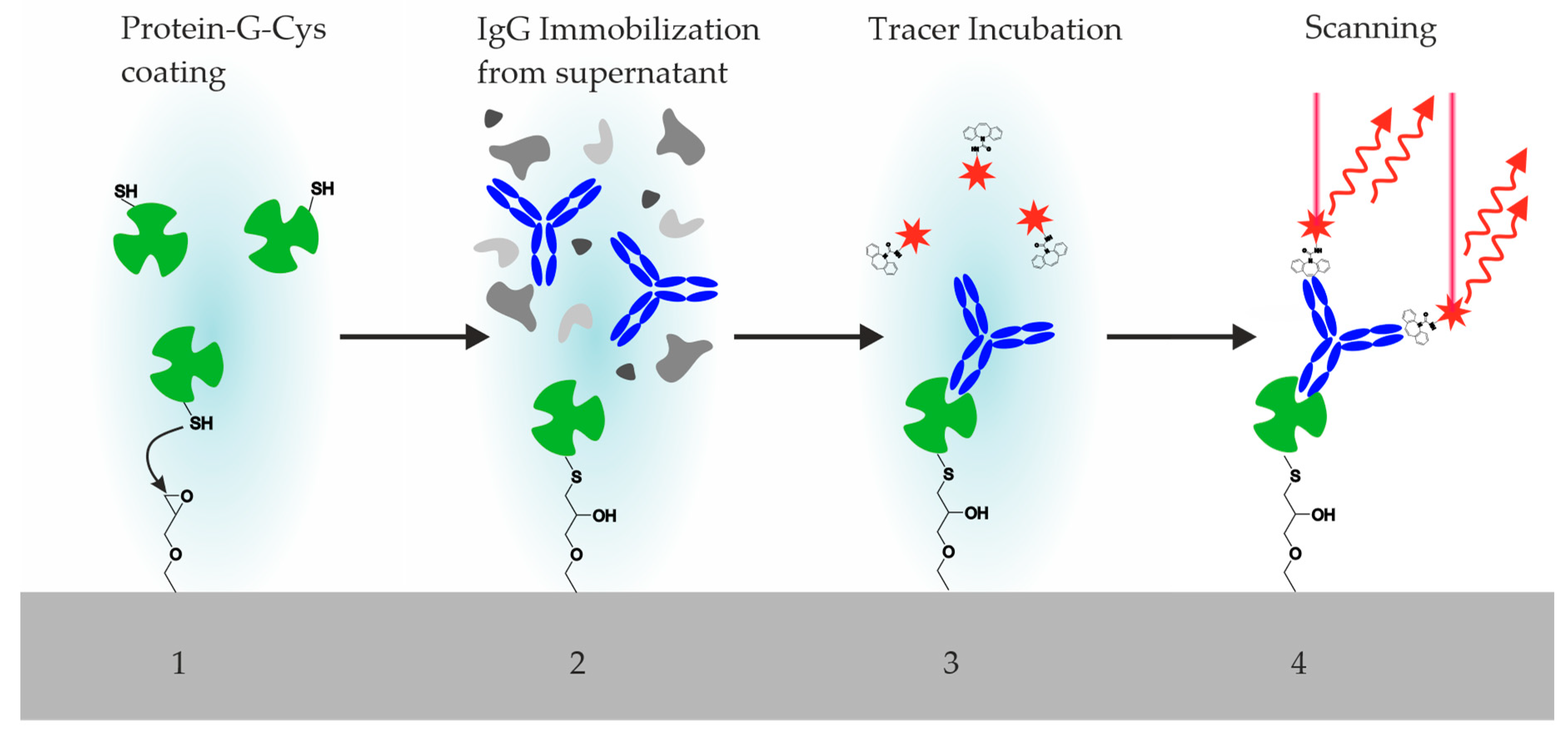
3.1. The Coating of Microarray Slides
3.2. Antibody Printing
3.3. Design and Synthesis of Hapten–Fluorophore Conjugates
3.4. Incubation Steps of Reagents and Hybridoma Supernatants
- (a)
- The dye conjugate is monovalent, which avoids confusing avidity (multivalency) effects, which are often misinterpreted.
