In Situ Atomic Force Microscopy Studies on Nucleation and Self-Assembly of Biogenic and Bio-Inspired Materials
Abstract
:1. Introduction
2. Soft Biogenic Materials
2.1. Peptide and Protein
2.2. Nucleic Acid
2.3. Lipid
3. Soft Bio-Inspired Materials
3.1. Peptoid
3.2. Other Polymers
4. Hard Materials
4.1. Calcium Phosphate
4.2. Calcite
4.3. Calcium Oxalate Monohydrate
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Wainwright, S.A. Mechanical Design in Organisms; Princeton University Press: Princeton, NJ, USA, 1982. [Google Scholar]
- Vincent, J.F. Structural Biomaterials; Princeton University Press: Princeton, NJ, USA, 2012. [Google Scholar]
- Vollrath, F. Biology of spider silk. Int. J. Biol. Macromol. 1999, 24, 81–88. [Google Scholar] [CrossRef]
- Heim, M.; Keerl, D.; Scheibel, T. Spider silk: From soluble protein to extraordinary fiber. Angew. Chem. Int. Ed. 2009, 48, 3584–3596. [Google Scholar] [CrossRef] [PubMed]
- Gosline, J.M.; DeMont, M.E.; Denny, M.W. The structure and properties of spider silk. Endeavour 1986, 10, 37–43. [Google Scholar] [CrossRef]
- Rammensee, S.; Slotta, U.; Scheibel, T.; Bausch, A. Assembly mechanism of recombinant spider silk proteins. Proc. Natl. Acad. Sci. USA 2008, 105, 6590–6595. [Google Scholar] [CrossRef] [PubMed]
- Shao, Z.; Vollrath, F. Materials: Surprising strength of silkworm silk. Nature 2002, 418, 741. [Google Scholar] [CrossRef] [PubMed]
- Hinman, M.B.; Jones, J.A.; Lewis, R.V. Synthetic spider silk: A modular fiber. Trends Biotechnol. 2000, 18, 374–379. [Google Scholar] [CrossRef]
- Lee, S.-M.; Pippel, E.; Gösele, U.; Dresbach, C.; Qin, Y.; Chandran, C.V.; Bräuniger, T.; Hause, G.; Knez, M. Greatly increased toughness of infiltrated spider silk. Science 2009, 324, 488–492. [Google Scholar] [CrossRef] [PubMed]
- O'Brien, J.P.; Fahnestock, S.R.; Termonia, Y.; Gardner, K.H. Nylons from nature: Synthetic analogs to spider silk. Adv. Mater. 1998, 10, 1185–1195. [Google Scholar] [CrossRef]
- Lewis, R.V. Spider silk: Ancient ideas for new biomaterials. Chem. Rev. 2006, 106, 3762–3774. [Google Scholar] [CrossRef] [PubMed]
- Whitesides, G.M.; Boncheva, M. Beyond molecules: Self-assembly of mesoscopic and macroscopic components. Proc. Natl. Acad. Sci. USA 2002, 99, 4769–4774. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S. Fabrication of novel biomaterials through molecular self-assembly. Nat. Biotechnol. 2003, 21, 1171–1178. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Woodhouse, K.A.; Yip, C.M. Substrate-facilitated assembly of elastin-like peptides: Studies by variable-temperature in situ atomic force microscopy. J. Am. Chem. Soc. 2002, 124, 10648–10649. [Google Scholar] [CrossRef] [PubMed]
- Zhong, J.; Ma, M.; Zhou, J.; Wei, D.; Yan, Z.; He, D. Tip-induced micropatterning of silk fibroin protein using in situ solution atomic force microscopy. ACS Appl. Mater. Interfaces 2013, 5, 737–746. [Google Scholar] [CrossRef] [PubMed]
- Zhao, N.; Wang, Z.; Cai, C.; Shen, H.; Liang, F.; Wang, D.; Wang, C.; Zhu, T.; Guo, J.; Wang, Y. Bioinspired materials: From low to high dimensional structure. Adv. Mater. 2014, 26, 6994–7017. [Google Scholar] [CrossRef] [PubMed]
- Chworos, A.; Smitthipong, W. Bio-inspired materials. In Bio-Based Composites for High-Performance Materials: From Strategy to Industrial Application; CRC Press: Boca Raton, FL, USA, 2014; Volume 43. [Google Scholar]
- Barron, A.E.; Zuckerman, R.N. Bioinspired polymeric materials: In-between proteins and plastics. Curr. Opin. Chem. Biol. 1999, 3, 681–687. [Google Scholar] [CrossRef]
- Sanchez, C.; Arribart, H.; Guille, M.M.G. Biomimetism and bioinspiration as tools for the design of innovative materials and systems. Nat. Mater. 2005, 4, 277–288. [Google Scholar] [CrossRef] [PubMed]
- Wegst, U.G.; Bai, H.; Saiz, E.; Tomsia, A.P.; Ritchie, R.O. Bioinspired structural materials. Nat. Mater. 2015, 14, 23–36. [Google Scholar] [CrossRef] [PubMed]
- Dujardin, E.; Mann, S. Bio-inspired materials chemistry. Adv. Mater. 2002, 14, 775. [Google Scholar] [CrossRef]
- Aizenberg, J.; Fratzl, P. Biological and biomimetic materials. Adv. Mater. 2009, 21, 387–388. [Google Scholar] [CrossRef]
- Sarikaya, M.; Tamerler, C.; Jen, A.K.-Y.; Schulten, K.; Baneyx, F. Molecular biomimetics: Nanotechnology through biology. Nat. Mater. 2003, 2, 577–585. [Google Scholar] [CrossRef] [PubMed]
- Lindsey, J.S. Self-assembly in synthetic routes to molecular devices. Biological principles and chemical perspectives: A review. New J. Chem. 1991, 15, 153–179. [Google Scholar] [CrossRef]
- Mayer, M.; Tebbe, M.; Kuttner, C.; Schnepf, M.J.; König, T.A.; Fery, A. Template-assisted colloidal self-assembly of macroscopic magnetic metasurfaces. Faraday Discuss. 2016, 191, 159–176. [Google Scholar] [CrossRef] [PubMed]
- Pan, H.M.; Seuss, M.; Neubauer, M.P.; Trau, D.W.; Fery, A. Tuning the mechanical properties of hydrogel core–shell particles by inwards interweaving self-assembly. ACS Appl. Mater. Interfaces 2016, 8, 1493–1500. [Google Scholar] [CrossRef] [PubMed]
