Periplasmic Binding Proteins in Thermophiles: Characterization and Potential Application of an Arginine-Binding Protein from Thermotoga maritima: A Brief Thermo-Story
Abstract
:1. Introduction
1.1. ABC Transporter System and Periplasmic Binding Proteins
1.2. Arginine-Binding Protein from Thermotoga Maritima and Its Potential Applications
2. TmArgBP Structure
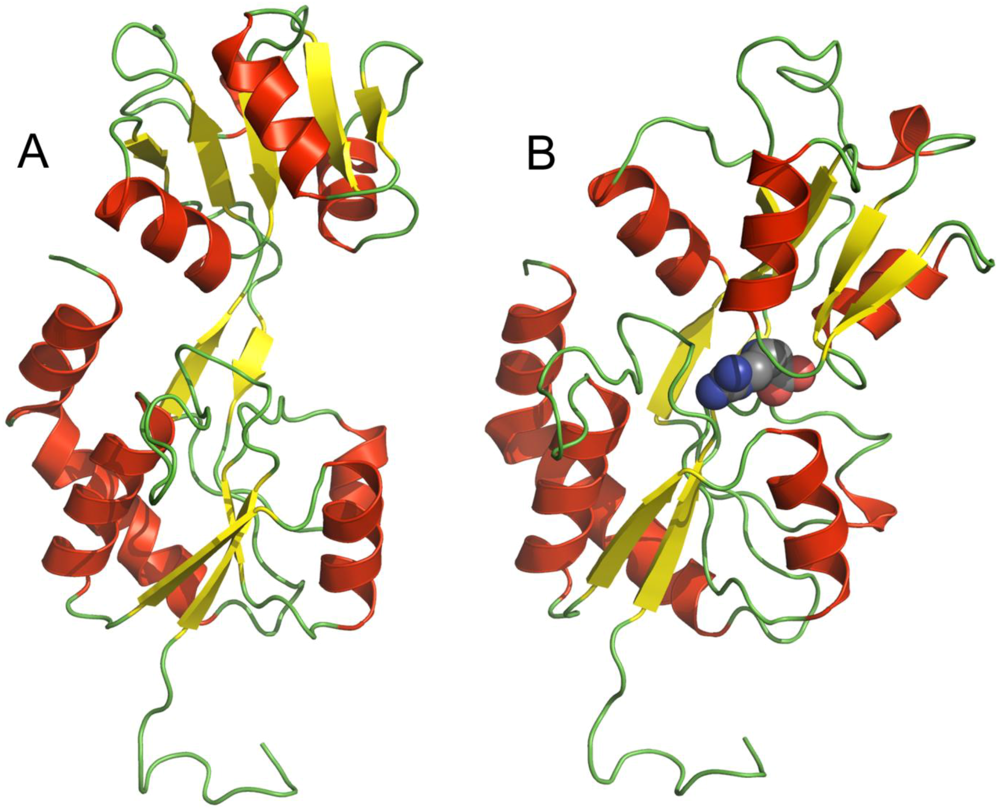
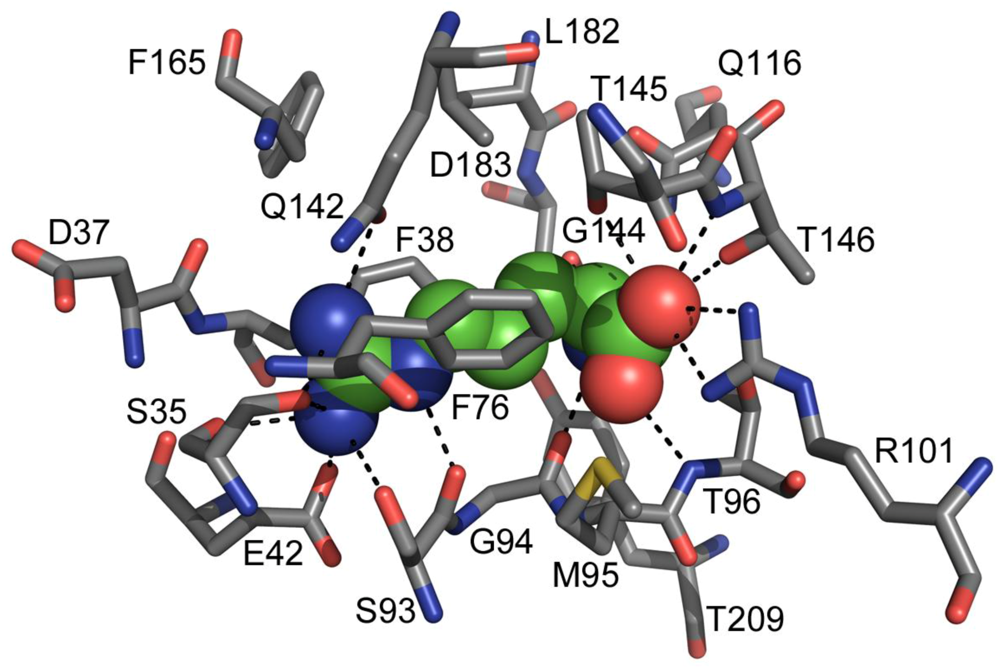
3. Stability of TmArgBP
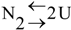
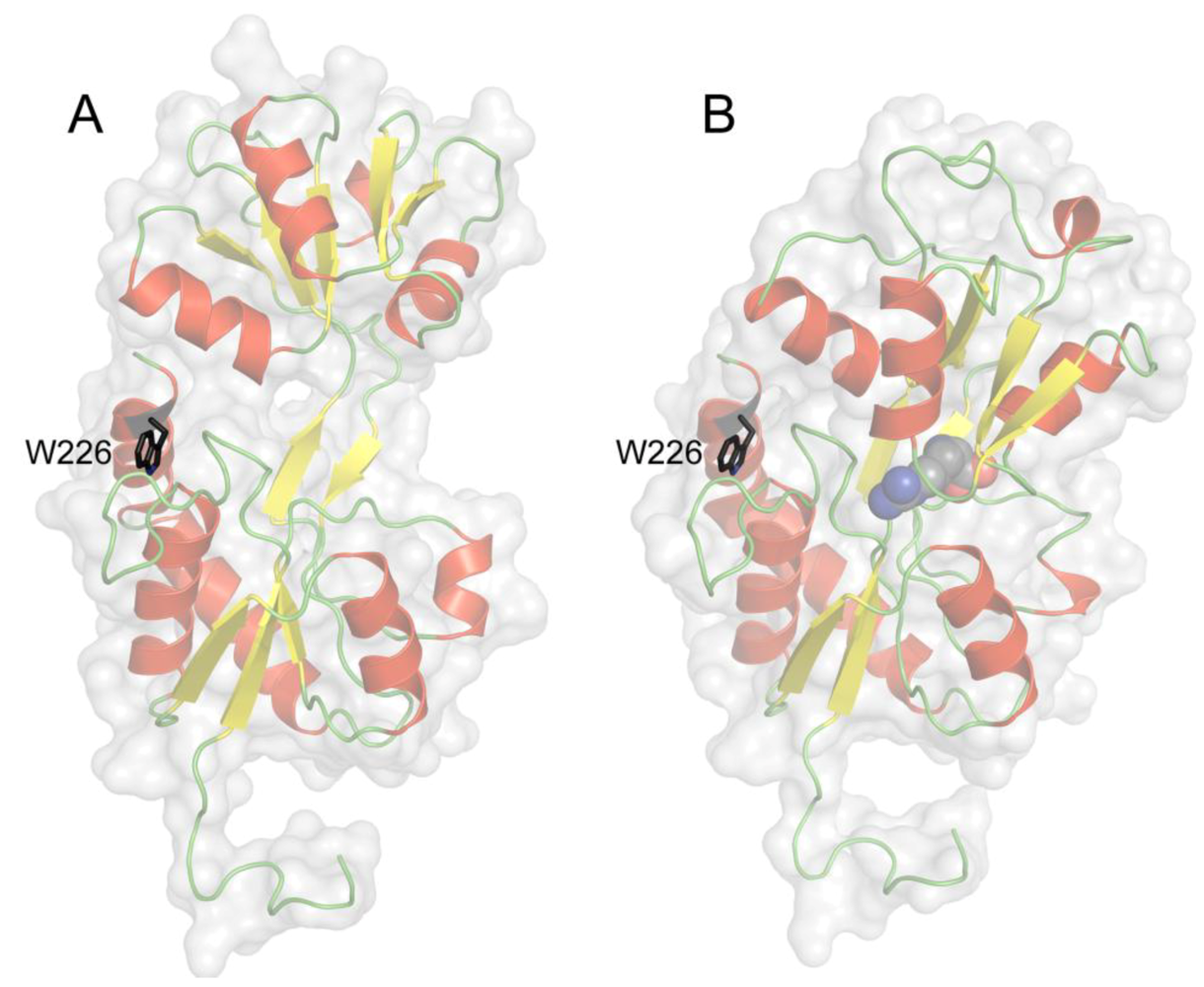
4. Tryptophan Microenvironment

5. Conclusions
Acknowledgements
References
- Higgins, C.F. ABC transporters: From microorganisms to man. Annu. Rev. Cell Biol. 1992, 8, 67–113. [Google Scholar] [CrossRef]
- Jones, P.M.; George, A.M. The ABC transporter structure and mechanism: Perspectives on recent research. Cell Mol. Life Sci. 2004, 61, 682–699. [Google Scholar] [CrossRef]
- Linton, K.J. Structure and function of ABC transporters. Physiology 2007, 22, 122–130. [Google Scholar] [CrossRef]
- Zolnerciks, J.K.; Andress, E.J.; Nicolaou, M.; Linton, K.J. Structure of ABC transporters. Essays Biochem. 2011, 50, 43–61. [Google Scholar] [CrossRef]
- Kos, V.; Ford, R.C. The ATP-binding cassette family: a structural perspective. Cell Mol. Life Sci. 2009, 66, 3111–3126. [Google Scholar] [CrossRef]
- Davidson, A.L.; Dassa, E.; Orelle, C.; Chen, J. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol. Mol. Biol. Rev. 2008, 72, 317–364. [Google Scholar] [CrossRef]
- Rees, D.C.; Johnson, E.; Lewinson, O. ABC transporters: the power to change. Nat. Rev. Mol. Cell. Biol. 2009, 10, 218–227. [Google Scholar] [CrossRef]
- Oldham, M.L.; Davidson, A.L.; Chen, J. Structural insights into ABC transporter mechanism. Curr. Opin. Struct. Biol. 2008, 18, 726–733. [Google Scholar] [CrossRef]
- Oldham, M.L.; Khare, D.; Quiocho, F.A.; Davidson, A.L.; Chen, J. Crystal structure of a catalytic intermediate of the maltose transporter. Nature 2007, 450, 515–521. [Google Scholar] [CrossRef]