- (b)
- This monovalent binding restricts the signals to high-affinity antibodies. With weakly binding antibodies, the tracer (labeled antigen) is washed away. The washing duration might modulate the cutoff of the detected clones according to their off rate.
- (c)
- The tracer binding is highly reversible, which makes it possible to reuse the chip without strong regeneration steps.
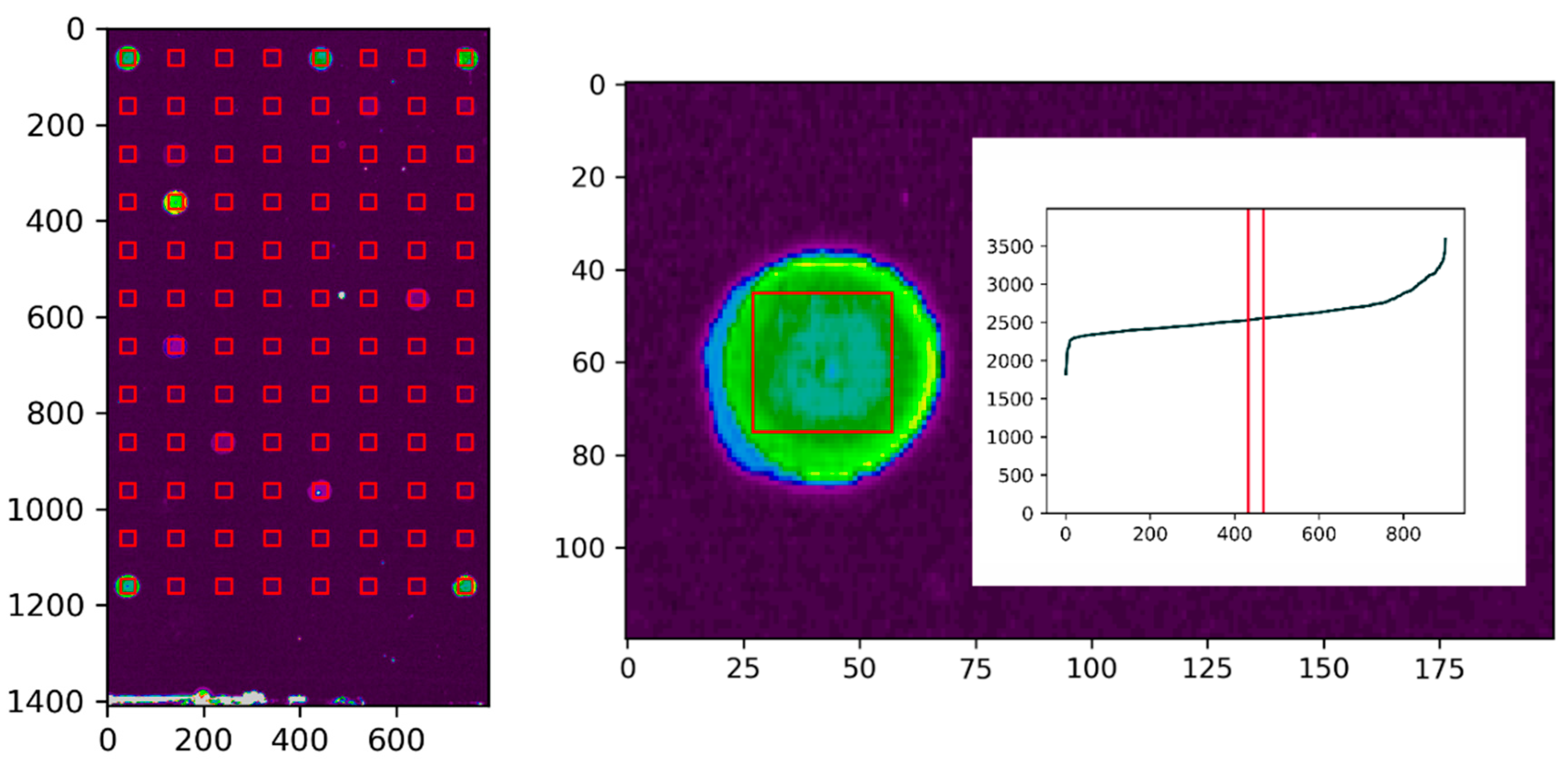
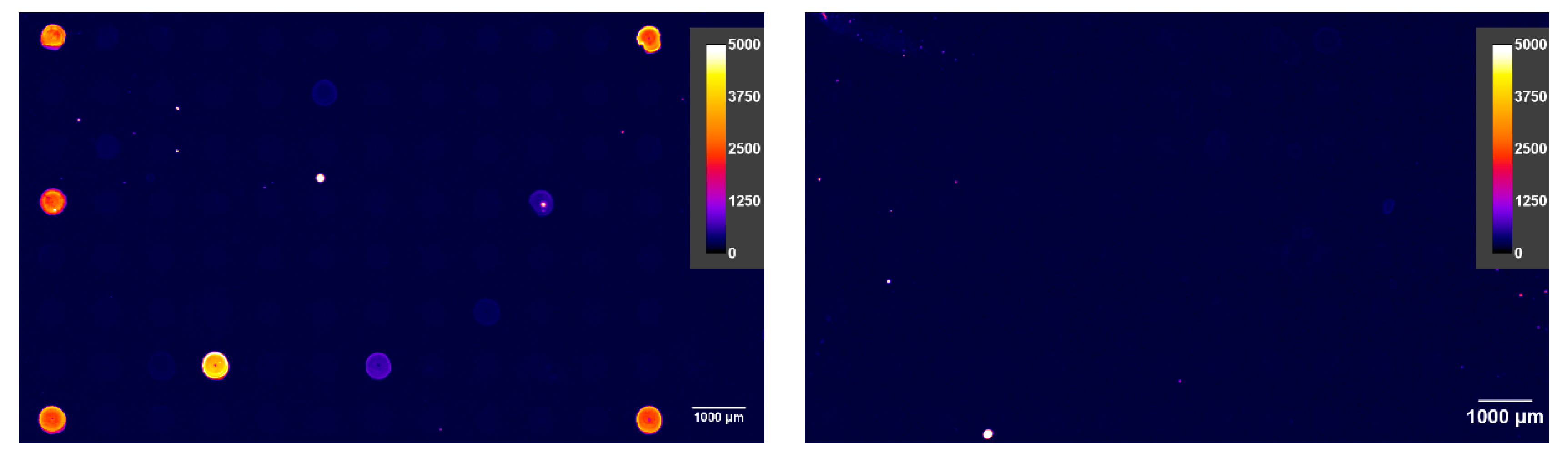
3.5. Data Evaluation
3.6. Identification and Ranking of Hybridoma Clones
3.7. Competition Experiments
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Broto, M.; McCabe, R.; Galve, R.; Marco, M.P. A high throughput immunoassay for the therapeutic drug monitoring of tegafur. Analyst 2017, 142, 2404–2410. [Google Scholar] [CrossRef] [PubMed]
- Marco, M.P.; Gee, S.; Hammock, B.D. Immunochemical Techniques for Environmental-Analysis 2. Antibody-Production and Immunoassay Development. TrAC Trend. Anal. Chem. 1995, 14, 415–425. [Google Scholar] [CrossRef]
- Zeck, A.; Weller, M.G.; Niessner, R. Characterization of a monoclonal TNT-antibody by measurement of the cross-reactivities of nitroaromatic compounds. Fresen. J. Anal. Chem. 1999, 364, 113–120. [Google Scholar] [CrossRef]
- Kim, Y.J.; Cho, Y.A.; Lee, H.S.; Lee, Y.T.; Gee, S.J.; Hammock, B.D. Synthesis of haptens for immunoassay of organophosphorus pesticides and effect of heterology in hapten spacer arm length on immunoassay sensitivity. Anal. Chim. Acta 2003, 475, 85–96. [Google Scholar] [CrossRef]
- Kusharyoto, W.; Pleiss, J.; Bachmann, T.T.; Schmid, R.D. Mapping of a hapten-binding site: Molecular modeling and site-directed mutagenesis study of an anti-atrazine antibody. Protein. Eng. 2002, 15, 233–241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weller, M.G.; Niessner, R. Affinity patterns of enzyme tracers for triazine immunoassays. Proc. Soc. Photo-Opt. Ins. 1997, 3105, 341–352. [Google Scholar] [CrossRef]
- Vandewater, C.; Haagsma, N.; Vankooten, P.J.S.; Vaneden, W. An Enzyme-Linked-Immunosorbent-Assay for the Determination of Chloramphenicol Using a Monoclonal-Antibody—Application to Residues in Swine Muscle-Tissue. Z. Lebensm. Unters. 1987, 185, 202–207. [Google Scholar]
- Harrison, R.O.; Goodrow, M.H.; Gee, S.J.; Hammock, B.D. Hapten Synthesis for Pesticide Immunoassay Development. ACS Symp. Ser. 1991, 451, 14–27. [Google Scholar]
- Xu, Z.L.; Shen, Y.D.; Beier, R.C.; Yang, J.Y.; Lei, H.T.; Wang, H.; Sun, Y.M. Application of computer-assisted molecular modeling for immunoassay of low molecular weight food contaminants: A review. Anal. Chim. Acta 2009, 647, 125–136. [Google Scholar] [CrossRef]