- Kuttner, C.; Hanisch, A.; Schmalz, H.; Eder, M.; Schlaad, H.; Burgert, I.; Fery, A. Influence of the polymeric interphase design on the interfacial properties of (fiber-reinforced) composites. ACS Appl. Mater. Interfaces 2013, 5, 2469–2478. [Google Scholar] [CrossRef] [PubMed]
- Kuttner, C.; Tebbe, M.; Schlaad, H.; Burgert, I.; Fery, A. Photochemical synthesis of polymeric fiber coatings and their embedding in matrix material: Morphology and nanomechanical properties at the fiber-matrix interface. ACS Appl. Mater. Interfaces 2012, 4, 3484–3492. [Google Scholar] [CrossRef] [PubMed]
- Fratzl, P.; Burgert, I.; Gupta, H.S. On the role of interface polymers for the mechanics of natural polymeric composites. Phys. Chem. Chem. Phys. 2004, 6, 5575–5579. [Google Scholar] [CrossRef]
- Kuttner, C. Macromolecular Interphases and Interfaces in Composite Materials; Verlag Dr. Hut: München, Germany, 2014. [Google Scholar]
- Ma, X.; Zhang, S.; Jiao, F.; Newcomb, C.J.; Zhang, Y.; Prakash, A.; Liao, Z.; Baer, M.D.; Mundy, C.J.; Pfaendtner, J. Tuning crystallization pathways through sequence engineering of biomimetic polymers. Nat. Mater. 2017. [Google Scholar] [CrossRef] [PubMed]
- Robertson, E.J.; Battigelli, A.; Proulx, C.; Mannige, R.V.; Haxton, T.K.; Yun, L.; Whitelam, S.; Zuckermann, R.N. Design, synthesis, assembly, and engineering of peptoid nanosheets. Acc. Chem. Res. 2016, 49, 379–389. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Jiao, F.; Daily, M.D.; Chen, Y.; Yan, F.; Ding, Y.-H.; Zhang, X.; Robertson, E.J.; Baer, M.D.; Chen, C.-L. Highly stable and self-repairing membrane-mimetic 2d nanomaterials assembled from lipid-like peptoids. Nat. Commun. 2016, 7, 12252. [Google Scholar] [CrossRef] [PubMed]
- Wagner, R.W.; Lindsey, J.S. A molecular photonic wire. J. Am. Chem. Soc. 1994, 116, 9759–9760. [Google Scholar] [CrossRef]
- Nam, Y.S.; Shin, T.; Park, H.; Magyar, A.P.; Choi, K.; Fantner, G.; Nelson, K.A.; Belcher, A.M. Virus-templated assembly of porphyrins into light-harvesting nanoantennae. J. Am. Chem. Soc. 2010, 132, 1462–1463. [Google Scholar] [CrossRef] [PubMed]
- Calzaferri, G.; Bossart, O.; Brühwiler, D.; Huber, S.; Leiggener, C.; Van Veen, M.K.; Ruiz, A.Z. Light-harvesting host–guest antenna materials for quantum solar energy conversion devices. C. R. Chim. 2006, 9, 214–225. [Google Scholar] [CrossRef]
- Choi, M.S.; Yamazaki, T.; Yamazaki, I.; Aida, T. Bioinspired molecular design of light—Harvesting multiporphyrin arrays. Angew. Chem. Int. Ed. 2004, 43, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Guo, Z. Biomimetic photonic materials with tunable structural colors. J. Coll. Interface Sci. 2013, 406, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Andrews, D.L. Energy Harvesting Materials; World Scientific: Singapore, 2005. [Google Scholar]
- Jacobs, M.; Lopez-Garcia, M.; Phrathep, O.-P.; Lawson, T.; Oulton, R.; Whitney, H.M. Photonic multilayer structure of begonia chloroplasts enhances photosynthetic efficiency. Nat. Plants 2016, 2, 16162. [Google Scholar] [CrossRef] [PubMed]
- Calver, C.F.; Schanze, K.S.; Cosa, G. Biomimetic light-harvesting antenna based on the self-assembly of conjugated polyelectrolytes embedded within lipid membranes. ACS Nano 2016, 10, 10598–10605. [Google Scholar] [CrossRef] [PubMed]
- Henry, S.L.; Withers, J.M.; Singh, I.; Cooper, J.M.; Clark, A.W.; Burley, G.A.; Cogdell, R.J. DNA-directed spatial assembly of photosynthetic light-harvesting proteins. Organ. Biomol. Chem. 2016, 14, 1359–1362. [Google Scholar] [CrossRef] [PubMed]
- Hemmig, E.A.; Creatore, C.; Wünsch, B.; Hecker, L.; Mair, P.; Parker, M.A.; Emmott, S.; Tinnefeld, P.; Keyser, U.F.; Chin, A.W. Programming light-harvesting efficiency using DNA origami. Nano Lett. 2016, 16, 2369–2374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kundu, S.; Patra, A. Nanoscale strategies for light harvesting. Chem. Rev. 2017, 117, 712–757. [Google Scholar] [CrossRef] [PubMed]
- Parker, A.R.; Lawrence, C.R. Water capture by a desert beetle. Nature 2001, 414, 33–34. [Google Scholar] [CrossRef] [PubMed]
- White, B.; Sarkar, A.; Kietzig, A.-M. Fog-harvesting inspired by the stenocara beetle—An analysis of drop collection and removal from biomimetic samples with wetting contrast. Appl. Surface Sci. 2013, 284, 826–836. [Google Scholar] [CrossRef]
- Garrod, R.; Harris, L.; Schofield, W.; McGettrick, J.; Ward, L.; Teare, D.; Badyal, J. Mimicking a stenocara beetle’s back for microcondensation using plasmachemical patterned superhydrophobic-superhydrophilic surfaces. Langmuir 2007, 23, 689–693. [Google Scholar] [CrossRef] [PubMed]