- Gilson, E.; Alloing, G.; Schmidt, T.; Claverys, J.P.; Dudler, R.; Hofnung, M. Evidence for high affinity binding-protein dependent transport systems in gram-positive bacteria and in Mycoplasma. EMBO J. 1988, 7, 3971–3974. [Google Scholar]
- Montesinos, M.L.; Herrero, A.; Flores, E. Amino acid transport in taxonomically diverse cyanobacteria and identification of two genes encoding elements of a neutral amino acid permease putatively involved in recapture of leaked hydrophobic amino acids. J. Bacteriol. 1997, 179, 853–862. [Google Scholar]
- Quiocho, F.A.; Ledvina, P.S. Atomic structure and specificity of bacterial periplasmic receptors for active transport and chemotaxis: variation of common themes. Mol. Microbiol. 1996, 20, 17–25. [Google Scholar] [CrossRef]
- Tam, R.; Saier, M.H., Jr. Structural, functional, and evolutionary relationships among extracellular solute-binding receptors of bactera. Microbiol. Rev. 1993, 57, 320–346. [Google Scholar]
- Dwyer, M.A.; Hellinga, H.W. Periplasmic binding proteins: a versatile superfamily for protein engineering. Curr. Opin. Struct. Biol. 2004, 14, 495–504. [Google Scholar] [CrossRef]
- Fukami-Kobayashi, K.; Tateno, Y.; Nishikawa, K. Domain dislocation: a change of core structure in periplasmic binding proteins in their evolutionary history. J. Mol. Biol. 1999, 286, 279–290. [Google Scholar] [CrossRef]
- Lee, Y.H.; Dorwart, M.R.; Hazlett, K.R.; Deka, R.K.; Norgard, M.V.; Radolf, J.D.; Hasemann, C.A. The crystal structure of Zn(II)-free Treponema pallidum TroA, a periplasmic metal-binding protein, reveals a closed conformation. J. Bacteriol. 2002, 184, 2300–2304. [Google Scholar] [CrossRef]
- Karpowich, N.K.; Huang, H.H.; Smith, P.C.; Hunt, J.F. Crystal structures of the BtuF periplasmic-binding protein for vitamin B12 suggest a functionally important reduction in protein mobility upon ligand binding. J. Biol. Chem. 2003, 278, 8429–8434. [Google Scholar]
- Wissenbach, U.; Six, S.; Bongaerts, J.; Ternes, D.; Steinwachs, S.; Unden, G. A third periplasmic transport system for L-arginine in Escherichia coli: molecular characterization of the artPIQMJ genes, arginine binding and transport. Mol. Microbiol. 1995, 17, 675–686. [Google Scholar]