- Peng, D.P.; Zhang, L.Y.; Situ, C.; Pan, Y.H.; Tao, Y.F.; Wang, Y.L.; Yuan, Z.H. Development of Monoclonal Antibodies and Indirect Competitive Enzyme-Linked Immunosorbent Assay Kits for the Detection of Clenbuterol and Salbutamol in the Tissues and Products of Food-Producing Animals. Food Anal. Method 2017, 10, 3623–3633. [Google Scholar] [CrossRef]
- Birkert, O.; Tunnernann, R.; Jung, G.; Gauglitz, G. Label-free parallel screening of combinatorial triazine libraries using reflectometric interference spectroscopy. Anal. Chem. 2002, 74, 834–840. [Google Scholar] [CrossRef] [PubMed]
- Hosoda, H.; Kobayashi, N.; Ishii, N.; Nambara, T. Bridging Phenomena in Steroid Immunoassays—The Effect of Bridge Length on Sensitivity in Enzyme-Immunoassay. Chem. Pharm Bull. 1986, 34, 2105–2111. [Google Scholar] [CrossRef] [Green Version]
- Tiefenauer, L.X.; Bodmer, D.M.; Frei, W.; Andres, R.Y. Prevention of Bridge Binding in Immunoassays—A General Estradiol Tracer Structure. J. Steroid Biochem. 1989, 32, 251–257. [Google Scholar] [CrossRef]
- Suarez-Pantaleon, C.; Mercader, J.V.; Agullo, C.; Abad-Somovilla, A.; Abad-Fuentes, A. Forchlorfenuron-mimicking haptens: From immunogen design to antibody characterization by hierarchical clustering analysis. Org. Biomol. Chem. 2011, 9, 4863–4872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, Y.; Sanders, M.; Galvita, A.; Heyerick, A.; Deforce, D.; Bracke, M.; Eremin, S.A.; De Saeger, S. Heterologous screening of hybridomas for the development of broad-specific monoclonal antibodies against deoxynivalenol and its analogues. World Mycotoxin J. 2014, 7, 257–265. [Google Scholar] [CrossRef] [Green Version]
- Weller, M.G. Quality Issues of Research Antibodies. Anal. Chem. Insights 2016, 11, 21–27. [Google Scholar] [CrossRef] [Green Version]
- Chames, P.; Van Regenmortel, M.; Weiss, E.; Baty, D. Therapeutic antibodies: Successes, limitations and hopes for the future. Br. J. Pharm. 2009, 157, 220–233. [Google Scholar] [CrossRef]
- Thurber, G.M.; Schmidt, M.M.; Wittrup, K.D. Antibody tumor penetration: Transport opposed by systemic and antigen-mediated clearance. Adv. Drug Deliv. Rev. 2008, 60, 1421–1434. [Google Scholar] [CrossRef] [Green Version]
- Weller, M.G. Structural and Kinetic Investigations for the Development and Optimization of Hapten Enzyme Immunoassays (ELISAs) Exemplified by the Determination of Triazine Herbicides (Transl.); Technische Universität München: Munich, Germany, 1992; pp. 209–217. [Google Scholar]