- Pang, C.; Kim, S.M.; Rahmawan, Y.; Suh, K.-Y. Beetle-inspired bidirectional, asymmetric interlocking using geometry-tunable nanohairs. ACS Appl. Mater. Interfaces 2012, 4, 4225–4230. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Guo, Z.; Liu, W. Biomimetic water-collecting materials inspired by nature. Chem. Commun. 2016, 52, 3863–3879. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Guo, Z. Hybrid engineered materials with high water-collecting efficiency inspired by namib desert beetles. Chem. Commun. 2016, 52, 6809–6812. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Qian, L.; Yuan, X.; Zhou, C.; Li, Z.; Cheng, J.; Xu, S.; Wang, S.; Pi, P.; Wen, X. Inspired by stenocara beetles: From water collection to high-efficiency water-in-oil emulsion separation. ACS Nano 2016, 11, 760–769. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Huang, J.; Chen, Z.; Lai, Y. Bioinspired special wettability surfaces: From fundamental research to water harvesting applications. Small 2016. [Google Scholar] [CrossRef] [PubMed]
- Espinosa, H.D.; Rim, J.E.; Barthelat, F.; Buehler, M.J. Merger of structure and material in nacre and bone—Perspectives on de novo biomimetic materials. Prog. Mater. Sci. 2009, 54, 1059–1100. [Google Scholar] [CrossRef]
- Finnemore, A.; Cunha, P.; Shean, T.; Vignolini, S.; Guldin, S.; Oyen, M.; Steiner, U. Biomimetic layer-by-layer assembly of artificial nacre. Nat. Commun. 2012, 3, 966. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Kotov, N.A.; Magonov, S.; Ozturk, B. Nanostructured artificial nacre. Nat. Mater. 2003, 2, 413–418. [Google Scholar] [CrossRef] [PubMed]
- Katti, K.S.; Mohanty, B.; Katti, D.R. Biomimetic Lessons Learnt from Nacre; INTECH Open Access: Rijeka, Croatia, 2010. [Google Scholar]
- Li, C.; Born, A.K.; Schweizer, T.; Zenobi-Wong, M.; Cerruti, M.; Mezzenga, R. Amyloid-hydroxyapatite bone biomimetic composites. Adv. Mater. 2014, 26, 3207–3212. [Google Scholar] [CrossRef] [PubMed]
- Thein-Han, W.; Misra, R. Biomimetic chitosan-nanohydroxyapatite composite scaffolds for bone tissue engineering. Acta Biomater. 2009, 5, 1182–1197. [Google Scholar] [CrossRef] [PubMed]
- Ng, J.; Spiller, K.; Bernhard, J.; Vunjak-Novakovic, G. Biomimetic approaches for bone tissue engineering. Tissue Eng. Part B Rev. 2017. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Heredia, M.A.; Łapa, A.; Mendes, A.C.; Balcaen, L.; Samal, S.K.; Chai, F.; Van der Voort, P.; Stevens, C.V.; Parakhonskiy, B.V.; Chronakis, I.S. Bioinspired, biomimetic, double-enzymatic mineralization of hydrogels for bone regeneration with calcium carbonate. Mater. Lett. 2017, 190, 13–16. [Google Scholar] [CrossRef]
- Binnig, G.; Quate, C.F.; Gerber, C. Atomic force microscope. Phys. Rev. Lett. 1986, 56, 930–933. [Google Scholar] [CrossRef] [PubMed]
- Bustamante, C.; Vesenka, J.; Tang, C.L.; Rees, W.; Guthold, M.; Keller, R. Circular DNA molecules imaged in air by scanning force microscopy. Biochemistry 1992, 31, 22–26. [Google Scholar] [CrossRef] [PubMed]
- Hansma, H.G.; Sinsheimer, R.L.; Li, M.-Q.; Hansma, P.K. Atomic force microscopy of single-and double-stranded DNA. Nucleic Acids Res. 1992, 20, 3585–3590. [Google Scholar] [CrossRef] [PubMed]
- Hansma, H.G.; Laney, D.E. DNA binding to mica correlates with cationic radius: Assay by atomic force microscopy. Biophys. J. 1996, 70, 1933–1939. [Google Scholar] [CrossRef]
- Ando, T.; Uchihashi, T.; Kodera, N. High-speed afm and applications to biomolecular systems. Annu. Rev. Biophys. 2013, 42, 393–414. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsov, Y.G.; Malkin, A.J.; Lucas, R.W.; Plomp, M.; McPherson, A. Imaging of viruses by atomic force microscopy. J. Gen. Virol. 2001, 82, 2025–2034. [Google Scholar] [CrossRef] [PubMed]
- De Pablo, P.J.; Carrión-Vázquez, M. Imaging biological samples with atomic force microscopy. Cold Spring Harb. Protoc. 2014, 2014, 167–177. [Google Scholar] [CrossRef] [PubMed]
- Butt, H.J.; Wolff, E.K.; Gould, S.A.C.; Dixon Northern, B.; Peterson, C.M.; Hansma, P.K. Imaging cells with the atomic force microscope. J. Struct. Biol. 1990, 105, 54–61. [Google Scholar] [CrossRef]
- Dufrêne, Y.F.; Ando, T.; Garcia, R.; Alsteens, D.; Martinez-Martin, D.; Engel, A.; Gerber, C.; Müller, D.J. Imaging modes of atomic force microscopy for application in molecular and cell biology. Nat. Nanotechnol. 2017, 12, 295–307. [Google Scholar] [CrossRef] [PubMed]
- Rugar, D.; Hansma, P. Atomic force microscopy. Phys. Today 1990, 43, 23–30. [Google Scholar] [CrossRef]
- Junno, T.; Anand, S.; Deppert, K.; Montelius, L.; Samuelson, L. Contact mode atomic force microscopy imaging of nanometer—Sized particles. Appl. Phys. Lett. 1995, 66, 3295–3297. [Google Scholar] [CrossRef]
- Le Grimellec, C.; Lesniewska, E.; Giocondi, M.-C.; Finot, E.; Vié, V.; Goudonnet, J.-P. Imaging of the surface of living cells by low-force contact-mode atomic force microscopy. Biophys. J. 1998, 75, 695–703. [Google Scholar] [CrossRef]
- Cleveland, J.; Anczykowski, B.; Schmid, A.; Elings, V. Energy dissipation in tapping-mode atomic force microscopy. Appl. Phys. Lett. 1998, 72, 2613–2615. [Google Scholar] [CrossRef]
- Hansma, P.; Cleveland, J.; Radmacher, M.; Walters, D.; Hillner, P.; Bezanilla, M.; Fritz, M.; Vie, D.; Hansma, H.; Prater, C. Tapping mode atomic force microscopy in liquids. Appl. Phys. Lett. 1994, 64, 1738–1740. [Google Scholar] [CrossRef]
- San Paulo, A.; Garcia, R. Unifying theory of tapping-mode atomic-force microscopy. Phys. Rev. B 2002, 66, 041406. [Google Scholar] [CrossRef]
