Pivotal Enzyme in Glutamate Metabolism of Poly-g-Glutamate-Producing Microbes
Abstract
:1. Introduction
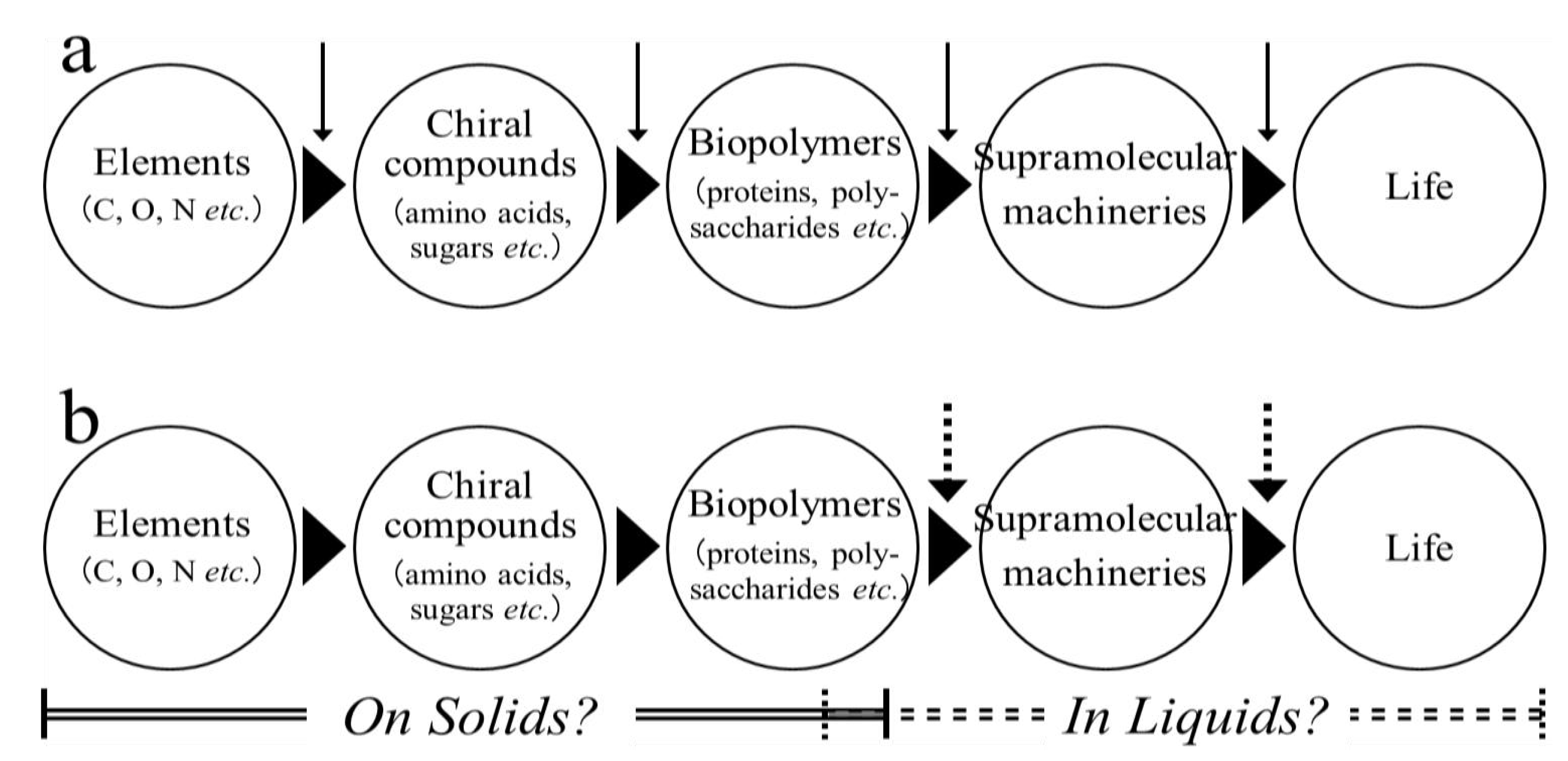
2. Results and Discussion
2.1. Determination of Sources of L-glutamate in the Production of L-PGA by N. aegyptiaca
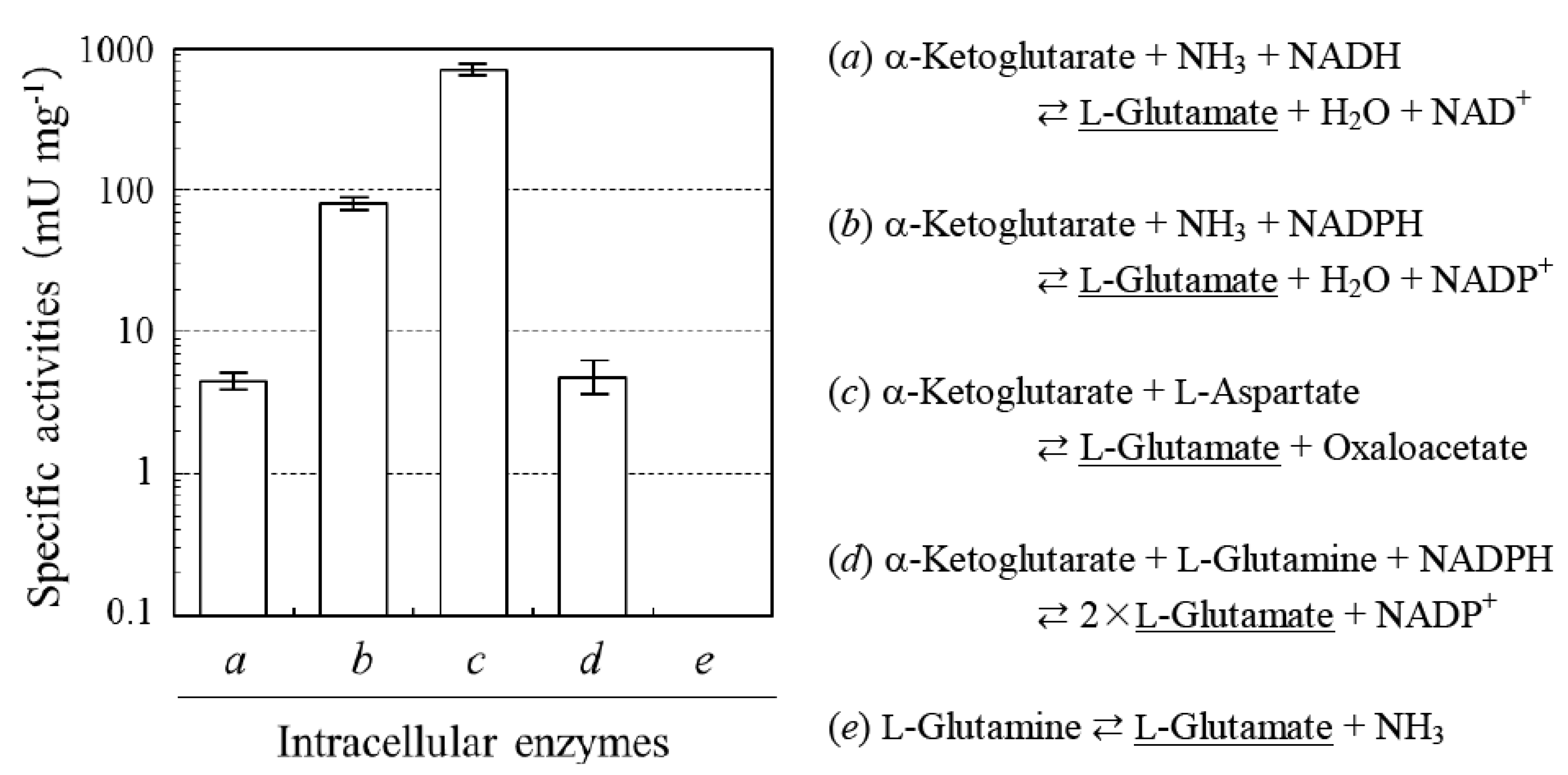
2.2. Absence of D-glutamate Suppliers in N. aegyptiaca
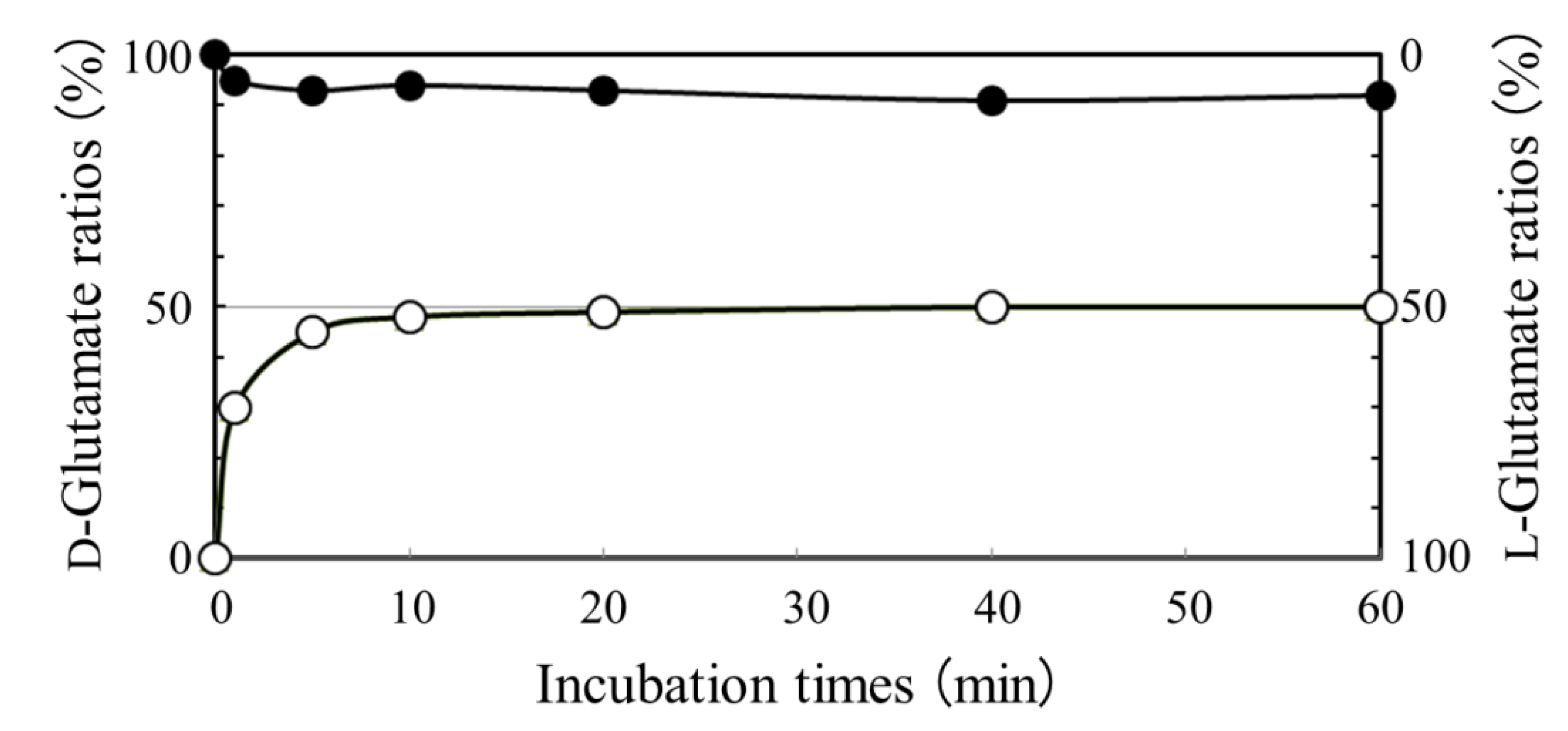
2.3. Analysis of a Pivotal D-glutamate Supplier in B. Subtilis Producing DL-PGA
3. Experimental Section
3.1. Microbes and Culture Conditions
3.2. Preparation of Cytosolic Fractions
3.3. Enzyme Assays
3.3.1. Sources of L-Glutamate
3.3.2. D-Glutamate Suppliers
3.3.3. Other Enzymes
3.4. Protein Assay
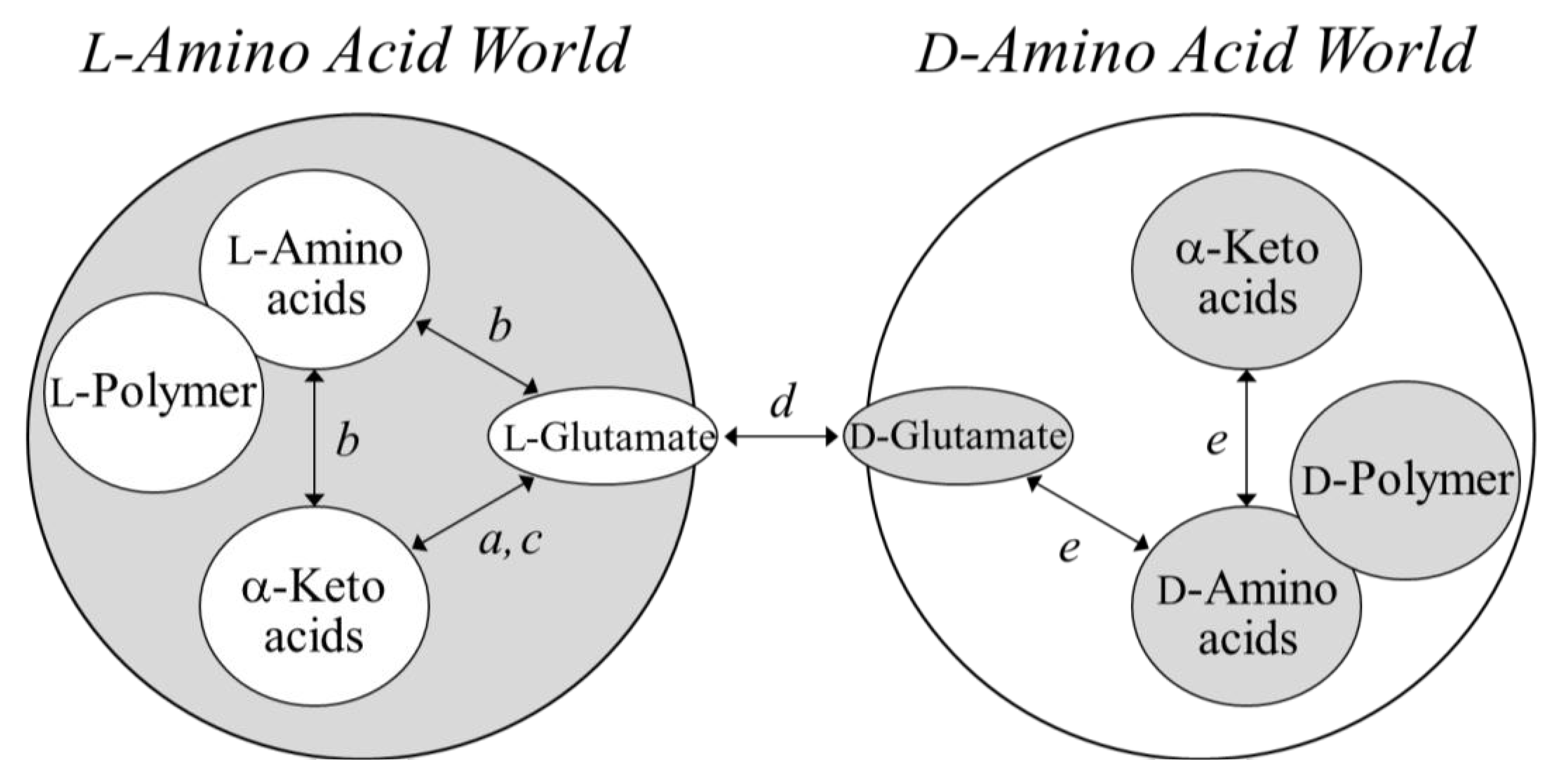
4. Concluding Remarks
References
- Shimada, A.; Ozaki, H. Flexible enantioselectivity of tryptophanase attributable to benzene ring in heterocyclic moiety of D-tryptophan. Life 2012, 2, 215–228. [Google Scholar] [CrossRef]