- Knecht, B.G.; Strasser, A.; Dietrich, R.; Martlbauer, E.; Niessner, R.; Weller, M.G. Automated microarray system for the simultaneous detection of antibiotics in milk. Anal. Chem. 2004, 76, 646–654. [Google Scholar] [CrossRef]
- Angenendt, P.; Wilde, J.; Kijanka, G.; Baars, S.; Cahill, D.J.; Kreutzberger, J.; Lehrach, H.; Konthur, Z.; Glokler, J. Seeing better through a MIST: Evaluation of monoclonal recombinant antibody fragments on microarrays. Anal. Chem. 2004, 76, 2916–2921. [Google Scholar] [CrossRef]
- Templin, M.F.; Stoll, D.; Schwenk, J.M.; Potz, O.; Kramer, S.; Joos, T.O. Protein microarrays: Promising tools for proteomic research. Proteomics 2003, 3, 2155–2166. [Google Scholar] [CrossRef] [PubMed]
- Ekins, R.P. Ligand assays: From electrophoresis to miniaturized microarrays. Clin. Chem. 1998, 44, 2015–2030. [Google Scholar] [PubMed]
- Weller, M.G. Classification of protein microarrays and related techniques. Anal. Bioanal. Chem. 2003, 375, 15–17. [Google Scholar] [CrossRef] [PubMed]
- Oberleitner, L.; Dahmen-Levison, U.; Garbe, L.A.; Schneider, R.J. Improved strategies for selection and characterization of new monoclonal anti-carbamazepine antibodies during the screening process using feces and fluorescence polarization immunoassay. Anal. Methods 2016, 8, 6883–6894. [Google Scholar] [CrossRef] [Green Version]
- Dippong, M.; Carl, P.; Lenz, C.; Schenk, J.A.; Hoffmann, K.; Schwaar, T.; Schneider, R.J.; Kuhne, M. Hapten-Specific Single-Cell Selection of Hybridoma Clones by Fluorescence-Activated Cell Sorting for the Generation of Monoclonal Antibodies. Anal. Chem. 2017, 89, 4007–4012. [Google Scholar] [CrossRef]
- Rieger, M.; Cervino, C.; Sauceda, J.C.; Niessner, R.; Knopp, D. Efficient Hybridoma Screening Technique Using Capture Antibody Based Microarrays. Anal. Chem. 2009, 81, 2373–2377. [Google Scholar] [CrossRef]
- Du, H.W.; Chen, G.Y.; Bian, Y.Z.; Xing, C.Z.; Ding, X.; Zhu, M.L.; Xun, Y.P.; Chen, P.; Zhou, Y.B.; Li, S.X. Screening hybridomas for anabolic androgenic steroids by steroid analog antigen microarray. Bioanalysis 2015, 7, 1201–1209. [Google Scholar] [CrossRef]
- Staudt, N.; Muller-Sienerth, N.; Wright, G.J. Development of an antigen microarray for high throughput monoclonal antibody selection. Biochem. Bioph. Res. Commun. 2014, 445, 785–790. [Google Scholar] [CrossRef] [Green Version]
- Poetz, O.; Ostendorp, R.; Brocks, B.; Schwenk, J.M.; Stoll, D.; Joos, T.O.; Templin, M.F. Protein microarrays for antibody profiling: Specificity and affinity determination on a chip. Proteomics 2005, 5, 2402–2411. [Google Scholar] [CrossRef]