- Tamayo, J.; Garcia, R. Deformation, contact time, and phase contrast in tapping mode scanning force microscopy. Langmuir 1996, 12, 4430–4435. [Google Scholar] [CrossRef]
- Kowalewski, T.; Holtzman, D.M. In situ atomic force microscopy study of alzheimer’s β-amyloid peptide on different substrates: New insights into mechanism of β-sheet formation. Proc. Natl. Acad. Sci. USA 1999, 96, 3688–3693. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, S.-I.; Iwatsuki, M. Observation of 7 × 7 reconstructed structure on the silicon (111) surface using ultrahigh vacuum noncontact atomic force microscopy. Jpn. J. Appl. Phys. 1995, 34, L145. [Google Scholar] [CrossRef]
- Morita, S.; Giessibl, F.J.; Meyer, E.; Wiesendanger, R. Noncontact Atomic Force Microscopy; Springer: Berlin, Germany, 2015; Volume 3. [Google Scholar]
- Ramachandran, T.; Baur, C.; Bugacov, A.; Madhukar, A.; Koel, B.; Requicha, A.; Gazen, C. Direct and controlled manipulation of nanometer-sized particles using the non-contact atomic force microscope. Nanotechnology 1998, 9, 237. [Google Scholar] [CrossRef]
- Sugimoto, Y.; Pou, P.; Abe, M.; Jelinek, P.; Pérez, R.; Morita, S.; Custance, O. Chemical identification of individual surface atoms by atomic force microscopy. Nature 2007, 446, 64–67. [Google Scholar] [CrossRef] [PubMed]
- Trtik, P.; Kaufmann, J.; Volz, U. On the use of peak-force tapping atomic force microscopy for quantification of the local elastic modulus in hardened cement paste. Cem. Concr. Res. 2012, 42, 215–221. [Google Scholar] [CrossRef]
- Alsteens, D.; Dupres, V.; Yunus, S.; Latgé, J.-P.; Heinisch, J.J.; Dufrêne, Y.F. High-resolution imaging of chemical and biological sites on living cells using peak force tapping atomic force microscopy. Langmuir 2012, 28, 16738–16744. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Song, Y.; Wang, S.; Dai, B.; Zhang, L.; Dong, Y.; Lü, J.; Hu, J. Mechanical mapping of nanobubbles by peakforce atomic force microscopy. Soft Matter 2013, 9, 8837–8843. [Google Scholar] [CrossRef]
- Foster, B. New atomic force microscopy(afm) approaches life sciences gently, quantitatively, and correctively. Am. Lab. 2012, 44, 24–28. [Google Scholar]
- Glatz, B.A.; Tebbe, M.; Kaoui, B.; Aichele, R.; Kuttner, C.; Schedl, A.E.; Schmidt, H.-W.; Zimmermann, W.; Fery, A. Hierarchical line-defect patterns in wrinkled surfaces. Soft Matter 2015, 11, 3332–3339. [Google Scholar] [CrossRef] [PubMed]
- De Yoreo, J.J.; Chung, S.; Friddle, R.W. In situ atomic force microscopy as a tool for investigating interactions and assembly dynamics in biomolecular and biomineral systems. Adv. Funct. Mater. 2013, 23, 2525–2538. [Google Scholar] [CrossRef]
- Schitter, G.; Astrom, K.J.; DeMartini, B.E.; Thurner, P.J.; Turner, K.L.; Hansma, P.K. Design and modeling of a high-speed afm-scanner. IEEE Trans. Control Syst. Technol. 2007, 15, 906–915. [Google Scholar] [CrossRef]
- Fantner, G.E.; Schitter, G.; Kindt, J.H.; Ivanov, T.; Ivanova, K.; Patel, R.; Holten-Andersen, N.; Adams, J.; Thurner, P.J.; Rangelow, I.W. Components for high speed atomic force microscopy. Ultramicroscopy 2006, 106, 881–887. [Google Scholar] [CrossRef] [PubMed]
- Sulchek, T.; Hsieh, R.; Adams, J.; Minne, S.; Quate, C.; Adderton, D. High-speed atomic force microscopy in liquid. Rev. Sci. Instrum. 2000, 71, 2097–2099. [Google Scholar] [CrossRef]
- Hartman, B.; Andersson, S.; Nagel, W.; Leang, K. Non-raster high-speed afm imaging of biopolymers. Biophys. J. 2017, 112, 587a. [Google Scholar] [CrossRef]
- Takahashi, H.; Miyagi, A.; Redondo-Morata, L.; Scheuring, S. Development of temperature-controlled high-speed afm. Biophys. J. 2017, 112, 587a. [Google Scholar] [CrossRef]
- Katan, A.J.; Dekker, C. High-speed afm reveals the dynamics of single biomolecules at the nanometer scale. Cell 2011, 147, 979–982. [Google Scholar] [CrossRef] [PubMed]
- Ando, T.; Kodera, N.; Takai, E.; Maruyama, D.; Saito, K.; Toda, A. A high-speed atomic force microscope for studying biological macromolecules. Proc. Natl. Acad. Sci. USA 2001, 98, 12468–12472. [Google Scholar] [CrossRef] [PubMed]
- Uchihashi, T.; Iino, R.; Ando, T.; Noji, H. High-speed atomic force microscopy reveals rotary catalysis of rotorless f1-atpase. Science 2011, 333, 755–758. [Google Scholar] [CrossRef] [PubMed]
- Ruan, Y.; Miyagi, A.; Wang, X.; Chami, M.; Boudker, O.; Scheuring, S. Direct visualization of glutamate transporter elevator mechanism by high-speed afm. Proc. Natl. Acad. Sci. USA 2017, 114, 1584–1588. [Google Scholar] [CrossRef] [PubMed]
- Kotani, N.; Kumaresan, R.; Kawamoto-Ozaki, Y.; Morii, T.; Okada, T. High-speed afm reveals advanced details on dynamic behavior of antibody. Biophys. J. 2017, 112, 587a. [Google Scholar] [CrossRef]
- Watanabe-Nakayama, T.; Ono, K.; Itami, M.; Takahashi, R.; Teplow, D.B.; Yamada, M. High-speed atomic force microscopy reveals structural dynamics of amyloid β1–42 aggregates. Proc. Natl. Acad. Sci. USA 2016, 113, 5835–5840. [Google Scholar] [CrossRef] [PubMed]