- Stamp, A.L.; Owen, P.; El Omari, K.; Lockyer, M.; Lamb, H.K.; Charles, I.G.; Hawkins, A.R.; Stammers, D.K. Crystallographic and microcalorimetric analyses reveal the structural basis for high arginine specificity in the Salmonella enterica serovar Typhimurium periplasmic binding protein STM4351. Proteins 2011, 79, 2352–2357. [Google Scholar] [CrossRef]
- Oh, B.H.; Pandit, J.; Kang, C.H.; Nikaido, K.; Gokcen, S.; Ames, G.F.; Kim, S.H. Three-dimensional structures of the periplasmic lysine/arginine/ornithine-binding protein with and without a ligand. J. Biol. Chem. 1993, 268, 11348–11355. [Google Scholar]
- Celis, R.T.; Leadlay, P.F.; Roy, I.; Hansen, A. Phosphorylation of the periplasmic binding protein in two transport systems for arginine incorporation in Escherichia coli K-12 is unrelated to the function of the transport system. J. Bacteriol. 1998, 180, 4828–4833. [Google Scholar]
- De Wolf, F.A.; Brett, G.M. Ligand-binding proteins: their potential for application in systems for controlled delivery and uptake of ligands. Pharmacol. Rev. 2000, 52, 207–236. [Google Scholar]
- Jain-Ghai, S.; Nagamani, S.C.; Blaser, S.; Siriwardena, K.; Feigenbaum, A. Arginase I deficiency: severe infantile presentation with hyperammonemia: more common than reported? Mol. Genet. Metab. 2011, 104, 107–111. [Google Scholar] [CrossRef]
- Brusilov, S.W.; Horwich, A.L. Urea cycle enzymes. In The Metabolic Basis of Inherited Disease; Scriver, C.R., Beaudet, A.L., Sly, W.S., Valle, D., Eds.; McGraw-Hill: New York, NY, USA, 1989; pp. 629–663. [Google Scholar]
- Rees, D.D.; Palmer, R.M.; Moncada, S. Role of endothelium-derived nitric oxide in the regulation of blood pressure. Proc. Natl. Acad. Sci. USA 1989, 86, 3375–3378. [Google Scholar] [CrossRef]
- Cooke, J.P. Does ADMA cause endothelial dysfunction? Arterioscler. Thromb. Vasc. Biol. 2000, 20, 2032–2037. [Google Scholar] [CrossRef]