- Ma, D.H.; Baruch, D.; Shu, Y.M.; Yuan, K.H.; Sun, Z.R.; Ma, K.Y.; Hoang, T.; Fu, W.; Min, L.; Lan, Z.S.; et al. Using protein microarray technology to screen anti-ERCC1 monoclonal antibodies for specificity and applications in pathology. BMC Biotechnol. 2012, 12, 88. [Google Scholar] [CrossRef] [Green Version]
- Charlermroj, R.; Oplatowska, M.; Kumpoosiri, M.; Himananto, O.; Gajanandana, O.; Elliott, C.T.; Karoonuthaisiri, N. Comparison of techniques to screen and characterize bacteria-specific hybridomas for high-quality monoclonal antibodies selection. Anal. Biochem. 2012, 421, 26–36. [Google Scholar] [CrossRef] [PubMed]
- Di Cristina, M.; Nunziangeli, L.; Giubilei, M.A.; Capuccini, B.; d’Episcopo, L.; Mazzoleni, G.; Baldracchini, F.; Spaccapelo, R.; Crisanti, A. An antigen microarray immunoassay for multiplex screening of mouse monoclonal antibodies. Nat. Protoc. 2010, 5, 1932–1944. [Google Scholar] [CrossRef] [PubMed]
- De Masi, F.; Chiarella, P.; Wilhelm, H.; Massimi, M.; Belinda, B.; Ansorge, W.; Sawyer, A. High throughput production of mouse monoclonal antibodies using antigen microarrays. Proteomics 2005, 5, 4070–4081. [Google Scholar] [CrossRef] [PubMed]
- Chiarella, P.; Fazio, V.M. Mouse monoclonal antibodies in biological research: Strategies for high-throughput production. Biotechnol. Lett. 2008, 30, 1303–1310. [Google Scholar] [CrossRef] [PubMed]
- Moller, I.; Marcus, S.E.; Haeger, A.; Verhertbruggen, Y.; Verhoef, R.; Schols, H.; Ulvskov, P.; Mikkelsen, J.D.; Knox, J.P.; Willats, W. High-throughput screening of monoclonal antibodies against plant cell wall glycans by hierarchical clustering of their carbohydrate microarray binding profiles. Glycoconj. J. 2008, 25, 37–48. [Google Scholar] [CrossRef] [Green Version]
- Oberleitner, L.; Dahmen-Levison, U.; Garbe, L.A.; Schneider, R.J. Application of fluorescence polarization immunoassay for determination of carbamazepine in wastewater. J. Environ. Manag. 2017, 193, 92–97. [Google Scholar] [CrossRef]
- Carl, P.; Sarma, D.; Gregorio, B.J.R.; Hoffmann, K.; Lehmann, A.; Rurack, K.; Schneider, R.J. Wash-Free Multiplexed Mix-and-Read Suspension Array Fluorescence Immunoassay for Anthropogenic Markers in Wastewater. Anal. Chem. 2019, 91, 12988–12996. [Google Scholar] [CrossRef]
- Bahlmann, A.; Weller, M.G.; Panne, U.; Schneider, R.J. Monitoring carbamazepine in surface and wastewaters by an immunoassay based on a monoclonal antibody. Anal. Bioanal. Chem. 2009, 395, 1809–1820. [Google Scholar] [CrossRef]