- Cohen, B.A.; Chyba, C.F. Racemization of meteoritic amino acids. Icarus 2000, 145, 272–281. [Google Scholar] [CrossRef]
- Bada, J.L.; Miller, S.L. Racemization and the origin of optically active organic compounds in living organisms. Biosystems 1987, 20, 21–26. [Google Scholar] [CrossRef]
- Ashiuchi, M.; Misono, H. Poly-γ-glutamic acid. In Biopolymers; Fahnestock, S.R., Steinbüchel, A., Eds.; Wiley-VCH: Weinheim, Germany, 2002; Volume 7, pp. 123–174, Chapter 6. [Google Scholar]
- Ashiuchi, M. Occurrence and biosynthetic mechanism of poly-γ-glutamic acid. In Microbiol. Monogr.: Amino-Acid Homopolymers Occurring in Nature; Hamano, Y., Ed.; Springer-Verlag: Heidelberg, Germany, 2010; Volume 15, pp. 77–94. [Google Scholar]
- Yamasaki, D.; Minouchi, Y.; Ashiuchi, M. Extremolyte-like applicability of an archaeal exo-polymer, poly-γ-L-glutamate. Environ. Technol. 2010, 31, 1129–1134. [Google Scholar] [CrossRef]
- Lentzen, G.; Schwarz, T. Extremolytes: natural compounds from extremophiles for versatile applications. Appl. Microbiol. Biotechnol. 2006, 72, 623–634. [Google Scholar] [CrossRef]