- Vallance, P.; Collier, J.; Moncada, S. Effects of endothelium-derived nitric oxide on peripheral arteriolar tone in man. Lancet 1989, 2, 997–1000. [Google Scholar]
- Dattelbaum, J.D.; Lakowicz, J.R. Optical determination of glutamine using a genetically engineered protein. Anal. Biochem. 2001, 291, 89–95. [Google Scholar] [CrossRef]
- Amiss, T.J.; Sherman, D.B.; Nycz, C.M.; Andaluz, S.A.; Pitner, J.B. Engineering and rapid selection of a low-affinity glucose/galactose-binding protein for a glucose biosensor. Protein Sci. 2007, 16, 2350–2359. [Google Scholar] [CrossRef]
- Staiano, M.; Bazzicalupo, P.; Rossi, M.; D'Auria, S. Glucose biosensors as models for the development of advanced protein-based biosensors. Mol. Biosyst. 2005, 1, 354–362. [Google Scholar] [CrossRef]
- Huber, R.; Langworthy, T.A.; König, H.; Thomm, M.; Woese, C.R.; Sleytr, U.B.; Stetter, K.O. Thermotoga maritima sp. nov. represents a new genus of unique extremely thermophilic eubacteria growing up to 90 °C. Arch. Microbiol. 1986, 144, 324–333. [Google Scholar] [CrossRef]
- Nelson, K.E.; Clayton, R.A.; Gill, S.R.; Gwinn, M.L.; Dodson, R.J.; Haft, D.H.; Hickey, E.K.; Peterson, J.D.; Nelson, W.C.; Ketchum, K.A.; et al. Evidence for lateral gene transfer between Archaea and bacteria from genome sequence of Thermotoga maritima. Nature 1999, 399, 323–329. [Google Scholar] [CrossRef]
- Worning, P.; Jensen, L.J.; Nelson, K.E.; Brunak, S.; Ussery, D.W. Structural analysis of DNA sequence: evidence for lateral gene transfer in Thermotoga maritima. Nucleic Acids Res. 2000, 28, 706–709. [Google Scholar] [CrossRef]
- Tian, Y.; Cuneo, M.J.; Changela, A.; Hocker, B.; Beese, L.S.; Hellinga, H.W. Structure-based design of robust glucose biosensors using a Thermotoga maritima periplasmic glucose-binding protein. Protein Sci. 2007, 16, 2240–2250. [Google Scholar] [CrossRef]
- Fox, J.D.; Routzahn, K.M.; Bucher, M.H.; Waugh, D.S. Maltodextrin-binding proteins from diverse bacteria and archaea are potent solubility enhancers. FEBS Lett. 2003, 537, 53–57. [Google Scholar] [CrossRef]