- Bahlmann, A.; Carvalho, J.J.; Weller, M.G.; Panne, U.; Schneider, R.J. Immunoassays as high-throughput tools: Monitoring spatial and temporal variations of carbamazepine, caffeine and cetirizine in surface and wastewaters. Chemosphere 2012, 89, 1278–1286. [Google Scholar] [CrossRef]
- Bahlmann, A.; Falkenhagen, J.; Weller, M.G.; Panne, U.; Schneider, R.J. Cetirizine as pH-dependent cross-reactant in a carbamazepine-specific immunoassay. Analyst 2011, 136, 1357–1364. [Google Scholar] [CrossRef]
- Calisto, V.; Bahlmann, A.; Schneider, R.J.; Esteves, V.I. Application of an ELISA to the quantification of carbamazepine in ground, surface and wastewaters and validation with LC-MS/MS. Chemosphere 2011, 84, 1708–1715. [Google Scholar] [CrossRef] [PubMed]
- Grandke, J.; Resch-Genger, U.; Bremser, W.; Garbe, L.A.; Schneider, R.J. Quality assurance in immunoassay performance-temperature effects. Anal. Methods 2012, 4, 901–905. [Google Scholar] [CrossRef]
- Grandke, J.; Oberleitner, L.; Resch-Genger, U.; Garbe, L.A.; Schneider, R.J. Quality assurance in immunoassay performance—Carbamazepine immunoassay format evaluation and application on surface and waste water. Anal. Methods 2013, 5, 3754–3760. [Google Scholar] [CrossRef] [Green Version]
- Oberleitner, L.; Eremin, S.A.; Lehmann, A.; Garbe, L.A.; Schneider, R.J. Fluorescence polarization immunoassays for carbamazepine—Comparison of tracers and formats. Anal. Methods 2015, 7, 5854–5861. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rueden, C.T.; Schindelin, J.; Hiner, M.C.; DeZonia, B.E.; Walter, A.E.; Arena, E.T.; Eliceiri, K.W. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinform. 2017, 18. [Google Scholar] [CrossRef]
- Kusnezow, W.; Hoheisel, J.D. Solid supports for microarray immunoassays. J. Mol. Recognit. 2003, 16, 165–176. [Google Scholar] [CrossRef]
- Domnanich, P.; Sauer, U.; Pultar, J.; Preininger, C. Protein microarray for the analysis of human melanoma biomarkers. Sens. Actuators B Chem. 2009, 139, 2–8. [Google Scholar] [CrossRef]
- Bjorck, L.; Kronvall, G. Purification and Some Properties of Streptococcal Protein-G, Protein-a Novel Igg-Binding Reagent. J. Immunol. 1984, 133, 969–974. [Google Scholar]
- Lu, B.; Smyth, M.R.; OKennedy, R. Oriented immobilization of antibodies and its applications in immunoassays and immunosensors. Analyst 1996, 121, R29–R32. [Google Scholar] [CrossRef]