- Ashiuchi, M.; Soda, K.; Misono, H. A poly-γ-glutamate synthetic system of Bacillus subtilis IFO 3336: Gene cloning and biochemical analysis of poly-γ-glutamate produced by Escherichia coli clone cells. Biochem. Biophys. Res. Commun. 1999, 263, 6–12. [Google Scholar] [CrossRef]
- Ruzheinikov, S.N.; Taal, M.A.; Sedelnikova, S.E.; Baker, P.J.; Rice, D.W. Substrate-induced conformational changes in Bacillus subtilis glutamate racemaseand their implications for drug discovery. Structure 2005, 13, 1707–1713. [Google Scholar] [CrossRef]
- Lundqvist, T.; Fisher, S.L.; Kern, G.; Folmer, R.H.A.; Xue, Y.; Newton, D.T.; Keating, T.A.; Alm, R.A.; De Jonge, B.L.M. Exploitation of structural and regulatory diversity in glutamate racemases. Nature 2007, 447, 817–822. [Google Scholar] [CrossRef]
- Kimura, K.; Tran, L.-S.P.; Itoh, Y. Roles and regulation of the glutamate racemase isogenes, racE and yrpC, in Bacillus subtilis. Microbiology 2004, 150, 2911–2920. [Google Scholar] [CrossRef]