- Milhiet, P.-E.; Yamamoto, D.; Berthoumieu, O.; Dosset, P.; Le Grimellec, C.; Verdier, J.-M.; Marchal, S.; Ando, T. Deciphering the structure, growth and assembly of amyloid-like fibrils using high-speed atomic force microscopy. PLoS ONE 2010, 5, e13240. [Google Scholar] [CrossRef] [PubMed]
- Zlotnikov, I.; Zolotoyabko, E.; Fratzl, P. Nano-scale modulus mapping of biological composite materials: Theory and practice. Prog. Mater. Sci. 2017, 87, 292–320. [Google Scholar] [CrossRef]
- Gal, A.; Wirth, R.; Kopka, J.; Fratzl, P.; Faivre, D.; Scheffel, A. Macromolecular recognition directs calcium ions to coccolith mineralization sites. Science 2016, 353, 590–593. [Google Scholar] [CrossRef] [PubMed]
- Casdorff, K.; Keplinger, T.; Bellanger, H.; Michen, B.; Schön, S.; Burgert, I. High-resolution adhesion mapping of the odd-even effect on a layer-by-layer coated biomaterial by atomic-force-microscopy. ACS Appl. Mater. Interfaces 2017, 9, 13793–13800. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Zhang, T.; Pan, Y.; Ciacchi, L.C.; Xu, B.; Wei, G. Afm-based force spectroscopy for bioimaging and biosensing. RSC Adv. 2016, 6, 12893–12912. [Google Scholar] [CrossRef]
- Alsteens, D.; Newton, R.; Schubert, R.; Martinez-Martin, D.; Delguste, M.; Roska, B.; Müller, D.J. Nanomechanical mapping of first binding steps of a virus to animal cells. Nat. Nanotechnol. 2017, 12, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Berne, C.C.; Ma, X.; Licata, N.A.; Neves, B.R.; Setayeshgar, S.; Brun, Y.V.; Dragnea, B. Physiochemical properties of caulobacter crescentus holdfast: A localized bacterial adhesive. J. Phys. Chem. B 2013, 117, 10492–10503. [Google Scholar] [CrossRef] [PubMed]
- Vaughan, R.; Tragesser, B.; Ni, P.; Ma, X.; Dragnea, B.; Kao, C.C. The tripartite virions of the brome mosaic virus have distinct physical properties that affect the timing of the infection process. J. Virol. 2014, 88, 6483–6491. [Google Scholar] [CrossRef] [PubMed]
- Ni, P.; Wang, Z.; Ma, X.; Das, N.C.; Sokol, P.; Chiu, W.; Dragnea, B.; Hagan, M.; Kao, C.C. An examination of the electrostatic interactions between the n-terminal tail of the brome mosaic virus coat protein and encapsidated rnas. J. Mol. Biol. 2012, 419, 284–300. [Google Scholar] [CrossRef] [PubMed]
- Kol, N.; Shi, Y.; Tsvitov, M.; Barlam, D.; Shneck, R.Z.; Kay, M.S.; Rousso, I. A stiffness switch in human immunodeficiency virus. Biophys. J. 2007, 92, 1777–1783. [Google Scholar] [CrossRef] [PubMed]
- Castellanos, M.; Pérez, R.; Carrasco, C.; Hernando-Pérez, M.; Gómez-Herrero, J.; de Pablo, P.J.; Mateu, M.G. Mechanical elasticity as a physical signature of conformational dynamics in a virus particle. Proc. Natl. Acad. Sci. USA 2012, 109, 12028–12033. [Google Scholar] [CrossRef] [PubMed]
- Marchetti, M.; Wuite, G.; Roos, W. Atomic force microscopy observation and characterization of single virions and virus-like particles by nano-indentation. Curr. Opin. Virol. 2016, 18, 82–88. [Google Scholar] [CrossRef] [PubMed]
- Denning, D.; Wuite, G.J.; Roos, W.H. A combined imaging and force spectroscopy approach reveals the material properties of viral nanoparticles. Biophys. J. 2016, 110, 500a. [Google Scholar] [CrossRef]
- Ramalho, R.; Rankovic, S.; Zhou, J.; Aiken, C.; Rousso, I. Analysis of the mechanical properties of wild type and hyperstable mutants of the hiv-1 capsid. Retrovirology 2016, 13, 17. [Google Scholar] [CrossRef] [PubMed]
- Korneev, D.; Popova, A.; Generalov, V.; Zaitsev, B. Atomic force microscopy-based single virus particle spectroscopy. Biophysics 2016, 61, 413–419. [Google Scholar] [CrossRef]
- Snijder, J.; Kononova, O.; Barbu, I.M.; Uetrecht, C.; Rurup, W.F.; Burnley, R.J.; Koay, M.S.; Cornelissen, J.J.; Roos, W.H.; Barsegov, V. Assembly and mechanical properties of the cargo-free and cargo-loaded bacterial nanocompartment encapsulin. Biomacromolecules 2016, 17, 2522–2529. [Google Scholar] [CrossRef] [PubMed]
- Quintana-Cataño, C.A.; Vives-Flórez, M.J.; Forero-Shelton, M. Force spectroscopy of t4 bacteriophage adhesion during infection. Biophys. J. 2017, 112, 588a. [Google Scholar] [CrossRef]
- Hernando-Perez, M.; Zeng, C.; Delalande, L.; Tsvetkova, I.; Bousquet, A.; Tayachi-Pigeonnat, M.; Temam, R.; Dragnea, B. Nanoindentation of isometric viruses on deterministically corrugated substrates. J. Phys. Chem. B 2016, 120, 340–347. [Google Scholar] [CrossRef] [PubMed]
- Zeng, C.; Moller-Tank, S.; Asokan, A.; Dragnea, B. Probing the link among genomic cargo, contact mechanics, and nanoindentation in recombinant adeno-associated virus 2. J. Phys. Chem. B 2017, 121, 1843–1853. [Google Scholar] [CrossRef] [PubMed]
- Delalande, L.; Tsvetkova, I.B.; Zeng, C.; Bond, K.; Jarrold, M.; Dragnea, B. Catching a virus in a molecular net. Nanoscale 2016, 8, 16221–16228. [Google Scholar] [CrossRef] [PubMed]
- Eeftens, J.; Katan, A.; Kschonsak, M.; Hassler, M.; Dief, E.; de Wilde, L.; Haering, C.; Dekker, C. Single-molecule experiments to resolve structural and mechanical properties of condensin. Biophys. J. 2016, 110, 528a. [Google Scholar] [CrossRef]