- Nanavati, D.M.; Thirangoon, K.; Noll, K.M. Several archaeal homologs of putative oligopeptide-binding proteins encoded by Thermotoga maritima bind sugars. Appl Environ. Microbiol. 2006, 72, 1336–1345. [Google Scholar] [CrossRef]
- Luchansky, M.S.; Der, B.S.; D'Auria, S.; Pocsfalvi, G.; Iozzino, L.; Marasco, D.; Dattelbaum, J.D. Amino acid transport in thermophiles: characterization of an arginine-binding protein in Thermotoga maritima. Mol. Biosyst. 2010, 6, 142–151. [Google Scholar] [CrossRef]
- Sun, Y.J.; Rose, J.; Wang, B.C.; Hsiao, C.D. The structure of glutamine-binding protein complexed with glutamine at 1.94 A resolution: Comparisons with other amino acid binding proteins. J. Mol. Biol. 1998, 278, 219–229. [Google Scholar] [CrossRef]
- Cuneo, M.J.; Changela, A.; Miklos, A.E.; Beese, L.S.; Krueger, J.K.; Hellinga, H.W. Structural analysis of a periplasmic binding protein in the tripartite ATP-independent transporter family reveals a tetrameric assembly that may have a role in ligand transport. J. Biol. Chem. 2008, 283, 32812–32820. [Google Scholar]
- Gonin, S.; Arnoux, P.; Pierru, B.; Lavergne, J.; Alonso, B.; Sabaty, M.; Pignol, D. Crystal structures of an Extracytoplasmic Solute Receptor from a TRAP transporter in its open and closed forms reveal a helix-swapped dimer requiring a cation for α-keto acid binding. BMC Struct. Biol. 2007. [Google Scholar] [CrossRef] [Green Version]
- Fang, Y.; Kolmakova-Partensky, L.; Miller, C. A bacterial arginine-agmatine exchange transporter involved in extreme acid resistance. J. Biol. Chem. 2007, 282, 176–182. [Google Scholar]
- Scire, A.; Marabotti, A.; Staiano, M.; Iozzino, L.; Luchansky, M.S.; Der, B.S.; Dattelbaum, J.D.; Tanfani, F.; D'Auria, S. Amino acid transport in thermophiles: characterization of an arginine-binding protein in Thermotoga maritima. 2. Molecular organization and structural stability. Mol. Biosyst. 2010, 6, 687–698. [Google Scholar] [CrossRef]
- Vahedi-Faridi, A.; Eckey, V.; Scheffel, F.; Alings, C.; Landmesser, H.; Schneider, E.; Saenger, W. Crystal structures and mutational analysis of the arginine-, lysine-, histidine-binding protein ArtJ from Geobacillus stearothermophilus. Implications for interactions of ArtJ with its cognate ATP-binding cassette transporter, Art(MP)2. J. Mol. Biol. 2008, 375, 448–459. [Google Scholar] [CrossRef]