- Schroeder, B.; Le Xuan, H.; Völzke, J.L.; Weller, M.G. Preactivation Crosslinking—An Efficient Method for the Oriented Immobilization of Antibodies. Methods Protoc. 2019, 2, 35. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.M.; Park, H.K.; Jung, Y.; Kim, J.K.; Jung, S.O.; Chung, B.H. Direct immobilization of protein G variants with various numbers of cysteine residues on a gold surface. Anal. Chem. 2007, 79, 2680–2687. [Google Scholar] [CrossRef] [PubMed]
- Kanno, S.; Yanagida, Y.; Haruyama, T.; Kobatake, E.; Aizawa, M. Assembling of engineered IgG-binding protein on gold surface for highly oriented antibody immobilization. J. Biotechnol. 2000, 76, 207–214. [Google Scholar] [CrossRef]
- Schuck, P. Use of surface plasmon resonance to probe the equilibrium and dynamic aspects of interactions between biological macromolecules. Annu. Rev. Biophys. Biom. 1997, 26, 541–566. [Google Scholar] [CrossRef]
- Schuck, P. Reliable determination of binding affinity and kinetics using surface plasmon resonance biosensors. Curr. Opin. Biotechnol. 1997, 8, 498–502. [Google Scholar] [CrossRef]
- MacKenzie, C.R.; Hirama, T.; Deng, S.J.; Bundle, D.R.; Narang, S.A.; Young, N.M. Analysis by surface plasmon resonance of the influence of valence on the ligand binding affinity and kinetics of an anti-carbohydrate antibody. J. Biol. Chem. 1996, 271, 1527–1533. [Google Scholar] [CrossRef] [Green Version]
- Nieba, L.; Krebber, A.; Plückthun, A. Competition BIAcore for measuring true affinities: Large differences from values determined from binding kinetics. Anal. Biochem. 1996, 234, 155–165. [Google Scholar] [CrossRef] [Green Version]
- Baker, M. Blame It on the Antibodies. Nature 2015, 521, 274–276. [Google Scholar] [CrossRef] [Green Version]







| Antibody | IC50 (µg/L) | Isotype | Rating | Test Result (1 mg/L) | Test Result (10 mg/L) | References |
|---|---|---|---|---|---|---|
| BAM-mab 01 (CBZ) * | 0.32 | IgG1 | good | + + | + + | [25,37,38] |
| B3212M | 0.15 | IgG1 | good | + + | + + | [25,39,40,41,42,43,44,45] |
| 6C5 | 23.0 | IgG1 | poor | − − | − − | [26] |
| 3B3 | 1700 | IgG1 | very poor | − − | − − | [26] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paul, M.; Weller, M.G. Antibody Screening by Microarray Technology—Direct Identification of Selective High-Affinity Clones. Antibodies 2020, 9, 1. https://doi.org/10.3390/antib9010001
Paul M, Weller MG. Antibody Screening by Microarray Technology—Direct Identification of Selective High-Affinity Clones. Antibodies. 2020; 9(1):1. https://doi.org/10.3390/antib9010001
Chicago/Turabian StylePaul, Martin, and Michael G. Weller. 2020. "Antibody Screening by Microarray Technology—Direct Identification of Selective High-Affinity Clones" Antibodies 9, no. 1: 1. https://doi.org/10.3390/antib9010001