- Heinze, K.; Sasaki, E.; King, N.; Baker, D.; Hilvert, D.; Wuite, G.; Roos, W. Protein nanocontainers from nonviral origin: Testing the mechanics of artificial and natural protein cages by afm. J. Phys. Chem. B 2016, 120, 5945–5952. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Madrid, F.; Martín-González, N.; Llauró, A.; Ortega-Esteban, A.; Hernando-Pérez, M.; Douglas, T.; Schaap, I.A.; de Pablo, P.J. Atomic force microscopy of virus shells. Biochem. Soc. Trans. 2017, 45, 499–511. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.F.; Xu, Y.; McKinnon, T.A.; Zhang, W. Biophysical mechanisms of von willebrand factor-collagen interactions. Biophys. J. 2017, 112, 455a. [Google Scholar] [CrossRef]
- Van Pattten, W.J.; Walder, R.; Adhikari, A.; Ravichandran, R.; Tinberg, C.E.; Baker, D.; Perkins, T.T. A computationally designed protein-ligand interaction is mechanically robust. Biophys. J. 2017, 112, 455a. [Google Scholar] [CrossRef]
- Yadav, A.; Paul, S.; Venkatramani, R.; Rama, S.; Ainavarapu, K. Examining the mechanical properties of copper binding azurin using single molecule force spectroscopy and steered molecular dynamics. Biophys. J. 2016, 110, 496a. [Google Scholar] [CrossRef]
- Hughes, M.L.; Dougan, L. The physics of pulling polyproteins: A review of single molecule force spectroscopy using the afm to study protein unfolding. Rep. Prog. Phys. 2016, 79, 076601. [Google Scholar] [CrossRef] [PubMed]
- Walder, R.; Van Patten, W.J.; Miller, T.W.; Perkins, T.T. Mechanical characterization of the hiv-1 rna hairpin using an atomic force microscope. Biophys. J. 2017, 112, 166a. [Google Scholar] [CrossRef]
- Shlyakhtenko, L.S.; Dutta, S.; Li, M.; Harris, R.S.; Lyubchenko, Y.L. Single-molecule force spectroscopy studies of apobec3a-single-stranded DNA complexes. Biochemistry 2016, 55, 3102–3106. [Google Scholar] [CrossRef] [PubMed]
- Fitzgibbon, C.J.; Josephs, E.A.; Marszalek, P.E. Resolving individual damage sites in DNA with afm using reengineered repair proteins. Biophys. J. 2016, 110, 496a. [Google Scholar] [CrossRef]
- Dutta, S.; Armitage, B.A.; Lyubchenko, Y.L. Probing of minipegγ-pna–dna hybrid duplex stability with afm force spectroscopy. Biochemistry 2016, 55, 1523–1528. [Google Scholar] [CrossRef] [PubMed]
- Camunas-Soler, J.; Ribezzi-Crivellari, M.; Ritort, F. Elastic properties of nucleic acids by single-molecule force spectroscopy. Annu. Rev. Biophys. 2016, 45, 65–84. [Google Scholar] [CrossRef] [PubMed]
- Fisher, G.J.; Shao, Y.; He, T.; Qin, Z.; Perry, D.; Voorhees, J.J.; Quan, T. Reduction of fibroblast size/mechanical force down-regulates tgf-β type ii receptor: Implications for human skin aging. Aging Cell 2016, 15, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Rianna, C.; Radmacher, M. Cell Mechanics As a Marker for Diseases: Biomedical Applications of Afm. In AIP Conference Proceedings; AIP Publishing: Melville, NY, USA, 2016; p. 020057. [Google Scholar]
- Ting, A.Y.P. Spatially resolved mapping of endogenous proteins and rna in living cells. Biophys. J. 2017, 112, 7a. [Google Scholar] [CrossRef]
- Burgert, I.; Keplinger, T. Plant micro-and nanomechanics: Experimental techniques for plant cell-wall analysis. J. Exp. Bot. 2013, 64, 4635–4649. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Nancollas, G.H. Pathways to biomineralization and biodemineralization of calcium phosphates: The thermodynamic and kinetic controls. Dalton Trans. 2009, 2665–2672. [Google Scholar] [CrossRef] [PubMed]
- Putnis, A.; Pina, C.M.; Astilleros, J.M.; Fernández-Díaz, L.; Prieto, M. Nucleation of solid solutions crystallizing from aqueous solutions. Phil. Trans. R. Soc. Lond. A 2003, 361, 615–632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, Y.; Wang, J.; Zhong, J.; Wang, Y.; Yin, Q.; Hou, B.; Hao, H. Application of atomic force microscopy in understanding crystallization process. Sci. Adv. Mater. 2017, 9, 89–101. [Google Scholar] [CrossRef]
- Jun, Y.-S.; Kim, D.; Neil, C.W. Heterogeneous nucleation and growth of nanoparticles at environmental interfaces. Acc. Chem. Res. 2016, 49, 1681–1690. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Lu, J.; Xu, F.; Zhang, F. Dynamics of crystallization and dissolution of calcium orthophosphates at the near-molecular level. Chin. Sci. Bull. 2011, 56, 713–721. [Google Scholar] [CrossRef]
- Motta, N.; Szkutnik, P.D.; Tomellini, M.; Sgarlata, A.; Fanfoni, M.; Patella, F.; Balzarotti, A. Role of patterning in islands nucleation on semiconductor surfaces. C. R. Phys. 2006, 7, 1046–1072. [Google Scholar] [CrossRef] [Green Version]
- Wesson, J.A.; Ward, M.D. Role of crystal surface adhesion in kidney stone disease. Curr. Opin. Nephrol. Hypertens. 2006, 15, 386–393. [Google Scholar] [CrossRef] [PubMed]
- Alamani, B.G.; Rimer, J.D. Molecular modifiers of kidney stones. Curr. Opin. Nephrol. Hypertens. 2017, 26, 256–265. [Google Scholar] [CrossRef] [PubMed]
- Rimer, J.D.; Kolbach-Mandel, A.M.; Ward, M.D.; Wesson, J.A. The role of macromolecules in the formation of kidney stones. Urolithiasis 2017, 45, 57–74. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Ohsuna, T.; Liu, Z.; Alfredsson, V.; Kjellman, T.; Asahina, S.; Suga, M.; Ma, Y.; Oleynikov, P.; Miyasaka, K. Structures of silica-based nanoporous materials revealed by microscopy. Z. Anorg. Allg. Chem. 2014, 640, 521–536. [Google Scholar] [CrossRef]