- Hsiao, C.D.; Sun, Y.J.; Rose, J.; Wang, B.C. The crystal structure of glutamine-binding protein from Escherichia coli. J. Mol. Biol. 1996, 262, 225–242. [Google Scholar] [CrossRef]
- Ausili, A.; Pennacchio, A.; Staiano, M.; Dattelbaum, J.D.; Fessas, D.; Schiraldi, A.; D'Auria, S. Amino acid transport in thermophiles: Characterization of an arginine-binding protein from Thermotoga maritima. 3. Conformational dynamics and stability. J. Photochem. Photobiol. B 2012, in press. [Google Scholar]
- Ruggiero, A.; Dattelbaum, J.D.; Pennacchio, A.; Iozzino, L.; Staiano, M.; Luchansky, M.S.; Der, B.S.; Berisio, R.; D'Auria, S.; Vitagliano, L. Crystallization and preliminary X-ray crystallographic analysis of ligand-free and arginine-bound forms of Thermotoga maritima arginine-binding protein. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2011, 67, 1462–1465. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Ausili, A.; Staiano, M.; Dattelbaum, J.; Varriale, A.; Capo, A.; D'Auria, S. Periplasmic Binding Proteins in Thermophiles: Characterization and Potential Application of an Arginine-Binding Protein from Thermotoga maritima: A Brief Thermo-Story. Life 2013, 3, 149-160. https://doi.org/10.3390/life3010149
Ausili A, Staiano M, Dattelbaum J, Varriale A, Capo A, D'Auria S. Periplasmic Binding Proteins in Thermophiles: Characterization and Potential Application of an Arginine-Binding Protein from Thermotoga maritima: A Brief Thermo-Story. Life. 2013; 3(1):149-160. https://doi.org/10.3390/life3010149
Chicago/Turabian StyleAusili, Alessio, Maria Staiano, Jonathan Dattelbaum, Antonio Varriale, Alessandro Capo, and Sabato D'Auria. 2013. "Periplasmic Binding Proteins in Thermophiles: Characterization and Potential Application of an Arginine-Binding Protein from Thermotoga maritima: A Brief Thermo-Story" Life 3, no. 1: 149-160. https://doi.org/10.3390/life3010149