- Kimura, K.; Tran, L.-S.P.; Uchida, I.; Itoh, Y. Characterization of Bacillus subtilis γ-glutamyltrans-ferase and its involvement in the degradation of capsule poly-γ-glutamate. Microbiology 2004, 150, 4115–4123. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 1989. [Google Scholar]
- Ashiuchi, M.; Kamei, T.; Baek, D.-H.; Shin, S.-Y.; Sung, M.-H.; Soda, K.; Yagi, T.; Misono, H. Isolation of Bacillus subtilis (chungkookjang), a poly-γ-glutamate producer with high genetic competence. Appl. Microbiol. Biotechnol. 2001, 57, 764–769. [Google Scholar] [CrossRef]
- Ashiuchi, M.; Shimanouchi, K.; Nakamura, H.; Kamei, T.; Soda, K.; Park, C.; Sung, M.-H.; Misono, H. Enzymatic synthesis of high-molecular-mass poly-γ-glutamate and regulation of its stereochemistry. Appl. Environ. Microbiol. 2004, 70, 4249–4255. [Google Scholar] [CrossRef]
- Ashiuchi, M. Analytical approaches to poly-γ-glutamate: Rapid quantification, molecular size determination, and stereochemistry investigation. J. Chromatogr. B 2011, 879, 3096–3101. [Google Scholar] [CrossRef]
- Wolosker, H.; Blackshaw, S.; Snyder, S.H. Serine racemase:A glial enzyme synthesizingD-serine to regulate glutamate-N-methyl-D-aspartateneurotransmission. Proc. Natl. Acad. Sci. USA 1999, 96, 13409–13414. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Ashiuchi, M.; Yamamoto, T.; Kamei, T. Pivotal Enzyme in Glutamate Metabolism of Poly-g-Glutamate-Producing Microbes. Life 2013, 3, 181-188. https://doi.org/10.3390/life3010181
Ashiuchi M, Yamamoto T, Kamei T. Pivotal Enzyme in Glutamate Metabolism of Poly-g-Glutamate-Producing Microbes. Life. 2013; 3(1):181-188. https://doi.org/10.3390/life3010181
Chicago/Turabian StyleAshiuchi, Makoto, Takashi Yamamoto, and Tohru Kamei. 2013. "Pivotal Enzyme in Glutamate Metabolism of Poly-g-Glutamate-Producing Microbes" Life 3, no. 1: 181-188. https://doi.org/10.3390/life3010181