- Glenner, G.G.; Wong, C.W. Alzheimer’s disease: Initial report of the purification and characterization of a novel cerebrovascular amyloid protein. Biochem. Biophys. Res. Commun. 1984, 120, 885–890. [Google Scholar] [CrossRef]
- Zhai, J.; Lee, T.-H.; Small, D.H.; Aguilar, M.-I. Characterization of early stage intermediates in the nucleation phase of aβ aggregation. Biochemistry 2012, 51, 1070–1078. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.S.; Ansaloni, A.; Mezzenga, R.; Lashuel, H.A.; Dietler, G. Novel mechanistic insight into the molecular basis of amyloid polymorphism and secondary nucleation during amyloid formation. J. Mol. Biol. 2013, 425, 1765–1781. [Google Scholar] [CrossRef] [PubMed]
- Karsai, A.; Slack, T.J.; Malekan, H.; Khoury, F.; Lin, W.F.; Tran, V.; Cox, D.; Toney, M.; Chen, X.; Liu, G.Y. Local mechanical perturbation provides an effective means to regulate the growth and assembly of functional peptide fibrils. Small 2016, 12, 6407–6415. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.-C.; Zhang, F.; Su, H.-N.; Li, H.; Zhang, Y.; Hu, J. Mechanical manipulation assisted self-assembly to achieve defect repair and guided epitaxial growth of individual peptide nanofilaments. ACS Nano 2010, 4, 5791–5796. [Google Scholar] [CrossRef] [PubMed]
- Dunlop, J.W.; Fratzl, P. Biological composites. Annu. Rev. Mater. Res. 2010, 40, 1–24. [Google Scholar] [CrossRef]
- Dunlop, J.W.; Weinkamer, R.; Fratzl, P. Artful interfaces within biological materials. Mater. Today 2011, 14, 70–78. [Google Scholar] [CrossRef]
- Tao, J.; Buchko, G.W.; Shaw, W.J.; De Yoreo, J.J.; Tarasevich, B.J. Sequence-defined energetic shifts control the disassembly kinetics and microstructure of amelogenin adsorbed onto hydroxyapatite (100). Langmuir 2015, 31, 10451. [Google Scholar] [CrossRef] [PubMed]
- Chung, S.; Shin, S.-H.; Bertozzi, C.R.; De Yoreo, J.J. Self-catalyzed growth of s layers via an amorphous-to-crystalline transition limited by folding kinetics. Proc. Natl. Acad. Sci. USA 2010, 107, 16536–16541. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.-H.; Chung, S.; Sanii, B.; Comolli, L.R.; Bertozzi, C.R.; De Yoreo, J.J. Direct observation of kinetic traps associated with structural transformations leading to multiple pathways of s-layer assembly. Proc. Natl. Acad. Sci. USA 2012, 109, 12968–12973. [Google Scholar] [CrossRef] [PubMed]
- Ido, S.; Kimura, K.; Oyabu, N.; Kobayashi, K.; Tsukada, M.; Matsushige, K.; Yamada, H. Beyond the helix pitch: Direct visualization of native DNA in aqueous solution. ACS Nano 2013, 7, 1817–1822. [Google Scholar] [CrossRef] [PubMed]
- Abel, G.R., Jr.; Josephs, E.A.; Luong, N.; Ye, T. A switchable surface enables visualization of single DNA hybridization events with atomic force microscopy. J. Am. Chem. Soc. 2013, 135, 6399–6402. [Google Scholar] [CrossRef] [PubMed]
- Thomson, N.H.; Kasas, S.; Smith, B.; Hansma, H.G.; Hansma, P.K. Reversible binding of DNA to mica for afm imaging. Langmuir 1996, 12, 5905–5908. [Google Scholar] [CrossRef]
- Burke, K.A.; Hensal, K.M.; Umbaugh, C.S.; Chaibva, M.; Legleiter, J. Huntingtin disrupts lipid bilayers in a polyq-length dependent manner. Biochim. Biophys. Acta (BBA)-Biomembr. 2013, 1828, 1953–1961. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Khadka, N.K. Kinetic defects induced by melittin in model lipid membranes: A solution atomic force microscopy study. J. Phys. Chem. B 2016, 120, 4625–4634. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, N.; Kobayashi, T. Visualization of lipid membrane reorganization induced by a pore-forming toxin using high-speed atomic force microscopy. ACS Nano 2015, 9, 7960–7967. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Wong, M.K.; Lin, L.E.; Yip, C.M. Nucleation and growth of elastin-like peptide fibril multilayers: An in situ atomic force microscopy study. Nanotechnology 2011, 22, 494018. [Google Scholar] [CrossRef] [PubMed]
- Charbonneau, C.; Kleijn, J.M.; Cohen Stuart, M.A. Subtle charge balance controls surface-nucleated self-assembly of designed biopolymers. ACS Nano 2014, 8, 2328–2335. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.-G.; Li, H.; Huynh, T.; Zhang, F.; Xia, Z.; Zhang, Y.; Zhou, R. Molecular mechanism of surface-assisted epitaxial self-assembly of amyloid-like peptides. ACS Nano 2012, 6, 9276–9282. [Google Scholar] [CrossRef] [PubMed]
- Dai, B.; Kang, S.-G.; Huynh, T.; Lei, H.; Castelli, M.; Hu, J.; Zhang, Y.; Zhou, R. Salts drive controllable multilayered upright assembly of amyloid-like peptides at mica/water interface. Proc. Natl. Acad. Sci. USA 2013, 110, 8543–8548. [Google Scholar] [CrossRef] [PubMed]
- Varongchayakul, N.; Johnson, S.; Quabili, T.; Cappello, J.; Ghandehari, H.; Solares, S.D.J.; Hwang, W.; Seog, J. Direct observation of amyloid nucleation under nanomechanical stretching. ACS Nano 2013, 7, 7734–7743. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Fung, S.-Y.; Pritzker, M.; Chen, P. Mechanical-force-induced nucleation and growth of peptide nanofibers at liquid/solid interfaces. Angew. Chem. Int. Ed. 2008, 47, 4397–4400. [Google Scholar] [CrossRef] [PubMed]
- LeGeros, R.Z. Calcium phosphate-based osteoinductive materials. Chem. Rev. 2008, 108, 4742–4753. [Google Scholar] [CrossRef] [PubMed]
- Tiselius, H.-G. A hypothesis of calcium stone formation: An interpretation of stone research during the past decades. Urol. Res. 2011, 39, 231–243. [Google Scholar] [CrossRef] [PubMed]
- Bazin, D.; Daudon, M.; Combes, C.; Rey, C. Characterization and some physicochemical aspects of pathological microcalcifications. Chem. Rev. 2012, 112, 5092–5120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.; Wang, L.; Zhang, W.; Putnis, C.V.; Putnis, A. Direct observation of spiral growth, particle attachment, and morphology evolution of hydroxyapatite. Cryst. Growth Des. 2016, 16, 4509–4518. [Google Scholar] [CrossRef]
- Wu, S.; Yu, M.; Li, M.; Wang, L.; Putnis, C.V.; Putnis, A. In situ afm imaging of octacalcium phosphate crystallization and its modulation by amelogenin’s c-terminus. Cryst. Growth Des. 2017, 17, 2194–2202. [Google Scholar] [CrossRef]
- Habraken, W.J.; Tao, J.; Brylka, L.J.; Friedrich, H.; Bertinetti, L.; Schenk, A.S.; Verch, A.; Dmitrovic, V.; Bomans, P.H.; Frederik, P.M. Ion-association complexes unite classical and non-classical theories for the biomimetic nucleation of calcium phosphate. Nat. Commun. 2013, 4, 1507. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Navarro, C.; Burgos Cara, A.; Elert, K.; Putnis, C.V.; Ruiz-Agudo, E. Direct nanoscale imaging reveals the growth of calcite crystals via amorphous nanoparticles. Cryst. Growth Des. 2016, 16, 1850–1860. [Google Scholar] [CrossRef]
- Wolf, S.L.P.; Caballero, L.; Melo, F.; Cölfen, H. Gel-like calcium carbonate precursors observed by in-situ afm. Langmuir 2016, 33, 158–163. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, S.; Voïtchovsky, K.; Spijker, P.; Schmidt, M.; Stumpf, T. Visualising the molecular alteration of the calcite (104)–water interface by sodium nitrate. Sci. Rep. 2016, 6, 21576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Qin, L.; Putnis, C.V.; Ruiz-Agudo, E.; King, H.E.; Putnis, A. Visualizing organophosphate precipitation at the calcite–water interface by in situ atomic-force microscopy. Environ. Sci. Technol. 2015, 50, 259–268. [Google Scholar] [CrossRef] [PubMed]
- So, C.R.; Liu, J.; Fears, K.P.; Leary, D.H.; Golden, J.P.; Wahl, K.J. Self-assembly of protein nanofibrils orchestrates calcite step movement through selective nonchiral interactions. ACS Nano 2015, 9, 5782–5791. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.R.; Salter, E.A.; De Yoreo, J.J.; Wierzbicki, A.; Elhadj, S.; Huang, Y.; Qiu, S.R. Growth inhibition of calcium oxalate monohydrate crystal by linear aspartic acid enantiomers investigated by in situ atomic force microscopy. CrystEngComm 2013, 15, 54–64. [Google Scholar] [CrossRef]
- Li, S.; Zhang, W.; Wang, L. Direct nanoscale imaging of calcium oxalate crystallization on brushite reveals the mechanisms underlying stone formation. Cryst. Growth Des. 2015, 15, 3038–3045. [Google Scholar] [CrossRef]
- Ando, T.; Uchihashi, T.; Fukuma, T. High-speed atomic force microscopy for nano-visualization of dynamic biomolecular processes. Prog. Surf. Sci. 2008, 83, 337–437. [Google Scholar] [CrossRef]
- Li, Y.; Bechhoefer, J. Model-free iterative control of repetitive dynamics for high-speed scanning in atomic force microscopy. Rev. Sci. Instrum. 2009, 80, 013702. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Wu, Y.; Zou, Q.; Su, C. An integrated approach to piezoactuator positioning in high-speed atomic force microscope imaging. Rev. Sci. Instrum. 2008, 79, 073704. [Google Scholar] [CrossRef] [PubMed]
- Heinisch, J.J.; Lipke, P.N.; Beaussart, A.; Chatel, S.E.K.; Dupres, V.; Alsteens, D.; Dufrêne, Y.F. Atomic force microscopy—Looking at mechanosensors on the cell surface. J. Cell Sci. 2012, 125, 4189–4195. [Google Scholar] [CrossRef] [PubMed]
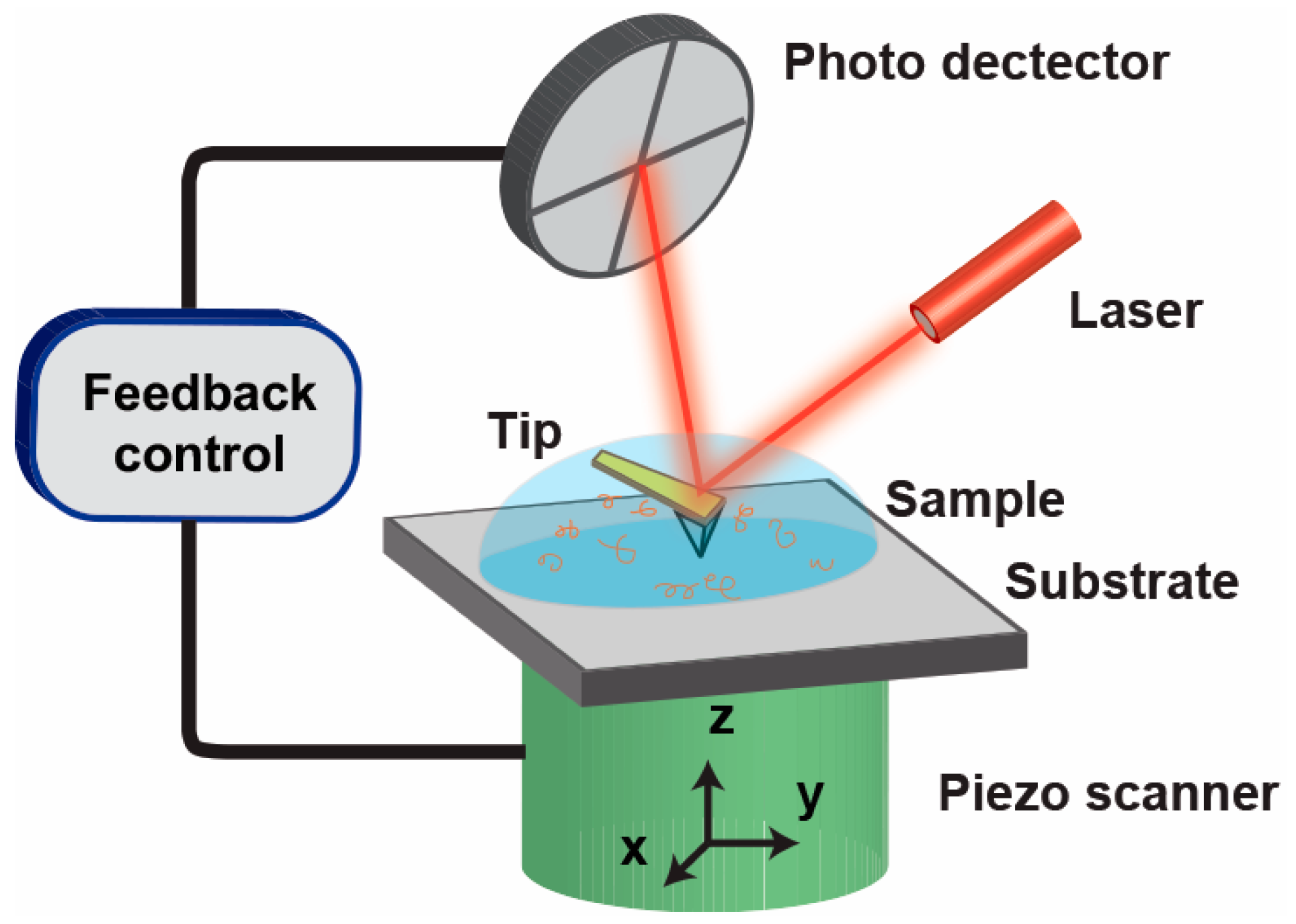
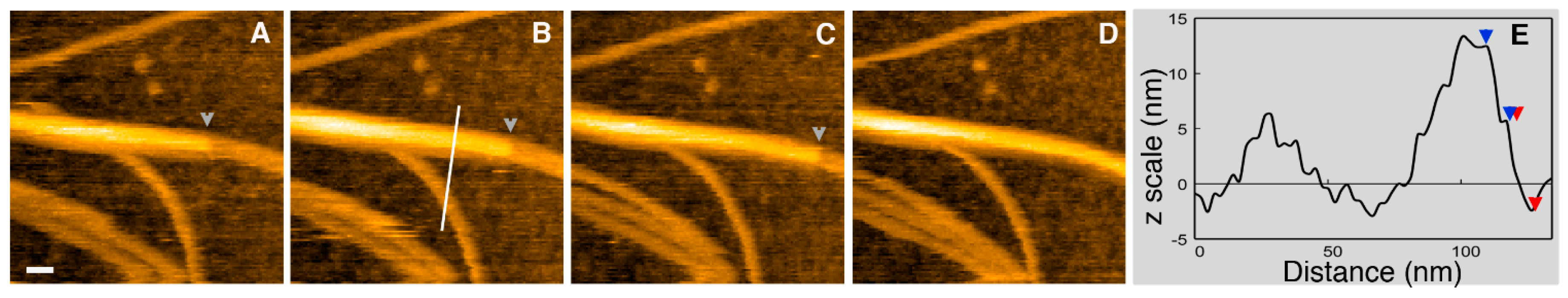
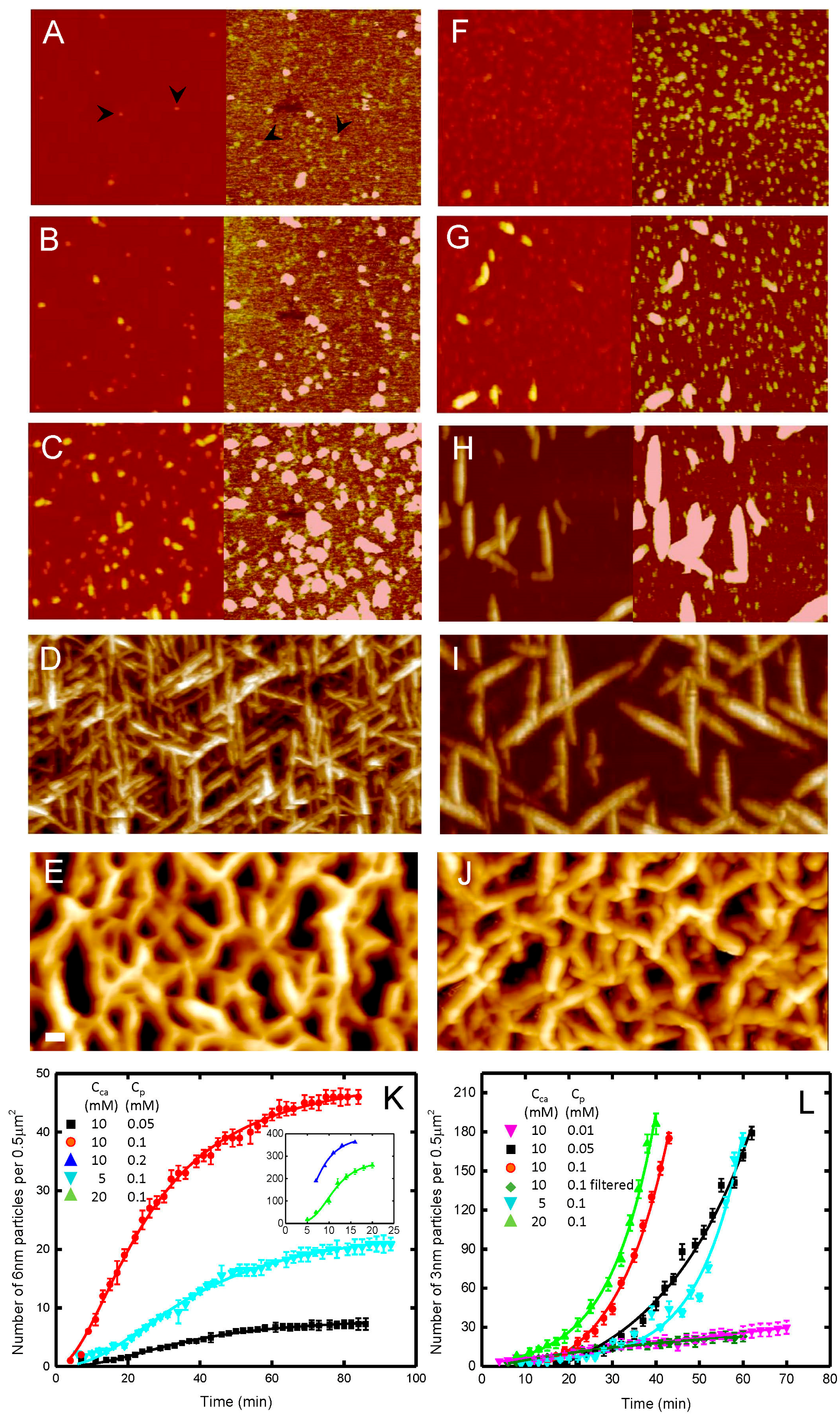

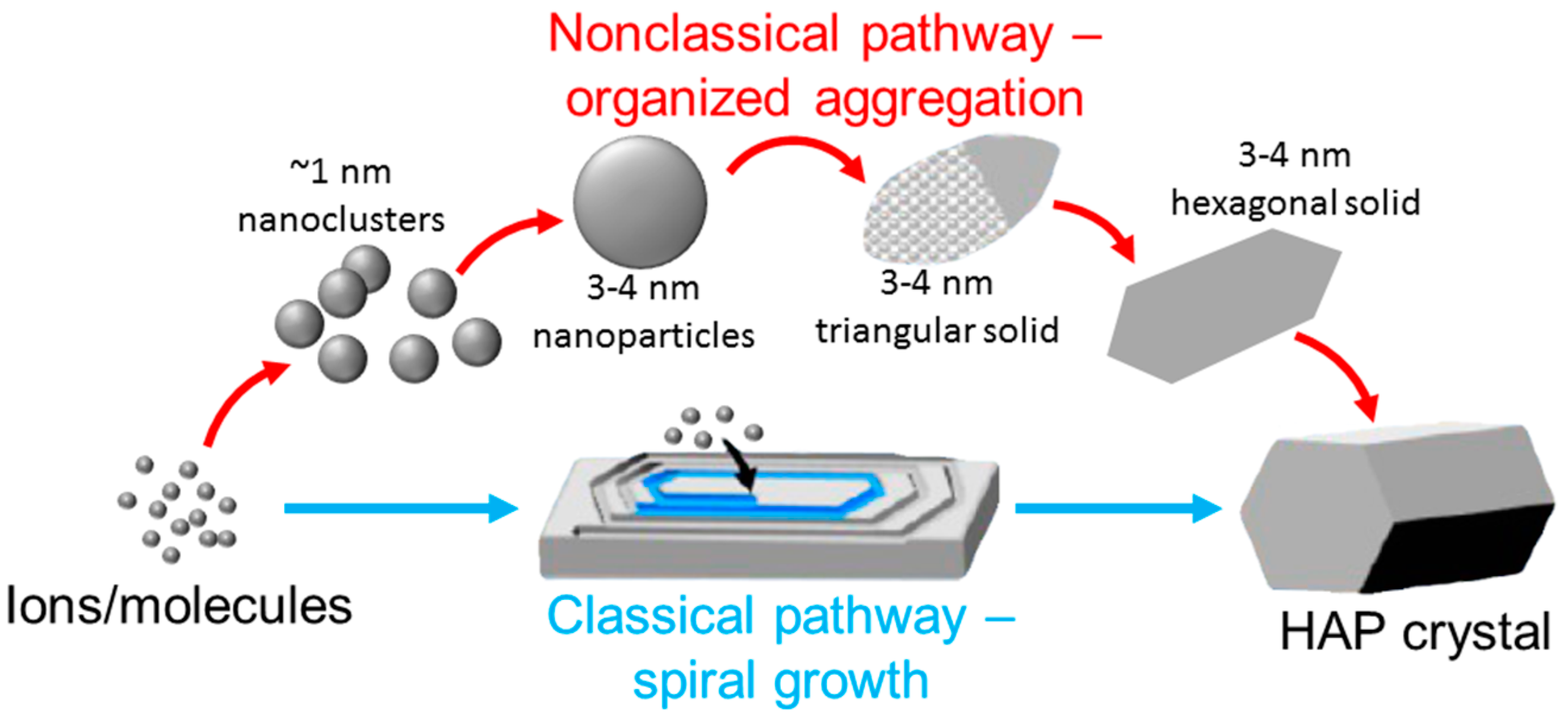
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeng, C.; Vitale-Sullivan, C.; Ma, X. In Situ Atomic Force Microscopy Studies on Nucleation and Self-Assembly of Biogenic and Bio-Inspired Materials. Minerals 2017, 7, 158. https://doi.org/10.3390/min7090158
Zeng C, Vitale-Sullivan C, Ma X. In Situ Atomic Force Microscopy Studies on Nucleation and Self-Assembly of Biogenic and Bio-Inspired Materials. Minerals. 2017; 7(9):158. https://doi.org/10.3390/min7090158
Chicago/Turabian StyleZeng, Cheng, Caitlin Vitale-Sullivan, and Xiang Ma. 2017. "In Situ Atomic Force Microscopy Studies on Nucleation and Self-Assembly of Biogenic and Bio-Inspired Materials" Minerals 7, no. 9: 158. https://doi.org/10.3390/min7090158



